Research Article Open Access
Biofilm-Mediated Heavy Metals Bioremediation in PGPR Pseudomonas
Amina Meliani1* and Ahmed Bensoltane2
1Department of Biology, University of Mustapha Stambouli, Mascara, Algeria
2Laboratory of Experimental Biotoxicology, Biodepollution and Phytoremediation, Department of Biology, University Oran (Es-senia), 31000 Oran, Algeria
- *Corresponding Author:
- Amina Meliani
Department of Biology, University of Mustapha Stambouli, Mascara, Algeria
Tel: 00213551805873
E-mail: ameliani2003@yahoo.fr
Received date: July 19, 2016; Accepted date: September 15, 2016; Published date: September 16, 2016
Citation: Meliani A, Bensoltane A (2016) Biofilm-Mediated Heavy Metals Bioremediation in PGPR Pseudomonas. J Bioremediat Biodegrad 7:370. doi: 10.4172/2155-6199.1000370
Copyright: © 2016 Meliani A, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
It is often reported that biofilm-grown cells exhibit enhanced tolerance toward adverse environmental stress conditions, and thus there has been a growing interest in recent years to use biofilms for biotechnological applications such as the uptake of heavy metal. We present in this study the promising application of Pseudomonas biofilms in heavy metal uptake. The main objective of this study is to investigate if these isolates can withstand metal toxicity, and concomitantly to evaluate the interaction between heavy metals and biofilm formation. Compared to control experiments, all Strains were found to produce a greasy-looking biofilm which varied in thickness from an ‘almost invisible film’ to a paper-thick structure depending on the presence of zinc and lead, they exhibited an important biofilm mass. These findings underline the robustness of biofilms under stress conditions and its potential to maintain a favorable niche in stressful environments with increased heavy metal concentrations. Statistically, the biofilms formation seems to be more correlated to the antibiotics resistance (r=0.73; P<0.05) than the heavy metals resistance (r=0.31; P<0.05). Surprisingly, stationary-phase growing was found to be more resistant than logarithmically growing. There is no direct evidence that links metal resistance in biofilms according to the statistical analysis.
Keywords
Pseudomonas; Biofilms; Heavy metal; PGPR; Resistance
Introduction
Bioremediation is an emerging in situ technology for the cleanup of environmental pollutants using microorganisms. The biological processes for treating toxic effluents are better than chemical and physical methods in terms of their efficiency and economy and the potential of biofilm communities for bioremediation processes has recently been realized [1].
Due to their extremely toxic nature even at trace concentrations and their non-biodegradability unlike organic pollutants, heavy metals are a persistent threat to our life and environment. Metal contamination has been linked to birth defects, cancer, skin lesions, mental and physical retardation, learning disabilities, liver and kidney damage, and a host of other diseases [2]. Heavy metals are the primary inorganic contaminants, which include cadmium, chromium, copper, lead, mercury, nickel and zinc etc. Heavy metal bioremediation can be achieved by immobilization, concentration and partitioning to an environmental compartment, thereby minimizing the anticipated hazards [3,4]. In White and Gadd [5] study by a simultaneous increase in the EPS content of the biofilm was also observed, which suggested the role of EPS and biofilms in the entrapment of metal precipitates. Biofilms can also affect the fate of other compounds in their vicinity as a consequence of their physiological response during the absorption of water and inorganic or organic solutes [6].
Therefore, the application of heavy metal solubilizing microorganisms is a promising approach for increasing heavy metal bioavailability in heavy metal amended soils. In addition, bacteria producing indole acetic acid, siderophores and 1-aminocyclopropane- 1-carbox-ylate deaminase and phosphate-solubilizing bacteria are capable of stimulating plant growth [7]. Generally, PGPR function in three different ways [8], synthesizing particular compounds for the plants, lessening or preventing the plants from diseases [9] and facilitating the uptake of certain nutrients from the environment [10]. In this context this study is undertaken, the feasibility of using a PGPR Pseudomonas, for the removal of heavy metals from a contaminated soil and sediments was evaluated by several scientific reports. This study deals with the use of PGPR Pseudomonas for the metals and antibiotics resistance combined with the formation of biofilm. It also discusses the importance of this microbial community in increasing the efficiency of the process of metal resistance.
