Blast resistance in Indian rice: Genetic dissection by gene markers
Received: 02-Jun-2022 / Manuscript No. rroa-22-67683 / Editor assigned: 04-Jun-2022 / PreQC No. rroa-22-67683 / Reviewed: 18-Jul-2022 / QC No. rroa-22-67683 / Revised: 24-Jun-2022 / Manuscript No. rroa-22-67683 / Published Date: 30-Jun-2022 DOI: 10.4172/2375-4338.1000306
Abstract
Understanding of genetic diversity is important to explore existing gene in any crop breeding program. Most of the diversity preserved in the landraces which are well–known reservoirs of important traits for biotic and abiotic stresses. In the present study, the genetic diversity at twenty-four most significant blast resistance gene loci using twenty-eight gene specific markers were investigated in landraces originated from nine diverse rice ecologies of India. Based on phenotypic evaluation, landraces were classified into three distinct groups: highly resistant (21), moderately resistant (70) and susceptible (70). The landraces harbour a range of five to nineteen genes representing blast resistance allele with the frequency varied from 4.96% to 100%. The cluster analysis grouped entire 161 landraces into two major groups. Population structure along with other parameters was also analyzed to understand the evolution of blast resistance gene in rice. The population structure analysis and principal coordinate analysis classified the landraces into two sub– populations. Analysis of molecular variance showed maximum (93%) diversity within the population and least (7%) between populations. Five markers viz; K3957, Pikh, Pi2–i, RM212and RM302 were strongly associated with blast disease with the phenotypic variance of 1.4% to 7.6%. These resistant landraces will serve as a valuable genetic resource for future genomic studies, host–pathogen interaction, identification of novel R genes and rice improvement strategies.
Introduction
Rice (Oryza sativa L.) is the staple food for more than half of the world’s population and depends on rice for more than 20% of their daily calorie intake. By 2035, it is expected that an extra 116 million tonnes of rice will be required to feed the world’s increasing population. This projected production has to be inevitably met with the expected water scarcity, less arable land, new emerging pathogens and pests and likely adverse effects of climate change [1].
The rice crop is affected by several diseases, of which blast disease caused by the fungus Magnaporthe oryzae is one of the most devastating disease causing enormous losses worldwide. M. oryzae can infect rice plant right from seedling to late vegetative/reproductive stages affecting leaves, nodes, collar, panicles, panicle neck, and roots. The fungal pathogen, M. oryzae has been placed among the top 10 fungal plant pathogens in the world based on its scientific and economic importance. Owing to its presence and survival in different environmental conditions in more than 85 countries, it causes a yield loss that is enough to feed more than 60 million people each year . Use of resistant cultivars, fungicides, optimum fertilizer applications and appropriate planting dates are some of the strategies to manage the disease. Though fungicide application to control the disease is feasible, it remains economically unprofitable for resource poor farmers and an possesses environmental risk at high application rates [2]. The utilization of R (resistant) genes is the most economically viable and environmentally friendly choice for the control of this disease. Resistance is generally conferred by either major R genes that provide complete protection against few races of the pathogen or minor genes, which conferred partial protection.
To date, more than 100 R genes, and more than 350 QTLs for resistance to rice blast have been identified, and 27 have been molecularly cloned and characterized viz., Pib, Pb1, Pita, Pi9, Pi2, Pizt, Pid2, Pi33, Pii, Pi36, Pi37, Pikm, Pit, Pi5, Pid3, Pid3–A4, Pikh, Pish, Pik, Pikp, Pia, PiCO39, Pi25, Pi1, pi21, Pi50 and Pi65(t). The rapid changes in virulence characteristics that take place in pathogen populations remains a constant challenge to the success of existing blast–resistant varieties of rice. However, the major blast resistance gene has been useful and should play a vital role in rice production if they are cautiously selected and deployed . Hence, there is an imperative need for mining the new R genes/alleles and minor resistance genes in landraces in which resistance has co–evolved along with fungus over thousands of years. Genetically diverse rice landraces are one of the most important sources for major blast resistance to be introgressed into rice cultivars for the control of the blast. Though breeding strategies focus on developing a durable and broad–spectrum resistant variety by pyramiding many R genes into popular rice cultivar through marker assisted selection , there is always need for a novel resistance allele to combat continuously evolving pathogen [3].
