BRCA1 Gene's EXON 11 and Breast Carcinoma: A Mutational Hot Spot for Familial Patients and Prone to Metastases in Northern India
Received: 09-Jan-2015 / Accepted Date: 11-Mar-2015 / Published Date: 15-Mar-2015 DOI: 10.4172/2161-0681.1000219
Abstract
Background: The prevalence of mutation in BRCA1 genes alarmingly augmented threat of Breast carcinoma among women. The occurrence of BRCA1 gene mutation in India is heterogeneous and varies according to geographical origin. Previous investigations have relied on subjective methods of recording prevalence and familial association. The influence of specific mutation biomarkers which may explain the link between, age, metastases, clinicopathological markers and risk of breast cancer has not been investigated prospectively in Uttar Pradesh (Northern India). Methods: This study was carried out on patients (N=381) diagnosed with breast cancer and further categorized into three groups according to family history. In the present work, blood/tissue samples were collected and mutations were detected using a PCR-SSCP (Single-strand conformation polymorphism) technique followed by sequencing. Results: In the study, 12 sequence variants out of which, eleven novels were identified in exon 11 of BRCA1 gene. BRCA1 mutations were detected in 4.7% (18/381) patients. Mutations in BRCA1 genes were significantly associated with family history and these mutations were found to be strongly associated with metastatic presentation (P=0.042, OR=6.567, 95% CI=1.073-40.174), younger age (P=0.032, OR=11.244, 95% CI=1.227-103.062), and negatively correlated with ER/PR/HER2. Thus, this can serve as important milestone in diagnosis of familial breast cancer. Conclusion: The higher prevalence of BRCA1 mutation among North Indian Breast cancer patients was associated with family history, metastases and younger age. The only alternate apart from early diagnosis is opting for a routine breast screening, which will prove to be a viable option for prevention in carcinoma of breast & better survival.
Keywords: Breast cancer; Metastases; BRCA; Mutation; SSCP
312549Introduction
Breast cancer (BC) is one of the most prevalent diseases affecting women [1]. It accounts for 23% of all cancers among women, and ranked second most common cancer overall when both sexes are considered together. Geographical variation plays a key role in breast cancer incidence. Among the Indian population, incidence of breast cancer varies widely. Regionally, incidence rates range from 18.8 per 105 in Trivandrum to 28.2 per 105 in Mumbai while amongst religious groups; it is highest among Parsi women [2].
Genetic susceptibility to cancer is triggered in several ways; the best understood causal mechanism being due to inactive germline mutations in tumor suppressor and DNA repair genes, which lead to an accumulation of mutations in oncogenes and cell-cycle checkpoints that leads to uncontrolled cell division [3]. Two major breast cancer susceptibility genes are BRCA1 (Genbank accession no- U14680) and BRCA2 (Genbank accession no-U43746). BRCA1 gene mutation is known to account for <5% of all breast cancers [4, 5]; and 10% of women of <40 years and around 30% of women with strong family history of breast or ovarian cancer or both [6]. Among the carriers of BRCA1 around 65% develop breast cancer and 32% ovarian cancer [7].
The spectrum of BRCA1 mutations has been characterized in different populations worldwide, with significant variation of the relative contribution of these genes to hereditary cancer among populations and examples of population specific founder mutations (BRCA1: 185delAG, 5382insC, BRCA2:6174delT in Jews [8].
However, the contribution of mutations in BRCA1 gene to breast cancer patients in the Indian population remains relatively unexplored apart from a few small studies [9-13]. Therefore this investigation is not only focused on understanding the frequency of BRCA1 gene mutation in patients with BC, but also to determine their association with, family history, metastases, demographic and clinico-pathological markers in North-Indian population.
