Research Article Open Access
Bioinformatic Characterization of Desaturase Partial Gene in Sesamum indicum
Spandana B1*, DSRS Prakash2 and Nagalakshmi S11Institute of Biotechnology (IBT), Acharya NG Ranga Agricultural University (ANGRAU), Guntur, Andhra Pradesh, India
2Department of Human Genetics, Andhra University, Visakhapatnam, Andhra Pradesh, India
- *Corresponding Author:
- Spandana B
Institute of Biotechnology (IBT)
Acharya NG Ranga Agricultural University (ANGRAU)
Guntur, Andhra Pradesh
India
Tel: 919440327774
E-mail: spandanabandila@gmail.com
Received date: February 10, 2017; Accepted date: February 20, 2017; Published date: February 27, 2017
Citation: Spandana B, Prakash DSRS, Nagalakshmi S (2017) Bioinformatic Characterization of Desaturase Partial Gene in Sesamum indicum. Adv Crop Sci Tech 5: 259. doi: 10.4172/2329-8863.1000259
Copyright: © 2017 Spandana B, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Advances in Crop Science and Technology
Abstract
By using Bioinformatic tools three sets of codehop, webprimer and manually, gene specific degenerate primers were designed for Sesamum desaturase. Through Gradient PCR approach maximum homology among desaturase sequences was observed to be with Arachis hypogaea than the other oil seed crops compared. Primers designed through Codehop and webprimershowed amplification of the gene sequence. The aim of this present study was to utilize bioinformatic tools for sesame improvement programs towards diversifying the fatty acid composition of the seed oil.
Keywords
Degenerate primers; Desaturase; Sesamum; Fatty acids; Conserved regions
Introduction
Fatty acid desaturases are the enzymes that are responsible for the insertion of cis-double bonds into pre-formed fatty acid chains in reactions that require oxygen and reducing equivalents. In the last few decades considerable work has been carried out in several plant species to alter the fatty acid composition of the oil by classical plant breeding methods and were able to generate low erucic acid. Brassica (canola oil) and sunflower oil containing high oleic acid. Currently Canola oil isthe lowest saturated fat vegetable oil. Fatty acids are synthesized from acetyl-Co A by a series of reactions that are localized in the plastids. Desaturases play a key role in polyunsaturated fatty acid homeostasis i.e., mostly the unsaturation which is the initial step in the essential fatty acid metabolism. Depending upon the intermediate product, Desaturases are of different types i.e., Stearoyl-ACP desaturases, oleate desaturases, linoleate desaturases etc. By understanding the enzymes involved in the synthesis and accumulation of seed- specific fatty acids, it may be possible to manipulate the composition of seed oil by generating transgenic plants by molecular biology techniques. It is necessary first to identify plant species which synthesize economically valuable oils and characterize the proteins/enzymes involved in the triacylglycerol biosynthesis.
Material and Methods
Genomic DNA Isolation: Total Genomic DNA of sesamum was extracted using a modified Cetyl trimethyl ammonium bromide (CTAB) method and quantified. All the reagents required for CTAB buffer preparation were obtained from Sigma-Aldrich (USA)
Designing of degenerate primers
The oil plant species containing the desaturase genes which have already been reported were collected from NCBI and EBI data banks. Desaturase nucleotide sequences of B. juncea, G.hirsutum, H.anus, S.indicum, A. hypogeae, R. communis, G. max. were taken from NCBI database and they are converted into protein sequences by web tool http://www.expasy.ch/tools/dna.html (Figures S1 and S2) The consensus amino acid and nucleotide sequences of desaturases (Fad D) were selected by using the Protein Domain database, Pro Dom (http:// prodes. toulouse. inra. Fr /prodom /doc/ prodom.html), NCBI (http:// ncbi.nlm.nih.gov) and EBI (www.ebi.ac.uk) tools and desaturase as the query. Clustal W software is used for multiple sequence alignment to identify the most conserved regions (Figures S3 and S4). The similarity matrix and the phylogenetic trees were produced using DNA Star (version 2004). The use of software in biological applications has given a new dimension to the field of bioinformatics. Many different programs for the design of primers are now available [1].
