Research Article Open Access
Bioremediation of Methomyl by Soil Isolate - Pseudomonas aeruginosa
| Amritha G Kulkarni and Kaliwal BB* | |
| Department of Studies in Biotechnology and Microbiology, Karnatak University, Dharwad-580 003, India | |
| Corresponding Author : | Kaliwal BB Department of Studies in Biotechnology and Microbiology, Karnatak University Dharwad-580 003, India Tel: 9449278648 E-mail: b_kaliwal@yahoo.com |
| Received December 15, 2014; Accepted February 24, 2015; Published February 27, 2015 | |
| Citation: Kulkarni AG, Kaliwal BB (2015) Bioremediation of Methomyl by Soil Isolate - Pseudomonas aeruginosa. J Bioremed Biodeg 6: 281.doi:10.4172/2155-6199.1000281 | |
| Copyright: © 2015 Kulkarni AG, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Bioremediation is emerging as one of the most promising technology for the removal of xenobiotics. Bioremediation of xenobiotic compounds by microorganisms is a crucial phenomenon by which these compounds can be detoxified or removed from the environment, thus preventing pollution, where screening of the potential degrading organism is one of the key step. Methomyl belonging to N-methyl carbamate group of insecticide is the most commonly used pesticide on fruits and vegetables. It is classified as a toxic, hazardous and restricted use pesticide by the World Health Organization (WHO), European Commission (EC) and Environmental Protection Act (EPA). The present study is therefore aimed to isolate a soil bacterium, characterize the most potential degrader of methomyl and further determine the role of plasmid in bioremediation. Soil samples were collected from different areas around Karnatak University campus, Dharwad and inoculated in minimal medium containing methomyl (10-3 M). Morphological characteristics of ten isolates were studied and subjected to HPLC analysis where methomyl degradation (%) was observed. The most potential strain with the ability to degrade methomyl was inoculated in synthetic medium containing methomyl (10-3 M) for 96 hrs. This strain was further identified morphologically, biochemically and genetically. The results indicated that the isolated strain was gram -ve rods and biochemical characteristics showed that indole, methyl red, vogues prauskauer and H2S test was negative and citrate, catalase, gelatin hydrolysis and oxidase test was positive. The 16Sr DNA reports revealed that the accession number was G01801181 and showed 99% similarity to Pseudomonas aeruginosa. The present study on HPLC analysis at regular intervals of 24 hrs of the soil isolate - Pseudomonas aeruginosa in synthetic medium containing methomyl (10-3 M) incubated for 96 hrs revealed that there was a significant decrease in the methomyl content in the treated group when compared with the initial control demonstrating its potential use in bioremediation. The plasmid isolation and curing study confirmed that the genes of the plasmid of Pseudomonas aeruginosa are involved in the methomyl degradation and such plasmids could be used as recombinant DNA technology for bioremediation.
| Keywords |
| Bioremediation; Methomyl; Pseudomonas aeruginosa; Plasmid |
| Introduction |
| A wide variety of synthetic chemicals have found their way into the ecosystem as a consequence of industrial activities, agricultural applications and use in domestic purpose [1]. Pesticides are organic compounds manufactured and used for pest control. When pesticides are dispersed in the environment, they become pollutants, with ecological effects that require remediation. Environmental pollution is caused by both excessive and continuous use of pesticides, and begins when these compounds enter the environment by various means (accidental spills, direct application, residues from cleaning of containers, state of equipment used and methods used to apply the products). The quality of soils, ground water, inland and coastal waters, and air are all affected by pesticide contamination [2,3]. The microbial degradation of hazardous waste offers a promising strategy by which such chemicals may be detoxified [4]. Methomyl belongs to a class of compounds known as oxime carbamates, and it is widely used for the control of insects and nematode pests by inhibiting the enzyme acetylcholinesterase which hydrolyzes the neurotransmitter acetylcholine. The IUPAC name of methomyl is S-methyl N-(methylcarbamoyloxy) thioacetimidate [5]. The WHO (World Health Organization), EPA (Environment Protection Agency) and ECC (European Chemical Classification) classify methomyl as a very toxic and hazardous pesticide. Methomyl is highly soluble in water (57.9 g/liter, at 25°C [5], and since the sorption affinity of soils for this pollutant is rather low, it can easily cause contamination of both ground and surface water resources [6]. The residual pesticide comes in contact with water causing surface/ ground water pollution leading to the toxicity of biotic environment. Therefore, bioremediation techniques for the treatment of pesticides are of paramount importance [7,8]. Bioremediation can be defined as any process that uses microorganisms (Bacteria,fungi), green plants or their enzymes to return the natural environment altered bycontaminants of original condition. Bioremediation constitutes an attractive alternative to physico-chemical methods of remediation, as it is less expensive and can selectively achieve complete destruction of organic pollutants [9]. Microorganisms are thought to play an important role in the removal and detoxification of these toxicants from the environment. Degradation of pesticides is usually beneficial, since the reactions that destroy pesticides, convert most pesticide residues in the environment to inactive, less toxic, harmless compounds [10]. |
| For many degradation studies Pseudomonas sp. was selected as it is a well-known biodegrader of agrochemicals [11]. Five bacterial isolates (Pseudomonas aeruginosa, Klebsiella Sp., Escherichia coli, Bacillus Sp. and Corynebacterium) isolated from brinjal cultivated field were found to degrade cypermethrin. It was confirmed that these isolated organisms were able to utilize and degrade cypermethrin. In this five different bacterial colonies, Psedomonas aeruginosa, Klebsiella Sp., Escherichia coli were found active in utilizing cypermethrin (1%) whereas Bacillus Sp. and Corynebacterium were moderately active in utilizing cypermethrin (0.1%) [12]. Numerous strains capable of degrading compounds such as 2,4-D and monocrotophus have been isolated from various environments and have been found to be distributed over many different phylogenetic groups [13-17]. Microbial biodegradation is a major process leading to transformation of organic chemicals in soil, and hence bioremediation is viewed as the most attractive approach to the ultimate destruction of exogenous agricultural chemicals added to cultivated soils. However the efficiency of bioremediation of agrochemicals in soils depends on soil conditions. Recombinant DNA technology is a relatively new and fast growing area where the bacterial plasmids are being relatively used as cloning vehicles, because of its small size and relaxed mode of replication, resulting in multiple copies in a cell [18]. Such plasmids increase the biochemical versatility of the host bacterium, extending the range of complex organic compounds used as sole sources of carbon and energy. Significantly, these plasmids have been isolated almost exclusively from species of the genus Pseudomonas, a group of bacteria known to play a major role in the breakdown and recycling of naturally occurring organic molecules in both soil and water [19]. Pesticide degrading genes in microbes have been found to be located on plasmids, transposons, and/ or on chromosomes. Some microbial strains possess genetic determinants that confer resistance. In bacteria, these determinants are often found on plasmids, which have facilitated their study at the molecular level [20]. Bacterial plasmid plays a role in the degradation of the pesticide [21,22]. |
| Therefore, the present investigation was undertaken to isolate different bacteria from various soil samples and characterize the isolated bacteria for its capability to utilize highest level of methomyl as a sole source of the carbon and energy Further, an attempt was also made to determine the role of plasmid or chromosome inVolved in the biodegradation of methomyl by the soil isolate-Pseudomonas aeruginosa. |
| Materials and Methods |
| Isolation of methomyl degrading bacteria from soil |