Materials and Methods
Study of population and clinical evaluation of patients
The study group included N=381 patients who underwent surgery for invasive breast cancer at the Surgical Oncology Department, Sir Sundarlal Hospital, Banaras Hindu University, UP, India from 2007. Exclusion criteria were based on the following factors: patients <18 years of age, who received prior treatment in the form of chemotherapy or radiotherapy and patients who were mentally incapable of giving their own consent. The patients subjects were further divided into following groups:
Group 1: Breast carcinoma patients with family history. Subjects were further subcategorized into first degree (Group 1A) and second degree relative (Group 1B).
Group 2: This group include Breast carcinoma patients without any family history. If a patient met the appropriate criteria, we visited the patient before surgery to explain the details of the study and ask for the patients consent and participation. Upon informed consent from the patient, we conducted a 30-minute interview with the patient. Data involving presentation, diagnosis, and staging were collected from office charts, hospital charts, and face-to-face interview with the patient. Informed consent was obtained from all participating patients and the study was carried out with the approval of Ethical Review Committee of Institute of Medical Sciences, Banaras Hindu University, Varanasi, India. Diagnosis was defined by the histological presence of invasive breast carcinoma tissue biopsy, generally by preoperative core biopsy.
Blood and tissue sample collection
Cancer tissue samples and peripheral blood samples were collected in Normal saline (0.9% w/v of NaCl) and EDTA vials respectively. The sample was frozen at -70°C till further use.
Isolation of genomic DNA from mammalian tissue
Genomic DNA was also extracted from breast tissue. The entire step was carried out at 4°C. The tissue was homogenized in ice cold SET (Sucrose 10.8 g, 0.5M EDTA 1ml, 1M Tris-Cl 2.5ml, pH 8.00 to 100 ml water, autoclaved and stored at 4°C) buffer and centrifuged at 4000 rpm for 10 minutes at a temperature of 4°C to pellet the nuclei and was washed once again with SET buffer. The pure nuclear pellet was suspended in TEN (1M Tris-Cl 2.0 ml, 0.5M EDTA 1ml, 5M NaCl 8 ml to 100 ml water, autoclaved and stored at 4°C) buffer, SDS was added to a final concentration of 1% and was mixed gently to lyses the nuclei. Proteinase K was added to the lysate (to a final concentration of 100 μg/ml and was incubated at 37°C overnight). The proteinase K treated lysate was mixed with equal volume of tris saturated phenol, pH 8.00, twice with phenol:chloroform (1:1) and once with chloroform:isoamyl alcohol (24:1) following centrifugation at 10,000 rpm after each extraction. The final aqueous phase was transferred to a fresh tube. To this aqueous phase, 1/10th volume 3M sodium acetate (pH 5.2) and two volume of ice-cold absolute alcohol was added. The DNA was precipitated and was washed with 70% ethanol, air dried and dissolved in appropriate volume of TE buffer (pH 8.00). The sample was kept at 37°C till the DNA dissolves completely.
Primers and PCR-SSCP analysis
For PCR amplification, 8 set of primer pairs were used to amplify exon 2, 5, 11.1, 11.2, 11.3, 11.4, 16 and 20 of BRCA1 gene. PCR was carried out in BiometraR at respective annealing temperature (Ta) using standard protocol (Table 1). Primers used have been chosen from the BIC database (http://www.nhgri.nih.gov/Intramural_research/Lab_transfer/Bic/). In addition primers for exon 11 of BRCA1 were designed during the course of study by using primer3 software.