Degenerate primers for conserved regions of desaturases were designed manually and by using few software programmes like Web Primer and Codehop [2,3]. The consensus of both these protein and DNA sequence alignments gave rise to 3 sets of primer pairs which are listed in Table 1. Primer sequence degeneracy was also taken into consideration. In the present study Apart from the above mentioned softwares, the universal site for most of the bioinformatics tools were also used. (www.expasy.org). All the sequences were given for multiple sequence analysis by Clustal W. The resulted aligned sequences were given to BLOCKMAKER (http://bioinformatics.weizmann.Ac.il/ blocks/blockmkr/www/make_blocks.html) with each block at a time to BLOCKMAKER. The blocks with maximum similarity and minimum degeneracy were selected from the results. These blocks were given to web programme Codehop (http://bioinformatics.weizmann.ac.il/ blocks/codehop.html) for designing of primers. Primers were selected based on minimum degeneracy minimum degeneracy, length and percentage of GC. With one DNA sample three sets of gene specific degenerate primers (Manual, Web primer, Codehop) synthesized by Biotech Desk, India Pvt. Ltd.) were tested for PCR amplification of the gene PCR reactions were carried out in a DNA thermocycler (Eppendorf, USA) with a heated lid. Each 20 μl reaction volume contained about 100 ng of template DNA, 1X PCR buffer (10 mM Tris-HCl pH 5.3, 50 mM KCl), 2 mM MgCl2 (Invitrogen, USA), 2 mM dNTP, (Sigma -Aldrich., USA), 5 pmol each of Forward and Reverse primer and 1U of Taq DNA polymerase (Stratagene, Germany) and programmed for an initial denaturation step of 5 min at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 50°C, extension was carried out at 72°C for 1 min and a final extension at 72°C for 10 min and a hold temperature of 14°C at the end. Negative controls were also run without template DNA to avoid non-specific amplification.
| Primer pair | Primer sequence (5’-3’) | bp | Product size(bp) |
|---|---|---|---|
| Des M | F -ACYGGNRTNTGGGTNMTNGCNCAYG R- NGCYTGYTAYTGNGGCATNGTNGAC |
25 25 |
400 |
| Des W | F -GGGCGTGTCACTAAGATTGAA R- TGTACCAGAGCACACCTTTGT |
21 21 |
400 |
| Des C1 | F- TGGACATCATGCTTTCTCTGAT-3' R- GGTACGATACCTCCGATGATTCC-5' |
24 24 |
400 |
| Des 2 | F -AGTGCTTTCACGCGATTTCT R -TCTCTCGAAGCAGTGTGGTG5’ |
20 20 |
400 |
Table 1: PCR primers designed and targeted fragments of the Desaturase gene in Sesamum indicum.
On 1.4% agarose gels PCR were electrophoresed, in 1X TAE Buffer at 50 V for 3 hrs and then stained with ethidium bromide (0.5 ug/ ul). To ascertain reproducibility of the raeaction PCR reactions were repeated three times reaction.
Gel elution
Fragments ranging from 400-700 bp were eluted by using Column method (Qiagen Kit)
Ligation
A 10 μl Ligation reaction volume was set up containing 1 μl of pGEM easy vector (Promega, USA); 5 μl of PCR enriched (gel eluted) product; 1 μl 10 X T4 DNA ligation buffer (New England Biolabs, USA), 2.4 U/μl of T4 DNA ligase (New England Biolabs, USA) and incubated at 14°C overnight. The samples were stored at - 20°C until used.
Transformation
Ligated product was transformed. The plasmid isolation was done by Alkaline lysis method. After PCR amplified plasmids amplification range (400 bp desaturase) was selected and was given for sequencing. Using Vecscreen (NCBI) the vector sequences were removed form the sequenced clones using and homology search was carried.
Sequence alignment and phylogenetic analysis
The nucleotide sequences and deduced aminoacid sequenceswere analysed using comparitive and bioinformatic analysis that are available online at the websites (http://www.ncbi.nlm.nih.gov/) and (http://cn.expasy.org/). The nucleotide sequence, deduced aminoacid sequence, and open reading frame were analyzed and the sequence comparision was conducted through a database search using BLAST programs (Figure S5). The phylogenetic analysis of desaturase gene from other oilcrop species was aligned with Clustal W program using default parameters [4].
From the edible oil yielding species Glycine max, Brassica rapa, Helianthus annuus, Arachis hypogaea the sequences were selected for characterization and converted to amino acid sequences to identify the conserved regions which are given Figure S1.