Materials and Methods
Organism and culture maintenance
The bacterial strains used in this study were: Pseudomonas aeruginosa (P8) isolated from waste water and three PGPR strains of P. fluorescens (P4, P9, P10) from different rhizospheres with PGPR traits like synthesis of amino-cyclopropane carboxylic acid (ACC) deaminase, siderophores, indole-3-acetic acid (IAA) and PO4 solubilzation. Stock cultures were stored at 80°C in Trypticase soy broth and 15% glycerol. Tests conducted for their identification have been based on physiological, nutritional tests [11] and by the use of the analytical Profile Index (API 20NE; bio Merieux Vitek). Prior to each experiment, a loopful of culture was grown in 10 ml of LB medium with incubation at 28 ± 2°C for 24 h.
Extracellular enzyme activity assays
Activities of extracellular enzymes of the strains were evaluated by the inoculation of 50 μl of cell-free, sterile-filtered supernatants from the stationary phase LB cultures (24 h, 37°C, 200 rpm) in holes (0.8 cm in diameter) stamped into substrate agar plates. The plates were incubated for 48 h at 37°C unless stated otherwise. Diameters of clear or turbid halos around the inoculate indicated a positive reaction.
Hemolytic activities were determined on LB agar supplemented with 5% sterile-filtered sheep blood. Protease activities were tested on 5% (w/v) skim milk agar plates. Clear zones around the inoculation hole indicated the production of hemolysins and proteases, respectively. For the estimation of phospholipases A and C activities, a clear halo after 24 h of incubation indicated phospholipase A activity, a white precipitate around the inoculum indicated phospholipase C activity. For biosurfactant production the drop collapsing and lipase test [12] was used as a qualitative test.
Screening for heavy metal resistance activity
Screening for heavy metal resistance was carried out using standard heavy metal solutions of Zinc (ZnSO47H2O) and lead (Pb (NO3) added to SLP agar medium as described by Sheng et al. [13]. The concentration of the standard heavy metal salts solutions was ranged 100 μL, 200 μL and 300 μL. The salt solutions were prepared with phosphate buffer saline PBS (pH 6.8).
Screening for antibiotic resistance
Antibiotic resistance was tested using LB agar containing Chloramphenicol (C30) (20 mg/ml), streptomycin (S10) (20 mg/ml), Ampicillin (AMP 10) (100 mg/ml) and Nalidixique acid (NA30) (20 mg/ml) which were added aseptically to the medium after autoclaving. All the antibiotics used in resistance tests were supplied from oxoid. Cultures were incubated at 30°C for 48 h.
Biofilm development analysis
Pseudomonas cells were harvested by centrifugation (9000 g, 10 min), washed twice with sterile water, and then resuspended in phosphate-buffered saline (PBS) to a OD580nm of 0,4 (~2.108 bacteria/ ml) for biofilm assay. The amount of surface-attached biofilm was determined by using a modified crystal violet method O’Toole and Kolter [14]. The bacterial suspensions were added to individual wells (0.2 ml) in tissue culture microtiter plates and incubated up to 72 h at 30°C. After incubation the biofilms were stained with 0.35% filtered crystal violet. Acetic acid (30% [vol/vol]) was added to each well, and absorbance was measured at 590 nm using a Magellan reader with a Tecan absorbance. Isolates were classified as biofilm producers if OD590 was >0.200 and further classified as strong, moderate, weak, or zero biofilm formers based on their final OD590 reading [15].
Statistics
The statistical processing of the data obtained from all studies was implemented by means of dispersion analysis with the STATISTICA 7 software. Data are expressed as means ± standard deviation (SD). Statistical analysis was performed with an analysis of variance (ANOVA) and correlations, a difference was considered statistically significant when p ≤ 0.05.