| Gene | Amplicon | Primer sequence | Annealing Temperature (Ta)°C | Product Size (bp) |
|---|---|---|---|---|
| BRCA1 | Exon 2 | (F)GAAGTTGTCATTTTATAAACCTTT (R)GTCTTTTCTTCCCTAGTATGT | 51.0 | 258 |
| BRCA1 | Exon 5 | (F)CTCTTAAGGGCAGTTGTGAG (R)ATGGTTTTATAGGAACGCTATG | 53.1 | 278 |
| BRCA1 | Exon 11.1 | (F)GACAATTCAGTTTTTGAGTACCTTG (R)TGTTATCCAAGGAACATCTTCAG | 55.0 | 497 |
| BRCA1 | Exon 11.2 | (F)CAGAAACTGCCATGCTCAGA (R)TATTTGTGAGGGGACGCTCT | 52.5 | 436 |
| BRCA1 | Exon 11.3 | (F)CTCCCCAACTTAAGCCATGT (R)GAAGACTTCCTCCTCAGCCTA | 54.5 | 437 |
| BRCA1 | Exon 11.4 | (F)TCCACAATTCAAAAGCACCT (R)GATCTTTGGGGTCTTCAGCA | 51.3 | 450 |
| BRCA1 | Exon 16 | (F) AATTCTTAACAGAGACCAGAAC (R)AAAACTCTTTCCAGAATGTTGT | 52.3 | 450 |
| BRCA1 | Exon 20 | (F)ATATGACGTGTCTGCTCCAC (R)AGTCTTACAAAATGAAGCGG | 52.3 | 259 |
Table 1: Primers used for screening different exons of BRCA1 gene.
PCR reaction was carried out in a volume of 25 μl with, 1 × PCR buffer (20 mM Tris-HCl pH 8.4, 50 mM KCl), 1.5 mM MgCl2, 5 mM dNTP mix, 10 μM of both forward and reverse primer and 1.25 U Taq (New England Biolabs) with the following conditions:
An initial de-naturation at 94°C for 3 minutes was followed by 35 cycles of amplification (30 s/94°C, 40 s/primer specific annealing temperature, and 50 s/72°C) and final elongation of 3 min/72°C. PCR products were analyzed on 2% Agarose gels (BIORON) in 1X TBE (890 mM Tris 890 mM Borate 24 mM EDTA.
SSCP analysis of PCR product was carried out on 7% nondenaturing polyacrylamide gel (PAG) utilizing either non-radioactive silver staining or radioactive procedures [14-16]. PCR products mixed in denaturing buffer (95% formamide, 10 mM NaOH, 0.05% xylenecyanol FF and 0.05% bromophenol blue) in 1:1 ratio were heat denatured at 95°C for 5 min, immediately cooled on ice for 5 min, 6 μl of which were loaded on 7% PAG and electrophoresed in 1X TBE buffer at ±17°C at 4 W constant power for 18-22 h. Gels were then silver stained.
A rapid silver staining method [17] was used to detect the bands. The gels were washed in deionized water and fixed in 10% ethanol for 30 minutes. They were then shaken in 0.1% silver nitrate for 10 minutes and thoroughly rinsed three times in deionized water. Staining was continued in partial darkness, using a solution of 1.5% sodium hydroxide and 0.05% formaldehyde until the desired band intensity was achieved. Development was stopped in 3% acetic acid and the gel was preserved in a solution of 10% ethanol and 5% glycerol.
Nucleotide sequencing
Purified PCR products of the samples showing mobility shift on SSCP analysis and product size having more than 350 bp were used for direct DNA sequencing using Automated DNA sequencer, genetic analyzer 3130. To minimize the sequencing artifacts induced by PCR, products from at least two different PCRs were sequenced using forward and reverse primers. Sequences were compared against the human BRCA1 (Genbank accession no- U14680).
Statistical analysis
For the propose of statistical analysis two-tailed tests were used at all times and statistical significance was set a priori at a P value of less than 0.05. Statistical analysis were performed with SPSS for Windows version 16.0 (SPSS Inc., Chicago, Illinois, USA). To determine all univariate analysis Student’s t-test or χ2 tests was used, as appropriate. Bivariate correlation was used to demonstrate the association and significance of the presence of mutation with various clinicopathological markers for BC such as estrogen receptor (ER), progesterone receptor (PR) and epidermal growth factor receptor-2 (HER-2). Statistical significance level was considered when it reached a P value of less than 0.05. Bivariate analysis, by logistic regression, was performed to evaluate age at diagnosis and the association of advanced nodal metastasis with the presence of mutation pattern.