With the discovery and fine mapping of blast resistance genes, many PCR–based markers have been developed and employed in the mining of blast resistance genes in the diverse elite germplasm which contain untapped resources of discrete alleles. The potential of elite germplasm will remain unknown unless the efforts are initiated to screen them intended for their possible use and function . Accurate identification of a specific R gene in diverse elite germplasm through DNA markers and differential blast races is an essential step in ensuring the accuracy in utilization of R gene in MAS for different rice breeding program . The introduction of modern rice cultivars may lead to the erosion of genetic resources like landraces and traditional varieties from the farmer’s fields which resulted in the loss of genetic diversity of rice as well. However, the genetic diversity is to some extent maintained and preserved in the gene banks in the form of landraces/wild collections.To date, seven blast epidemics have occurred from 1980 to 1987 in different states of India, viz. Andhra Pradesh, Himachal Pradesh, Haryana, and Tamil Nadu resulted in severe yield losses in the farmer’s fields which necessitates in assessing the genetic diversity of the blast resistance genes in the landraces/germplasm. Since, the landraces has provided a rich source for genetic improvement of rice for specific traits and represent rich sources of specific allelic variation. India has a rich genetic diversity of landraces due to the diverse agro–climates and growing conditions, therefor, the present study, aimed to investigate a) the genetic diversity of major blast resistant genes b) identify donors for blast resistance and c) genetic association of markers with the blast resistance traditional in rice landraces collected from nine major rice growing states of India. The outcome of present study will help in identification of novel valuable genetic resource for rice blast resistance genes for development of durable blast resistant varieties in India.
Materials and methods
Plant material
A set of 161 diverse Indian rice landraces obtained from the National Gene Bank, ICAR–National Rice Research Institute, Cuttack were used in this study was. These landraces were collections of nine states of India with diverse ecologies.
Disease reaction in uniform blast nursery
A total of 161 landraces from all over India, representing nine states was screened for their leaf blast resistance under natural condition in the Uniform Blast Nursery (UBN) at the experimental farm of ICAR– NRRI, Cuttack (85°55′48′E longitudes and 20°26′35′ N latitude) as described in Yadav et al. The leaf blast screening was conducted twice during two wet seasons of 2015 & 2016 with two replications. Each landrace (30 plants/test entry) was planted in 50 cm long rows in nursery beds at row spacing of 10 cm apart [4]. In addition, the known susceptible checks (HR12 and CO39) were sown in borderlines as spreader rows as well as after every five test entries for the uniform spread of the disease. Disease reaction was recorded twenty five days after sowing and continued up to the 40th day after sowing or the spreader row/checks achieved 85% of the disease symptom. Reactions of each landrace for leaf blast were scored as per the Standard Evaluation System (SES) of IRRI, (2002). The test entries with 0–3 scores were graded as highly resistant (HR), 4–5 as moderately resistant (MR), and 6–9 as susceptible (S). Whenever differences observed in score between the replications, the higher value was considered for scoring.
Location severity index (LSI) was calculated using following formula:
LSI=Score×Entries×100/Totalnumberofentries
Genomic DNA extraction
Young leaf tissues of all the test entries were collected from two weeks old seedlings and stored at –80°C for genomic DNA extraction. Genomic DNA was isolated using the cetyltrimethyl ammonium bromide (CTAB) method with slight modifications [5].
The DNA quality and quantity were estimated based on 0.8% agarose gel electrophoresis and NanoDrop ND–1000 Spectrophotometer (Thermofisher scientific, USA). The Genomic DNA samples were finally diluted with 1X TE buffer to 20 ng/μl and stored at –20°C for further uses.