Results
Surfactant production
Extracellular enzyme activity assays for the isolates exhibited similar characteristics, with a few noticeable exceptions for hemolytic activity, P. fluorescens strains (S4 and S9) were β-hemolytic, however P. aeruginosa (S8) and P. fluorescens (S10) were found α -hemolysin positive. They were positive for lecithinase, esterase and phospholipase C activity.
The selected strains were screened for production of surface active molecules. In the drop collapsing test a flat drop was observed around colonies for each isolates, with an important aspect for P. aeruginosa (S8) and P. fluorescens (S9) indicating a biosurfactant activity. Furthermore, the presence of lipase confirmed that these isolates are potential producers of surface active molecules.
Heavy metal resistance
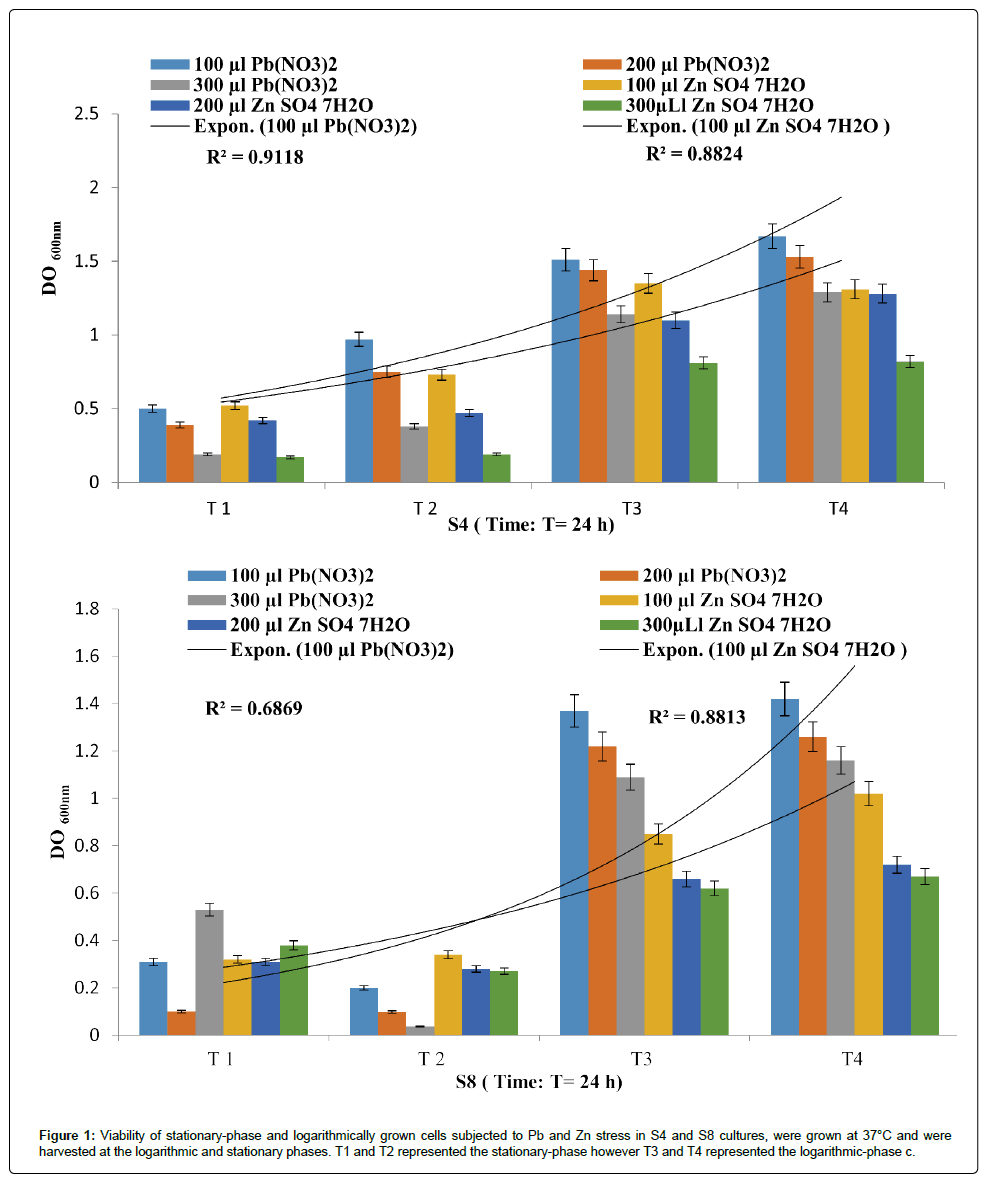
The isolates exhibited different multiple heavy metal resistance characteristics as described in Figures 1 and 2. The P. fluorescens strains (S4) seems to tolerate more PbNO3 than ZnSO4 with a R2 of 0.911 and 0.882 respectively. Eventually, P. aeruginosa strains (S8) was found to be completely resistant towards the tested metals with a R2 of 0.686 and 0.881.
Figure 1: Viability of stationary-phase and logarithmically grown cells subjected to Pb and Zn stress in S4 and S8 cultures, were grown at 37°C and were harvested at the logarithmic and stationary phases. T1 and T2 represented the stationary-phase however T3 and T4 represented the logarithmic-phase c.