Results
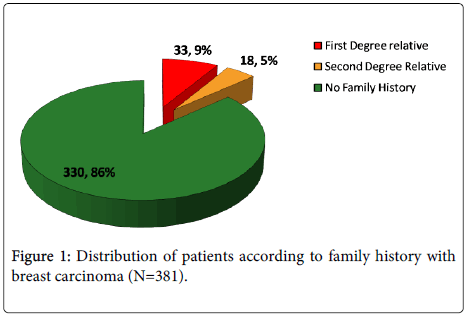
Evaluation of age of onset and family history among breast cancer patients was recorded from Sir Sundarlal Hospital, Banaras Hindu University, Varanasi. Mean age of onset for 381 women diagnosed with BC during study was 47.50 ± 10.407 years. Approximately 33.1% of cases were diagnosed under the age of 40 years. Out of the 381 cases, 33 (9%) belonged to first degree and 18 (5%) were second degree relative of breast cancer patient (Figure 1).
To determine the contribution of BRCA1 gene to breast cancer in local population of Varanasi and nearby cities in India, we screened for alterations in the coding sequences of gene in 381 breast cancer patients. There were 18 (4.7%) patients having BRCA1 gene mutation. When we compared BRCA1 gene mutation with family history, BRCA1 showed significant association (P<0.001) with patients belonging to first (9,50%) and second (6,33.3%) degree relative (Table 2).
| Family History | No Family History | P Value | ||
|---|---|---|---|---|
| BRCA1 Gene | First Degree Relative | Second Degree Relative | ||
| Mutated (N=18) Un-mutated (N=363) | 9(50%) 24(6.6%) | 6(33.3%) 12(3.3%) | 3(16.7%) 327(90.1%) | <0.001 |
Table 2: Association of mutation with family history.
Sequence variants in BRCA 1 genes
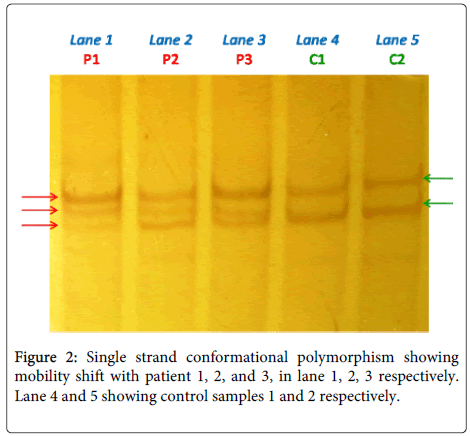
All sequence variants were confirmed by the Single strand conformational polymorphism followed by sequencing (Figure 2). In total, 12 sequence variants were identified in the study group, including 10 Frame Shift (FS), 1 missense (MS), 1 nonsense (Table 3). Sequencing result explored one previously reported 185delAG (Table 3) in exon 2 (deleterious frame-shift mutations) resulting in a premature termination codon that were identified in BRCA1 gene and 11 novel 1230delC, 1238delA, 1273insG, 1415insA, 1611delC, 1614insC, 1961delG, 2025insC, 2026TtoA, 2354AtoC, 2361insC, in exon11 (Table 3) with group 1 (BC with Family History) i.e. 15 in number out of 51 subjects (29.4%). Only one sequence variant (185delAG) was observed in Group 2 (BC without family history) i.e. 3 out of 330 subjects (0.9%). The BRCA1 185delAG mutation was identified in two early onset index case [age 26 Y and 28 Y] with family history and three case [age 46 Y, 54 Y, 58Y] without family history.