| Soil samples were collected from 25 different areas around the Karnatak University campus, Dharwad. 1 gm of soil sample was added to 10 ml of autoclaved minimal salt medium (MSM) containing methomyl (10-3 M) as a source of carbon and energy. Cultures were incubated in an orbital shaker at 37°C at 120 rpm for 24-48 hrs. After enrichment cultivation, 0.1 ml of the culture was spread on minimal salt agar plates (MSA) that contained methomyl 10-3 M as a sole carbon source. These plates were then incubated at 37° for 24-48 hrs. A luxuriantly grown colony from each isolate was then used to inoculate into 10 ml of minimal salt medium containing methomyl (10-3 M) and further incubated for 24 hrs in an orbital shaker under the same conditions in order to confirm the biodegradability. These strains were subjected to high performance liquid chromatography (HPLC) and the percentage degradation was assayed. |
| The isolated strain which showed maximum methomyl degradation was subjected and regularly sub cultured to ensure the maintenance of the degradative phenotype. |
| Characterization and identification of isolated bacterial strain |
| The selected, isolated, most potential bacterial strain was characterized and identified on the basis of morphological and biochemical parameters. This was sent to Bioserve technologies, Pvt India Ltd, where it was subjected to further molecular characterization by polymerase chain reaction (PCR) amplification and sequence analysis by 16S rDNA identification. |
| Biochemical characteristics of the soil isolate |
| The following biochemical tests were performed according to the methods mentioned by Aneja [23]. |
| Determination of methomyl (%) using HPLC |
| Degradation of methomyl by the isolated strain was confirmed by high performance liquid chromatography (HPLC). The first one (Standard sample) was non-inoculated sample containing methomyl (10-3 M) in 100 ml of minimal medium was kept as a control. In the test sample 5 ml of the culture was added to sterile minimal medium containing methomyl (10-3 M) incubated in orbital shaker at 120 rpm at 370 C and the percentage degradation was analyzed every 24 hrs upto 96 hrs. The HPLC analysis was carried out as per Tamimi et al. [24]. |
| High performance liquid chromatography (HPLC) analysis was performed at USIC Department, Karnatak University Dharwad with a Shimadzu HPLC system. The analytical column used was C-18 (150 mm x 3 mm, 3 μm). The flow rate was 0.5 ml min-1 and the injection Volume was 20 μl. the isocratic elution conditions were acetonitrile and water. The wavelength for detection was 231 nm. |
| Plasmid isolation of the soil isolate- Pseudomonas aeruginosa |
| Plasmid DNA was isolated and cured by following the method described by Brown [25]. 2 ml of fresh bacterial culture was centrifuged to harvest cells. The cell pellet was resuspend in 0.3 ml of solution I (50 mg glucose, 25 mM tris pH8.0, 10 mM EDTA), gently vortexed and cooled for 10 min. 0.6 ml of solution II (0.2 N NaOH, 1% SDS) was added, mixed well and incubated for 10 min at room temperature. 0.450 ml of solution III (5 M sodium acetate), added, mix gently and incubated for 10 min, it was then centrifuged at 12,000 rpm for 10 min at 4°C. To the supernatant 1 ml of isopropanol was added, mixed well and allowed the plasmid to precipitate by incubating for 10 min at 4°C. The plasmid was separated by centrifuging at 10, 000 rpm for 10 min. the dried pellet was dissolved in TE buffer and stored until use. |
| Agarose gel electrophoresis |
| The method described by Maniatis et al. [26] was followed for agarose gel electrophoresis. 200 mg of a agarose was weighed and dissolved in 25 ml of TAE buffer (4.84 g Tris base, pH 8.0, 0.05 M EDTA/1 ltr) by boiling. When the gel attained ~55°C, it was poured into the gel cassette fitted with comb and allowed the gel to solidify. The comb was carefully removed and the gel was place in the electrophoresis chamber flooded with TAE buffer. 20 μl of DNA sample was loaded (mixed with bromophenol blue dye @ 1:1 ratio), carefully into the wells, along with standard DNA marker and the constant 50 V of electricity was passed for around 30 min. The gel was removed and carefully stained with ETBR solution (10 μg/ml) for 10-15 min and the bands were observed under UV transilluminator. |
| Plasmid curing of soil isolate-Pseudomonas aeruginosa |
| The LD-50 values were determined using a curing agent and then the cultures were subjected to plasmid curing. |
| Determination of LD-50 |