Genotyping of rice blast R genes
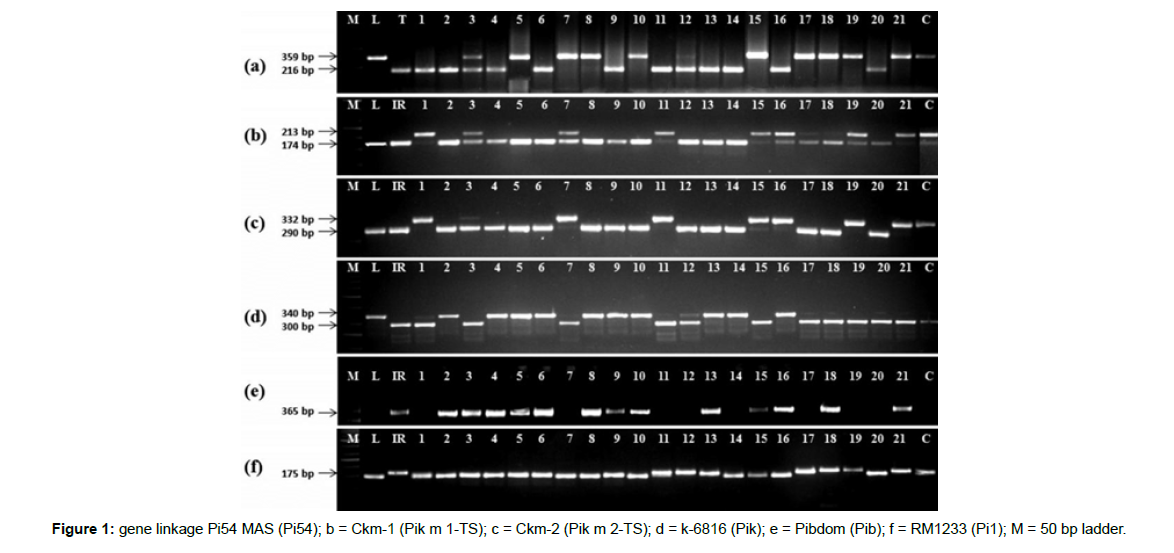
The entire set of 161 landraces were genotyped using markers specific to 24 different blast resistance genes viz. Pib, Pb1, Pita, Pi9, Pi2, Pizt, Pid2, Pi33, Pi36, Pi37, Pikm, Pit, Pi5, Pi54, Pish, Pik, Pikp, Pia, Pi25, Pi1, pi21, Pi56, Pi65(t) and Piz. A total of twenty eight functional/ linked markers corresponding to the twenty four R genes were used for screening of the blast resistance genes. The details of the markers used in the present study are shown in Table 1 with the physical locations on the corresponding chromosomes in (Figure 1).
PCR amplification
Polymerase chain reaction was carried out in a 25 μl reaction volume with the following composition: 20 ng of genomic DNA, 0.2 μM of dNTP (25 mM), 0.2 μM of primers, 1.5 mM of MgCl2, 1X Taq buffer and 1U of Taq DNA polymerase (Thermo Scientific, USA). The PCR program was conducted as: initial denaturation at 94°C for 5 min; followed by 35 cycles of denaturation for 30 sec at 94°C, primers annealing for 30 sec at different temperatures (Table 1), and elongation at 72°C for 1 min, with 10 min final elongation at 72°C. The PCR amplified products were separated in 2.5–3% agarose gels along with a 100 bp DNA ladder (BR Biochem Life Sciences, India) and visualized through staining with ethidium bromide. The gel pictures were taken under UV light in a gel documentation system (AlphaImager, USA). All PCR reactions were repeated twice to confirm the results.
Allele scoring and diversity analysis
The amplified PCR products of 28 markers were scored based on the presence (1) or absence (0) to create binary matrix for each marker [6]. The genetic distance and similarity coefficients were estimated using the binary matrix of 28 markers. Major allele frequency, gene diversity and Polymorphism information content (PIC) value of each marker were estimated using Powermarker program Ver3.25 software . An unweighted Neighbor Joining tree was constructed by the calculated NEI coefficient of dissimilarity index using DARwin5 software.
Association analysis
The hypothesis of genetic association between blast resistance genes and blast disease was tested through a general linear model [7] (GLM) function using TASSEL5 software. The TASSEL 5 software was run with one thousand permutations of data and it represents a significant association only if the P–value was observed in <5% of the permutations for the most significant polymorphism in a region.
Population structure
Population structure analysis of 161 landraces based on 28 markers was investigated using STRUCTURE version 2.3.4 [8]. The number of subpopulations (K) was estimated using the programme at different K value by setting at K = 1 to 10, with 5 independent iterations per K using the admixture model and allele frequencies correlated. Each run, was based on 200,000 Markov chain Monte Carlo (MCMC) iterations after 200 000 burn–in phase. The peak value of ΔK was estimated to determine the optimal K as explained by Evanno et al using the STRUCTURE HARVESTER programme . The binary data matrix of twenty eight markers was used to develop pairwise individual genetic distance, to compute the PCoA (Principal Coordinate Analysis) in GenAlEx 6.502. All the other genetic analyses such as Analysis of Molecular Variance (AMOVA) and pairwise FST were performed using the GenAlEx version 5.0 software [9].
Results
Disease reactions of landraces
Based on the blast disease scoring data for consecutive two seasons in the UBN, 161 landraces were categorized into three groups; twenty one (HR; 13.04%) were highly resistant (score 0–3), seventy (MR; 43.47%) exhibited moderate resistance (score 4–5) and seventy (S; 43.47%) showed susceptible reaction (score 6–9) . The location severity index (LSI) of the two seasons was calculated to know the disease reaction of landraces and it was found to be 5.04. A total of 91 landraces (56.52%) showed resistant (HR and MR) against the blast disease while 70 landraces were found susceptible (S) to disease reaction (43.47%) (S1 Fig). Out of the 21 HR landraces, the maximum number of HR landraces was the collections of Sikkim (7) followed by Maharashtra (5), Tripura (4), Gujarat (4) and Punjab (1) It shows that more than 50% landraces showing HR (11) were the collections of North–Eastern states of India (Sikkim and Tripura).