| Gene | E | NT | Codan | Base change | AA Change | Designation | Mutation Type | C.I. | BIC Entry | Age/Sex | Family history |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BRCA1 | 2 | 185 | 23 | del AG | Stop 39 | 185del AG | Frameshift | Yes | Reported | 28/F, 46/F | No, Yes |
| BRCA1 | 11A | 1230 | 371 | del C | Stop 373 | 1230 del C | Frameshift | Yes | Novel | 30/F | Yes |
| BRCA1 | 11A | 1238 | 373 | del A | Stop 375 | 1238 del A | Frameshift | Yes | Novel | 30/F | Yes |
| BRCA1 | 11A | 1273 | 385 | Ins G | Stop 390 | 1273 ins G | Frameshift | Yes | Novel | 30/F | Yes |
| BRCA1 | 11A | 1415 | 432 | Ins A | Stop 435 | 1415 ins A | Frameshift | Yes | Novel | 30/F | Yes |
| BRCA1 | 11A | 1611 | 498 | Del C | Stop 502 | 1611 del C | Frameshift | Yes | Novel | 30/F | Yes |
| BRCA1 | 11A | 1614 | 498 | Ins C | Stop 500 | 1614 ins C | Frameshift | Yes | Novel | 30/F | Yes |
| BRCA1 | 11B | 2025 | 636 | Ins C | Stop 638 | 2025 ins C | Frameshift | Yes | Novel | 40/F | Yes |
| BRCA1 | 11B | 2361 | 747 | Ins C | Stop 761 | 2361 ins C | Frameshift | Yes | Novel | 40/F | Yes |
| BRCA1 | 11B | 1961 | 614 | Del G | Stop 625 | 1961 del G | Frameshift | Yes | Novel | 45/F | Yes |
| BRCA1 | 11B | 2354 | 745 | A to C | Arg-Ser | 2354 A to C | Missense | Yes | Novel | 45/F | Yes |
| BRCA1 | 11B | 2026 | 636 | T to A | Leu-stop | 2026 T to A | Nonsense | Yes | Novel | 40/F,40/F | Yes |
Table 3: BRCA1 and BRCA2 sequence variants.
Association of mutation with demographic and pathological factor
BRCA1 mutation was more frequently observed in tumor sample obtained from women who were ≤ 40 years of age. Out of the 126 cases ≤ 40 years old included in the study 11.9% were mutated whereas only 1.2% of the 255 cases from women >40 years old were mutated.
Multivariate analysis (Table 4) suggested, a strong significant association of mutation and age at diagnosis i.e. the age group of ≤ 40 have more higher incidence than the age group >40 (OR=11.244, 95% CI=1.227-103.062). BRCA1 mutation that was found most frequently with family history to be found independently associated with metastatic presentation. The increased frequency of metastatic presentation persisted on multinomial logistic regression analysis (Table 4), suggesting an independent association between BRCA1 mutation (OR=6.567, 95% CI=1.073-40.174). BRCA1 mutation was negatively correlated with ER, PR and HER-2 with the significant level of P=0.051, P=0.074, P=0.028 respectively.
| Gene | Variable | OR | 95% Confidence Interval (CI) | P value |
|---|---|---|---|---|
| BRCA1 | Age at diagnosis (years) | 0.032 | ||
| >40 | 1 | - | ||
| ≤40 | 11.244 | 1.227-103.062 | ||
| Metastases | 0.042 | |||
| No | 1 | - | ||
| Yes | 6.567 | 1.073-40.174 |
Table 4: Multivariate regression analysis shows association of mutation in BRCA1 gene with age at diagnosis and metastatic presentation.
Discussion
The incidence of breast cancer in India has been increased in topical years and high penetrance reported in metropolitan cities. It is probable to create an ever increasing health concern and burden, as socio-economic changes bring increased exposure to standard of living enhancing risk factors. The earlier average age of onset among Indian women compared to Western populations and the increased likelihood of early-onset disease being attributable to genetic susceptibility, suggests the existence of a strong genetic component in this population. Despite the increasing demand for genetic testing roused from the patients and their relatives, it can be offered only to individuals belonging to high-risk families. This is defined by the high probability of germline mutation in a BRCA gene and thus cancer occurrence is likely the expression of a highly penetrant genetic predisposition. In this study, we explored the mutation frequency, age of onset, metastases, family history association and role of pathological factors in North India hospital based study. We found that mutation in BRCA1 is more common in early age (≤ 40). These observations were supported by the study carried out at Safdarjung Hospital, New Delhi [3].