| Pseudomonas aeruginosa culture was grown in different concentrations of acryflavin (0-50 μg/ml) for 24 h in nutrient broth and the OD of biomass were observed at 660 nm against autoclaved media as blank. The OD of cultures was compared with control OD of culture (culture grown in absence of acryflavin) and the concentration of acryflavin giving nearly 50% less OD was considered as LD-50. The LD-50 values obtained in our results for Pseudomonas aeruginosa was 20 μg respectively. |
| Procedure for plasmid curing |
| 25 ml of nutrient broth was prepared, autoclaved and inoculated with test organism and incubated at 37°C for 18 hrs. 5 tubes of nutrient broth containing LD-50 concentrations of acryflavin were prepared and inoculated with 18 hr old test organism (1%) in tube no.1. The control tubes without acryflavin were also prepared and inoculated, as mentioned earlier. The tubes were incubated at 37°C for 24 hrs. From tube no.1, 1% culture was inoculated to tube no.2 and incubated further for 24 hrs. This serial inoculation was continued for 5 generations. At every generation, the plasmid was isolated and run on 1% agarose gel to observe for the presence/absence of plasmid and simultaneously the sample was analyzed by HPLC for methomyl degradation. |
| Statistical analysis |
| The statistical analysis has been performed according to routine formulae found in standard work on biological statistics [27-29]. |
| Statistical significance between the control and experimental data were subjected to analysis of variance (ANOVA) together with Dunnett’s test (P<0.05). |
| Results |
| Identification and characterization of methomyl degrading bacteria |
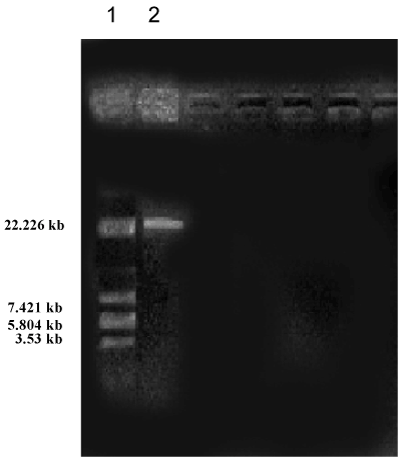
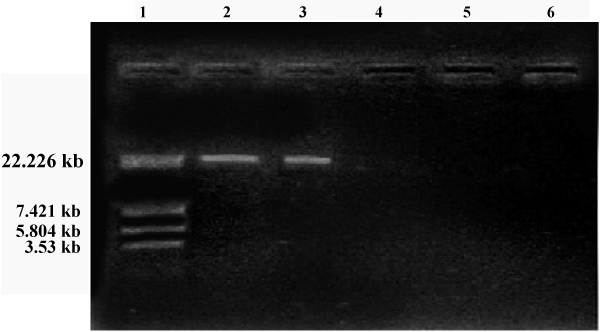
| Soil samples were collected from various areas around the University campus of which ten isolated were observed. Morphological characteristics of the isolates were studied (Table 1). All the isolates were subjected to HPLC analysis and the degradation of methomyl (%) observed for the Isolates No s. I, II, III, IV, V, VI, VII, VIII, IX and X were 46.43, 28.80, 56.42, 43.39, 47.38, 46.93, 45.82, 44.43, 45.27 and 44.62 % respectively, from the initial control of 62.26 %. The present study revealed that there was a significant degradation of methomyl, observed with all the isolates. However, maximum degradation of methomyl was observed by the isolate No.II (Table 2). This isolate (Isolate-II) which showed maximum methomyl degradation was observed morphologically, biochemically and sent for 16s rDNA identification. The selected isolate was gram negative rods, the biochemical characteristics showed that indole formation, methyl red test Voges-Proskauer tests were negative, citrate test was positive, H2S was not produced, catalase, gelatin hydrolysis and oxidase tests were positive (Table 3). The 16s rDNA reports revealed that the accession number was GO180118.1 (Tables 3 and 4) and the culture showed 99% similarity to Pseudomonas aeruginosa (Figures 1 and 2). |
| Methomyl degradation (%) using Pseudomonas aeruginosa |
| Pseudomonas aeruginosa: On treatment with 10-3 M of methomyl the degradation of methomyl was 74.42, 70.10, 68.59 and 66.34% at a given duration of 24, 48, 72 and 96 hrs respectively from an initial concentration of 94.66%. |
| The results of the present study revealed that, there was a significant decrease in the methomyl content in the treated group when compared with that of the control of Pseudomonas aeruginosa. But in comparison to Escherichia coli cells, the% degradation of methomyl observed was lesser with Pseudomonas aeruginosa cells (Tables 4 and 5). |
| Plasmid curing and methomyl degradation (%) using Pseudomonas aeruginosa (Table 2): The size of the plasmid was estimated on the basis of electrophoretic mobility of the isolated fragments as compared to the sizes of marker. The isolated plasmid DNA pattern of Pseudomonas aeruginosa showed that our strain harbours a plasmid. |
| The normal and plasmid cured strains were analyzed for methomyl degradation and it was observed that the normal cells of Pseudomonas aeruginosa showed a degradation of 56.29, 52.92, 50.61, 43.22 and 37.39% from an initial amount of 62.18% of methomyl on the other hand the plasmid cured cells reported a degradation of 57.90, 56.94, 56.48, 56.34 and 56.23% at 24, 48, 72, 96 and 120 hrs respectively. |
| In the present study it was observed that the plasmid was cured in the third generation and the cured strain was unable to degrade methomyl further (Table 5) which indicates that the degradative property of methomyl was encoded in the plasmid of Pseudomonas aeruginosa. |