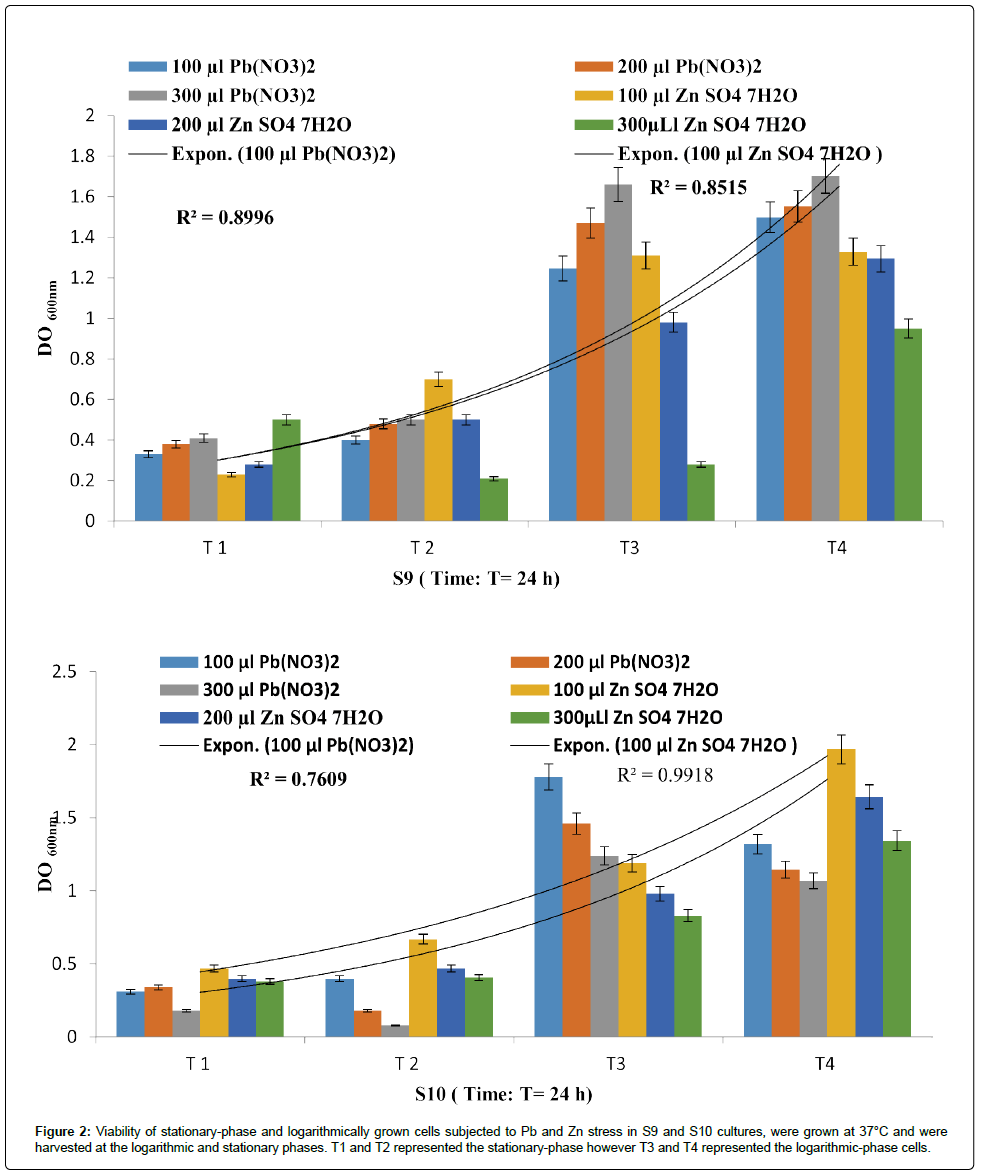
Figure 2: Viability of stationary-phase and logarithmically grown cells subjected to Pb and Zn stress in S9 and S10 cultures, were grown at 37°C and we re harvested at the logarithmic and stationary phases. T1 and T2 represented the stationary-phase however T3 and T4 represented the logarithmic-phase cells.
It is interesting to point out that P. fluorescens (S10) showed the best resistance towards Zn (R²=0.991) more than Pb (R²=0.760), despite the fact that P. fluorescens strains (S9) had recorded a R²=0.899 and R²=0.851 for PbNO3 and ZnSO4 respectively.
Statistically, it appears that there is a relationship between the metal resistances in function of time for each isolate. Even though the correlations obtained in the present study are not statistically significant, this disparity can be attributed to different intrinsic and extrinsic factors as signaled by Harrison et al. [16]. There is now some recognition that antibiotic-resistant bacteria can be maintained in the environment owing to co- or cross-resistance to toxic metals or the coregulation of resistance pathways.
Harrison et al. [16] suggested that both antibiotics and toxic metal species kill biofilm populations in a time- and concentration-dependent manner. This is observed as biphasic population killing, in which most of the growing population is rapidly killed by a low concentration of the antimicrobial. However, a larger portion of the biofilm population is able to withstand these lethal factors for exposure durations and at concentrations that exceed that which is lethal to the corresponding planktonic form, Harrison et al. [17] signaled that the concentrationdependent killing of microbial populations is exemplified by a plateau in the activity of the antimicrobial.
In accordance of the model described by Harrison et al. [16] about understanding biofilms strategy during exposure to toxic stressors, the kinetic of PbNO3 and ZnSO4 solubilization seems to be related to the bacterial growth, especially for the latent period where a significant solubilization was recorded for S4 (p=0.029) from T1 to T2, for S9 (p=0.03T1-T2) and for S10 (p=0.018T1-T2; p=0.52T3-T4 p=0.031). According to Gilbert et al. [18] the stationary-phase bacteria are more resistant to antibiotics that target actively growing cells than logarithmically growing cells has been known for some time. However, and in contrast to our results with comparing the data generated by Spoering and Lewis [19], it seems to be difficult to explain this variation. Teitzel and Parsek [20] determined that logarithmically growing cells were more resistant to copper and lead stress than stationary-phase cells. However, biofilms were observed to be more resistant to heavy metals than either stationary-phase or logarithmically growing planktonic cells.