In this research work, patients at high risk of carrying BRCA1 mutations were studied, 50% patients were belonging to first degree relative, 33.3% with second degree relative and only 16.7% showed the index case (Table 2). Recently, a meta-analysis has combined the data derived from 22 studies based on unselected series. Using a modified segregation analysis, it has been estimated that BRCA1 mutation carriers have 65% higher risk of breast cancer of by 70 years of age and 39% average cumulative risk of ovarian cancer [7]. In contrast to this only 2% of non-familial patients had pathologic germline mutations in BRCA1 and 2 genes in a group of English patients who were diagnosed with breast cancer at the age of 30 years or younger [18]
We screened for alterations in the coding sequences of 381 breast cancer patients. A total of 12 sequence variants were observed. The BRCA1 185delAG mutation was identified in five cases two in early onset case [age 26Y, 28Y] with family history and three in an index case [age 46Y, 54Y, 58Y]. This mutation is common in Ashkenazi Jews, having attained a 1% carrier frequency within the population [19] since the origin of the ancestral mutation [20]. Population studies have shown that the 185delAG mutation predates the separation of Sephardi and Ashkenazi Jewish populations and is probably 2000 years old [21]. In India, 185delAG has been reported in all populations studied so far [9-13]. This deleterious frame shift mutation was first reported in a family residing in a part of Trivandrum not far from the small towns with settlement of Jewish people [9]. It was later reported in two South Indian families from Kerala province [12] as well as in two sisters from Goa, where a multi-ethnic population exists, with a significant influence of Portuguese (potential introduction of the mutation through Sephardic Jews) [13]. Surprisingly, Sunita et al. also found 185delAG in a North Indian Hindu patient residing in New Delhi who claimed to have no Jewish ancestry. Similarly, Lakhotia et al., in their initial screening found the same mutation in four Indian breast cancer families [22].
The rest sequence variants (Frame-shift: 1230delC, 1238delA, 1273insG, 1415insA, 1611delC, 1614insC, 2025insC, 2361insC, 1961delG, Missense: 2354A→C and Nonsense: 2026T→A) were found to be novel and not reported anywhere else and Exon 11 was found to be the mutational hot spot for familial breast cancer patients of North India. The BRCA1 exon 11 mutation was found in patients of New Delhi India. Valarmathi MT explored 3672 G→T of exon 11 in a patient with age 36 years [23]. A recent study of Pandrangi S L from New Delhi, India established Two novel triple negative breast cancer cell lines, NIPBC-1 and NIPBC-2 from primary tumors of two young breast cancer patients aged 39 and 38 years respectively, diagnosed as infiltrating duct carcinoma of breast. Screening for mutations in BRCA gene revealed presence of three heterozygous polymorphisms in exon 11 of BRCA1 gene in both the cell lines [24]. The allocation of BRCA1 mutations recognized among Indian high-risk patients is similar to what has been observed in other populations. The most of the patients identified with breast cancer were under the age of 40 years. Factors like age at diagnosis and advanced metastatic presentation retained their statistical significance in the multivariate analysis of BRCA1 mutation. This result suggests that women with BRCA1 mutations in this series were at increased risk for death due to breast cancer because of presentation with more advanced disease. Alternatively, it was observed that the worst outcome may result from adverse biologic features that have been described in BRCA1 associated breast cancers. Such features include high histological grade and proliferation rates [25-29].
Molecular markers currently used to predict treatment response in both early and metastatic breast cancer include Estrogen receptor (ER), Progesterone receptor (PR), and HER2/neu. We found that BRCA1 mutation was negatively correlated with estrogen receptor, progesterone receptor and HER-2. These findings are supported by reports obtained from a detailed examination of 182 tumors samples, out of which 119 were BRCA1-mutated. Results showed that BRCA1 tumors were less likely to express ER (10%), PR (21%), and HER2/neu (3%) [30].