| Discussion |
| The discovery of microorganisms capable of tolerating or growing in high concentrations of pesticides provides a potentially interesting avenue for treating hazardous wastes [4]. Some investigations resulting in the identification of microbial isolates which are apparently responsible for the accelerated degradation of individual pesticides is necessary [30,31] of which the 16s rDNA analysis is considered to the most valid and widely used technique in the identification of microorganisms [8,32-34]. |
| In the present study, the most potential bacterial strain with the highest ability to degrade methomyl was identified morphologically, biochemically and genetically. They were gram negative, rod shaped bacteria, they were Indole, MR and VP negative, H2S negative, citrate, catalase, gelatin hydrolysis and oxidase tests were positive and the analysis of the 16S rDNA showed highest similarity to Pseudomonas aeruginosa. |
| Biodegradation of methomyl by Pseudomonas aeruginosa |
| The present study revealed that the methomyl was decreased significantly by Pseudomonas aeruginosa. However, different degradation rates were observed. All the organisms could utilize methomyl as a sole carbon source and had the bioremediation ability. Further it was observed that the soil isolate Pseudomonas aeruginosa, exhibited a good growth rate and higher biodegradation efficiency towards methomyl when compared to the other isolates demonstrating their potential use in bioremediation. It is suggested that the detoxification metabolism occurs when a microorganism uses the pesticide as a carbon and energy source and the process is facilitated by resistant microorganisms [34]. Earlier studies have reported that Pseudomonas is a versatile genus and suggested that this genus could degrade a number of chemicals like pesticides including carbaryl, malathion, p-nitrophenol and parathion, bethoxazin, it is widely present in soil and can be used to clean up different xenobiotics compounds like propiconazole [35]. Earlier, two microorganisms classified as Pseudomonas sp. [36] and Escherichia coli [37] have been isolated and described as possessing the ability to degrade lindane. The results obtained in this study were in agreement with earlier reports that indicated the inVolvement of different species of Enterobacteriaecae in the degradation of insecticides like chlorpyrifos, phosphonate and glyphosate [38]. Several Pseudomonas sps that metabolize organophosphorous and carbamate pesticides have been isolated from soil [30,31]. It has been reported that Stenotrophomonas maltophilia M1 strain is capable of methomyl degradation [39]. Bhagobaty and Malik [40] have reported of the Pseudomonas strains capable of degrading chlorpyrifos. Bioremediation of endosulfan was carried out by Staphylococcus sp and Bacillus circulans [1]. Murugesan et al. [12] reported that a marked negative effect on the rate of degradation was observed with increased concentration of cypermethrin which may be due to mineral nutrients which are required for the growth of Pseudomonas and biodegradation of cypermethrin may become rate limiting in the wastewater sample. Their findings suggest that the utilization of cypermethrin by Pseudomonas aeruginosa may be feasible and this treatment option for the removal of pesticide from the soil and degradation observed only in the presence of microorganisms. Invitro testing of cypermethrin act against E.coli, S.aureus, P.aeruginosa and B.subtilis culture, were examined. Cypermethrin cause increases the effect of plant growth and the soil components. It was natural, inexpensive, and eco-friendly microbes endowed with pesticide degrading potential could be an ecologically good alternative in detoxifying soil residues encourage the farmers to use natural pesticides rather than chemical pesticides [41]. |