Desaturase
The Degenerate primers were designed by manually, webprimer and codehop methods which are given in Tables 1 and 2.
| Accession | Organism | Max score |
|---|---|---|
| AB 262185.1 | Phanerochaete chrysosporium Pc-ole1 gene for delta9-fatty acid desaturase, complete cds | 87.8 |
| AY500378.1 | Lilium longiflorum phytoene desaturase mRNA, partial cds | 87.8 |
| AY 234124.1 | Primula farinosa sphingolipid delta-8 desaturase mRNA, complete cds | 41.0 |
| EU 424174.1 | Sorghum bicolor fatty acid desaturase mRNA, complete cds | 31.9 |
Table 2: Homology results of Desaturase.
Results
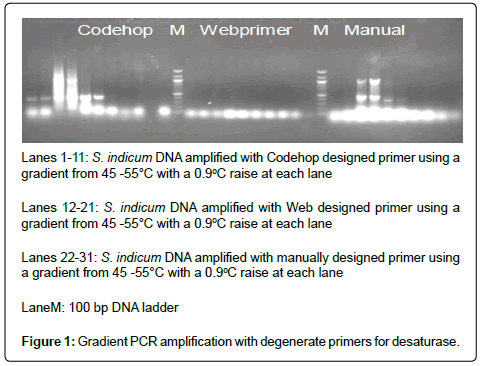
Three sets of gene specific degenerate primers (manual, Web primer, Codehop) synthesized by Biotech Desk, India Pvt. Ltd.) were tested for PCR amplification by gradient PCR with one sesame sample Figure 1 The amplified products of each primer is eluted and transformed and isolated plasmids from selected colonies are sequenced. A total of 10 positive clones of which 6 clones were sequenced. The sequences obtained was subjected to BLAST search.
Search results of desaturase in the NCBI for homologs
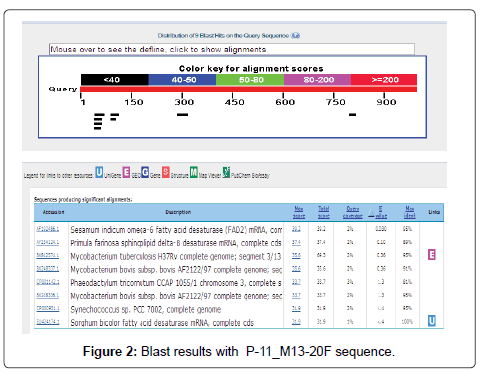
The Six sequenced desaturase sequences were submitted to the NCBI (National Centre for Biotechnology Information) website for BLAST-N (http://www. ncbi.nlm. nih.gov/ BLAST/) which showed high level of sequence homology both at the nucleotide and amino acid level with those from P. crysosporium, L. longiflorum, P. farinose and S. bicolor as shown in Figure 2.
Most of the sequences showed high homology with Sesamum indicum omega-6 fatty acid desaturase (FAD2) mRNA, complete cds and also with Phanerochaete chrysosporium Pc-ole1 gene for delta9-fatty acid desaturase, complete cds, Lilium longiflorum phytoene desaturase mRNA, partial cds, Primula farinosa sphingolipid delta-8 desaturase mRNA, complete cds Mortierella sp. strain M10 mRNA for delta-9 fatty acid desaturase, partial Sorghum bicolor fatty acid desaturase mRNA, complete cds, Chlamydomonas reinhardtii fatty acid desaturase (CHLREDRAFT_32523) mRNA, partial cds.
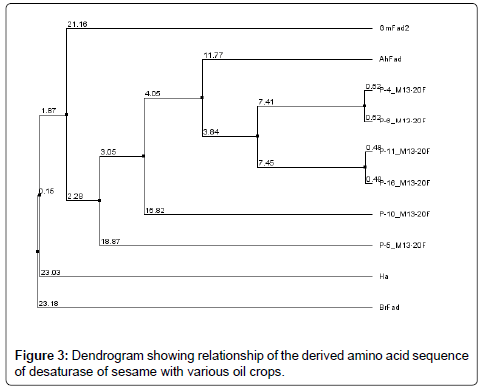
A comparison of Fatty acid desaturase sequences showing phylogenetic relationship at the deduced amino acid level with those of other oilseed crops is shown in the dendrogram (Figure 3). The homology was based on small portion of entire coding sequence, as the sequence length of the obtained sequence was 181 to 538 bp, where as the length of the selected sequences was 1149 to 1095 bp.The expected DNA fragment range was 400 bp and the primers were designed excluding N terminal region with C terminal region only as N terminal region was having more INDELS (Insertions and deletions).
A sequence homology search revealed that the putative protein had high homology with the desaturase sequences of Arachis hypogaea indicating that the desaturase isolated from S. indicum was closer to A. hypogaea than the other oil crops compared.