Conclusion
So we can conclude that, self-risk information could assist in taking preventive measures and also induce a high-risk woman to adopt breast screening that may promote early detection and improve chances of surviving breast cancer. This study found evidence for an overall association between the mutations in exon 11 of BRCA1 gene play a role in etiology of familial breast cancer with advanced metastatic presentation. Further experiment with large sample size and different geographical origin are warranted to confirm these findings.
Acknowledgements
Funding support of this work includes Dr. D. S. Kothari Post- Doctoral Fellowship from the University Grants Commission [No.F. 4/2006 (BSR)/13-581/2012(BSR)]. We thank the Institute of Medical Sciences, Banaras Hindu University, Varasnasi, India for providing the infrastructure, and other research facility.
Conflict of Interest
Accordingly there is no-conflict of interest arising whatsoever with this article. This study was supported by a research grant from University Grants Commission, India, availed through Banaras Hindu University DSK-Post Doctoral Fellowship. There is neither medical writing nor editorial assistance was used for the preparation of the article. The authors declare that all of them have made substantial contribution towards the writing of this article.
References
- Marcus JN, Watson P, Page DL, Narod SA, Lenoir GM, et al. (1996) Hereditary breast cancer: pathobiology, prognosis, and BRCA1 and BRCA2 gene linkage. Cancer 77: 697-709.
- Jussawalla DJ, Jain DK (1977) Breast cancer and religion in greater Bombay women: an epidemiological study of 2130 women over a 9-year period. Br J Cancer 36: 634-638.
- Saxena S, Chakraborty A, Kaushal M, Kotwal S, Bhatanager D, et al. (2006) Contribution of germline BRCA1 and BRCA2 sequence alterations to breast cancer in Northern India. BMC Med Genet 7: 75.
- Blackwood MA, Weber BL (1998) BRCA1 and BRCA2: from molecular genetics to clinical medicine. J ClinOncol 16: 1969-1977.
- [No authors listed] (2000) Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Anglian Breast Cancer Study Group. Br J Cancer 83: 1301-1308.
- Ligtenberg MJ, Hogervorst FB, Willems HW, Arts PJ, Brink G, et al. (1999) Characteristics of small breast and/or ovarian cancer families with germline mutations in BRCA1 and BRCA2. Br J Cancer 79: 1475-1478.
- Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, et al. (2003) Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72: 1117-1130.
- Szabo CI, King MC (1997) Population genetics of BRCA1 and BRCA2. Am J Hum Genet 60: 1013-1020.
- Kumar BV, Lakhotia S, Ankathil R, Madhavan J, Jayaprakash PG, et al. (2002) Germline BRCA1 mutation analysis in Indian breast/ovarian cancer families. Cancer BiolTher 1: 18-21.
- Saxena S, Szabo CI, Chopin S, Barjhoux L, Sinilnikova O, et al. (2002) BRCA1 and BRCA2 in Indian breast cancer patients. Hum Mutat 20: 473-474.
- Valarmathi MT, A A, Deo SS, Shukla NK, Das SN (2003) BRCA1 germline mutations in Indian familial breast cancer. Hum Mutat 21: 98-99.
- Valarmathi MT, Sawhney M, Deo SS, Shukla NK, Das SN (2004) Novel germline mutations in the BRCA1 and BRCA2 genes in Indian breast and breast-ovarian cancer families. Hum Mutat 23: 205.
- Hedau S, Jain N, Husain SA, Mandal AK, Ray G, et al. (2004) Novel germline mutations in breast cancer susceptibility genes BRCA, BRCA2 and p53 gene in breast cancer patients from India. Breast Cancer Res Treat 88: 177-186.
- Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T (1989) Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. ProcNatlAcadSci U S A 86: 2766-2770.
- Bassam BJ, Caetano-Anollés G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196: 80-83.