| The plasmid curing results in the present study reveals that the degrading ability for methomyl is encoded in the plasmid for Pseudomonas aeruginosa. Many xenobiotic degradation genes present in soil bacteria have been shown to reside on plasmid, a common location for other degradation genes [42]. Soil microbial populations, particularly members of the genus Pseudomonas, have evolved the considerable nutritional versatility and are capable of the degradation of a range of complex, naturally occurring aromatic and aliphatic compounds an interesting feature of these degradative plasmids are that they have been isolated almost exclusively from species of the genus Pseudomonas [13]. Significantly these plasmids have been isolated most exclusively from species of the genus Pseudomonas, a group of bacteria known to play a major role in the breakdown and recycling of organic molecules both in soil and water [19]. Similar result of plasmid encoded ampicillin resistance, obtained from Pseudomonas species was isolated from water and soil samples [43]. Plasmid conferring resistance to heavy metals and antibiotics was recorded in bacteria such as Pseudomonas fluorescence and Alcaligenes eutrophus from aquatic environments [44]. Plasmid-mediated dimethoate degradation was reported earlier by Deshpande et al. [21], employing Pseudomonas aeruginosa. Mandal et al. [4] reported that a single plasmid of Bacillus licheniformis was involved in dimethoate degradation and the genes encoding resistance to antibiotic and heavy metal were also located on the plasmid. |
| Plasmid curing results of the study by Raja and Selvam [45] suggested that nickel and ampicillin resistance was conferred by plasmid DNA, while cadmium, chromium and lead resistance seems to be encoded by genes of the bacterial chromosome of Pseudomonas aeruginosa. It has been suggested that the development of the resistant population in a contaminated soil can result from vertical gene transfer or horizontal gene transfer, including transposons and broad host range plasmids and selection pressures on spontaneous mutants due to the presence of xenobiotics. It has also been reported that pesticidedegrading plasmids are wide spread in soil microbial populations. The isolation and characterization of a variety of such plasmids should form the basis for the construction by either In-vivo or In-vitro genetic engineering techniques of strains of bacteria capable of the degradation of what were previously considered to be environmental pollutants [13]. Plasmids are thought to play an important role in the eVolution of pesticide degrading ability in microorganisms. Guha et al. [46] reported the plasmid borne genes of a soil bacterium Micrococcus to be responsible for chlorpyrifos degradation. The first ever documented organophosphorus biodegrading gene was identified in Pseudomonas diminuta. Serdar et al. [47] and Singh et al. [48] reported a bacterial strain Enterobacter that could catabolically degrade chlorpyrifos. Bhagobaty and Malik [40] have reported the Pseudomonas strains harboured a single plasmid with a role in chlorpyrifos biodegradation. |
| Plasmid coded biodegradation of methomyl observed in the present study may be due to the broad host range plasmids and selection pressures on spontaneous mutants due to the presence of xenobiotics, vertical gene transfer or horizontal gene transfer, including transposons and broad host range plasmids and selection pressures on spontaneous mutants due to the presence of xenobiotics or due to strains that harbour a single plasmid with a role in pesticide biodegradation [40]. |
| The present results confirm that the genes of the plasmid of Pseudomonas aeruginosa are involved in methomyl degradation and such bacterial plasmids can be used as cloning vehicles in recombinant DNA technology. |
| Conclusion |
| This study demonstrated that the isolated Pseudomonas aeruginosa possesses a strong ability for methomyl degradation. This strain contains a plasmid; and is believed to be responsible for the degradation of methomyl carrying the degrading gene. This plasmid could be transferred to another bacterial strain in the environment and provide it with methomyl pesticide degradative ability and potentially a selective advantage under a given environmental state. |
| Acknowledgements |
| The authors are grateful to the Post Graduate Department of Studies in Microbiology and Biotechnology, Karnatak University Dharwad for providing the necessary facilities. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 | Table 4 | Table 5 |
Figures at a glance
 |
 |
| Figure 1 | Figure 2 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 15198
- [From(publication date):
March-2015 - Apr 07, 2025] - Breakdown by view type
- HTML page views : 10429
- PDF downloads : 4769