Discussion
Degenerate primers based on the amino acid sequence of conserved regions were also used to search for members of a gene family [2]. Computer programs have also been developed specifically for degenerate primer design [3].
Bioinformatics approaches to lipid research have recently begun using large amounts of mass spectrometry and microarray data. Phylogenetic analysis of protein families related to fatty acids has also been performed. However, to our knowledge, there is no report that describes the investigation of fatty acid structures based on the comprehensive analysis of the gene contents in the genomes. The fatty acid structures is hard to predict from genomic information due to the the functional diversity of the key enzymes. Many studies in plant systems were also conducted, which eventually led to the purification of a soluble stearoyl-acyl carrier protein (ACP) 9-desaturase from safflower [5]. Later studies allowed for the full nucleotide sequence of stearoyl- ACP 9-desaturases from castor and cucumber to be reported [6]. Various genes encoding enzymes involved in fatty acid synthesis have been isolated from the species and characterized. Carotene desaturase gene was cloned and characterized from chlorella protothecoides cs- 41 to elucidate lutein biosynthesis pathway [7]. Cloning of Psy1, Pds and Zds cDNAs encoding the enzymes responsible for lycopene biosynthesis, namely phytoene synthase 1 (PSY1), phytoene desaturase (PDS) and ζ -carotene desaturase (ZDS), respectively, from high lycopene tomato cultivar, Solanum lycopersicum KKU-T34003 [8]. Although the pathway for sesame is not documented, the fatty acid profile suggests synthesis via the known route common to most major oil crops Desaturase, This enzyme in fatty acid biosynthesis in sesame appear to be showing homology to different oilseed helianthus and arachis. The reasons are unknown but a more detailed study of these enzymes across the different oilcrops will through some light on the fatty acid biosynthesis pathway.
In the present study after analysis of available sequences of desaturase related proteins, retrieved desaturase subgroup clusters and selected six representative sequences of most members of desaturase families which specifically belonged to dicots as Sesamum is a eudicot species. In the present study codehop and Webprimer designed primers showed amplification for desaturase whereas manual designed primer gave no amplification. Desaturase derived sequences showed high homology with desaturase genes of Phanerochaete, lillium, primula sp.
References
- Kamel A, Abd-Elsalam A (2003) Bioinformatic tools and guideline for PCR primer design. African Journal of Biotechnology 2: 91-95.
- Wilks AF, Kurban RR, Hovens CM, Ralph SJ (1989) The application of the polymerase chain reaction to cloning members of the protein tyrosine kinase family. Gene 85: 67-74.
- Chen H, Zhu G (1997) Computer program for calculating the melting temperature of degenerate oligonucleotides used in PCR or hybridization. Biotechniques 22: 1158-1160.
- Thompson JD, GibsonTJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876-4882.
- McKeon TA, Stumpf PK (1982) Purification and characterization of the stearoyl-acyl carrier protein desaturase and the acyl-acyl carrier protein thioesterase from maturing seeds of safflower. Journal of Biological Chemistry 257: 1214-12147.
- Shanklin J, Sommerville CH (1991) Stearoyl- acyl-carrier-protein desaturase from higher plants is structurally unrelated to the animal and fungal homologs. Proc Natl Acad Sci88: 2510-2514.
- Meiya Li, Zhibing Gan, Yan Cui, Chunlei Shi, Xianming Shi (2011) Cloning and Characterization of the ζ-Carotene Desaturase Gene from Chlorella protothecoides CS-41. Journal of Biomedicine and Biotechnology 2011: 731542.
- Krittaya S, Preekamol K (2013) cDNA cloning and expression analyses of phytoene synthase 1, phytoene desaturase and ζ -carotene desaturase genes from Solanum lycopersicumKKU-T34003 Songklanakarin. Journal of Science and Technology 35: 517-527.
Relevant Topics
- Agricultural science
- Agronomy
- Climate impact on crops
- Crop Productivity
- Crop Sciences
- Crop Technology
- Field Crops Research
- Hybrid Seed Technology
- Irrigation Technology
- Organic Cover Crops
- Organic Crops
- Pest Management
- Plant Genetics
- Plant Breeding
- Plant Nutrition
- Seed Production
- Seed Science and Technology
- Soil Fertility
- Weed Control
Recommended Journals
Article Tools
Article Usage
- Total views: 3066
- [From(publication date):
February-2017 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 2250
- PDF downloads : 816