- Bosari S, Marchetti A, Buttitta F, Graziani D, Borsani G, et al. (1995) Detection of p53 mutations by single-strand conformation polymorphisms (SSCP) gel electrophoresis. A comparative study of radioactive and nonradioactive silver-stained SSCP analysis. DiagnMolPathol 4: 249-255.
- Neilan BA, Leigh DA, Rapley E, McDonald BL (1994) Microsatellite genome screening: rapid non-denaturing, non-isotopic dinucleotide repeat analysis. Biotechniques 17: 708, 710, 712.
- Lalloo F, Varley J, Ellis D, Moran A, O'Dair L, et al. (2003) Prediction of pathogenic mutations in patients with early-onset breast cancer by family history. Lancet 361: 1101-1102.
- Struewing JP, Abeliovich D, Peretz T, Avishai N, Kaback MM, et al. (1995) The carrier frequency of the BRCA1 185delAG mutation is approximately 1 percent in Ashkenazi Jewish individuals. Nat Genet 11: 198-200.
- Neuhausen SL, Mazoyer S, Friedman L, Stratton M, Offit K, et al. (1996) Haplotype and phenotype analysis of six recurrent BRCA1 mutations in 61 families: results of an international study. Am J Hum Genet 58: 271-280.
- Bar-Sade RB, Kruglikova A, Modan B, Gak E, Hirsh-Yechezkel G, et al. (1998) The 185delAG BRCA1 mutation originated before the dispersion of Jews in the diaspora and is not limited to Ashkenazim. Human Molecular Genetics 7: 801-805.
- Lakhotia S, Somasundaram K (2003) Conformation-sensitive gel electrophoresis for detecting BRCA1 mutations. Methods MolBiol 223: 403-412.
- Valarmathi MT, Sawhney M, Deo SS, Shukla NK, Das SN (2004) Novel germline mutations in the BRCA1 and BRCA2 genes in Indian breast and breast-ovarian cancer families. Hum Mutat 23: 205.
- Pandrangi SL, RajuBagadi SA, Sinha NK, Kumar M, Dada R, et al. (2014) Establishment and characterization of two primary breast cancer cell lines from young Indian breast cancer patients: mutation analysis. Cancer Cell Int 14: 14.
- [No authors listed] (1997) Pathology of familial breast cancer: differences between breast cancers in carriers of BRCA1 or BRCA2 mutations and sporadic cases. Breast Cancer Linkage Consortium. Lancet 349: 1505-1510.
- Eisinger F, Stoppa-Lyonnet D, Longy M, Kerangueven F, Noguchi T, et al. (1996) Germ line mutation at BRCA1 affects the histoprognostic grade in hereditary breast cancer. Cancer Res 56: 471-474.
- Eisinger F, Noguès C, Birnbaum D, Jacquemier J, Sobol H (1998) BRCA1 and medullary breast cancer. JAMA 280: 1227-1228.
- Robson M, Rajan P, Rosen PP, Gilewski T, Hirschaut Y, et al. (1998) BRCA-associated breast cancer: absence of a characteristic immunophenotype. Cancer Res 58: 1839-1842.
- Jóhannsson OT, Idvall I, Anderson C, Borg A, Barkardóttir RB, et al. (1997) Tumour biological features of BRCA1-induced breast and ovarian cancer. Eur J Cancer 33: 362-371.
- Lakhani SR, Van De Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, et al. (2002) The pathology of familial breast cancer: Predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-, and p53 in patients with mutations in BRCA1 and BRCA2. J ClinOncol 20: 2310-2318.
Citation: Singh AK, Pandey A, Tewari M, Pandey P, Pandey HP, et al. (2015) BRCA1 Gene's EXON 11 and Breast Carcinoma: A Mutational Hot Spot for Familial Patients and Prone to Metastases in Northern India. J Clin Exp Pathol 5:219. DOI: 10.4172/2161-0681.1000219
Copyright: © 2015 Singh AK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 17287
- [From(publication date): 4-2015 - Apr 04, 2025]
- Breakdown by view type
- HTML page views: 12591
- PDF downloads: 4696