Discussion
The widespread use of high–yielding varieties has significantly lowered the genetic base of plant breeding material of agriculturally important food crops, which restricts their future improvements . Cultivation of genetically uniform varieties over large scale imposed high selection pressure on the pathogen populations which leads the cultivars highly prone to biotic stress. Changing climate and the emergence of new virulent races imposed a continuous threat to the rice production and global food security. Accordingly, a protection measure necessitates constant progress to keep pace with the evolving pathogen. This needs the identification of new resistance genes and alleles from landraces and wild relatives [10]. The genetic diversity of the majority crop plants is being stored in the form of germ plasm/ accessions in the gene banks. However, the genotypic diversity of most of the accessions has not been fully explored and understood . In this study, we investigated the genetic diversity of geographically diverse Indian landraces which are unique, unexplored and untapped germplasm for blast resistance genes that originated from nine major rice growing states of India with diverse ecology using major blast resistance genes.
Conclusion
The phenotypic screening and molecular characterization of blast resistance genes will help in identification of potential germplasm for leaf blast. Our results offered an outline of the genotypic diversity of Indian landraces representing nine major rice growing states of India with diverse ecologies. Besides, the precise screening of leaf blast for the identification of resistance genes in landraces along with the identified associated marker could be used for the selection of parental materials and the development of resistant breeding lines. Identification of resistant landraces from diverse ecologies will help in better utilization of these landraces as a donor for improvement of existing varieties with blast resistance. Potential landraces for blast resistance could be utilized for mapping to discover novel blast R gene(s) and identification of potential donors for their use in rice breeding. Additionally, the genotyping and phenotyping data of 161 landraces generated in this study could be quiet useful to identify the novel blast R gene(s) using association mapping in a precise way.
References
- Khush GS (2005) What it will take to feed 5.0 billion rice consumers in 2030.Plant Mol Biol 59:1-6.
- 5.Zhu YY, Fang H, Wang YY, Fan JX, Yang SS, Mew TW, et al. (2005)Panicleblast and canopy moisture in rice. cultivar mixturesPhytopathology 95:433-438.
- Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, et al. (2012) The top 10 fungal pathogens in molecular plant pathology.Mol Plant Pathol. 13: 414-430.
- Talbot NJ, (2003) on the trail of a cereal killer: exploring the biology ofMagnaporthe grisea.Annu. Rev Microbiol57: 177-202.
- Yadav MK, Aravindan S, Ngangkham U, Shubudhi HN, Bag MK, Adak T, et al. (2017)Use of molecular markers in identification and characterization of resistance to rice blast in India.PloS one 12:176-236
- SBarik,E Pandit,SPradhan,SMohanty,TMohapatara (2019) Genetic mapping of morpho-physiological traits involved during reproductive stage drought tolerance in rice. PLoS One 214-979.
- Bernier J, Kumar A , Venuprasad R, Spaner D, Atlin G (2007) A large-effect QTL for grain yield under reproductive-stage drought stress in upland rice. Crop Sci 47:pp.507-516.
- Bhattacharjee S, Dey N (2018), Redox metabolic and molecular parameters for screening drought tolerant indigenous aromatic rice cultivars. Physiol Mol Biol Plants241: pp7-23.
- Bin Rahman and Zhang (2016), Flood and drought tolerance in rice: Opposite but may coexist. Food Energy Secur pp.76-88.
- BCheah,KNadarajah,M D Divate,R Wickneswari (2015), Identification of four functionally important microRNA families with contrasting differential expression profiles between drought-tolerant and susceptible rice leaf at vegetative stage BMC Genom,16(1) p.692
Indexed at, Crossref, Google Scholar
Indexed at, Crossref, Google Scholar
Indexed at, Crossref, Google Scholar
Indexed at, Crossref, Google Scholar
Citation: Chaturvedi AK (2022) Blast Resistance in Indian Rice: Genetic Dissection by Gene Markers. J Rice Res 10: 306. DOI: 10.4172/2375-4338.1000306
Copyright: © 2022 Chaturvedi AK. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 1015
- [From(publication date): 0-2022 - Apr 02, 2025]
- Breakdown by view type
- HTML page views: 689
- PDF downloads: 326