Spoering and Lewis [19] examined the relative effects of antimicrobial agents on stationary and logarithmic phase cells of P. aeruginosa and found that stationary-phase cells were more resistant to a variety of different antimicrobial agents. In the same study the researchers suggested that the resistance of biofilms to antimicrobial agents can be primarily attributed to the stationary phase or slow growth and the presence of a small resistant subpopulation of cells termed persistors. Our findings are consistent with these reports and attributed this resistance to the stationary phase and not to the logarithmic phase.
Studies have also shown that the S8 resistance was statistically no significant p ≥ 0.05. The R2 of the regression is relatively high for all the strains with the exception of S10. The results described above revealed that S4 and S8 were able to biosorbe or to bioprecipitate more Pb NO3 than S9 and S10 which had affinity for Zn SO4. It is important to note S4 had exhibited a significant resistance via a Cross-resistance. It has been reported that the heavy metal driven co-selection of antibiotic resistance. As it is pointed out in these lines, there is now some recognition that antibiotic-resistant bacteria can be maintained in the environment owing to co- or cross-resistance to toxic metals or the co-regulation of resistance pathways. Cross-resistance describes mechanisms that provide tolerance to more than one antimicrobial agent such as antibiotics and heavy metals [21]. As an example, several multi drug efflux pumps are known to mediate decreased susceptibility toward antibiotics and heavy metals by rapid extrusion of the toxins out of the cell [22]. Further well-characterized cross-resistance mechanisms were reviewed by Baker-Austin et al. [23]. Co-resistance is defined as two or more genetically linked resistance genes, meaning that genes responsible for two or more resistances are located next to each other on one mobile genetic element [21].
Moreover, environmental pollution by heavy metals not only triggers co-selection processes, but also increases the level of tolerance to antibiotics due to co-regulation of resistance genes. Heavy metal ions are known to co-regulate genes responsible for antibiotic resistance and decrease antibiotic susceptibility [23]. Furthermore, Studies investigating co-selection in the environment frequently show the correlation of increased heavy metal concentrations with increased phenotypic or genotypic antibiotic resistance as described in some articles. However, some studies indicate that increasing heavy metal concentrations lead to a decrease of antibiotic resistance [24]. These contradicting results were investigated by Hölzel et al. [24].
Our values were closer to those of Teitzel and Parsek [20] who found that free-swimming P. aeruginosa cells were susceptible to the heavy metals tested in the following order of susceptibility: Zn2+<Cu2+<Pb2+. The measured MICs of lead and zinc were lower than those previously reported for P. aeruginosa (24 to 48 mM for Zn and 15 mM for Pb) [25]. The discrepancy between previously reported values and the values obtained in our study is most likely due to different growth medium (Mueller-Hinton broth) used by de Vicente et al. [25], which probably resulted in a high level of complexation between the metal cations and components of the growth medium. Our values were closer to those of Bender and Cooksey [26] who reported a MIC of CuSO4 of 100 M for P. syringae.
Antibiotic resistance
Results in Figure 3 revealed that the four isolates displayed different response of resistance to the tested antibiotics, however S9 seems to be more susceptible to Chloramphenicol (C30), Streptomycin (S10), Ampicillin (AMP 10) and Nalidixique acid (NA30) with a R2 of 0.90. Taken together, our data also indicate that S4 had exhibited a significant resistance.
Unsurprisingly, biofilm antibiotic susceptibility has been the subject of intense research and has been the focus of several excellent reviews [16]. By contrast, there have been many recent advances in the study of biofilm metal susceptibility, but until now the topic has not been reviewed. Other components of biofilm matrices are membrane vesicles, which bind to and sequester the positively charged aminoglycoside antibiotics [27].
The resistance described above may also explain, in part the role of the other components of biofilm matrices, which bind to and sequester the positively charged aminoglycoside antibiotics as described by the same author. Similar arguments have previously been made for the fluoroquinolone antibiotic ciprofloxacin, which otherwise quickly penetrates these highly tolerant biofilms [28].
Biofilms formation
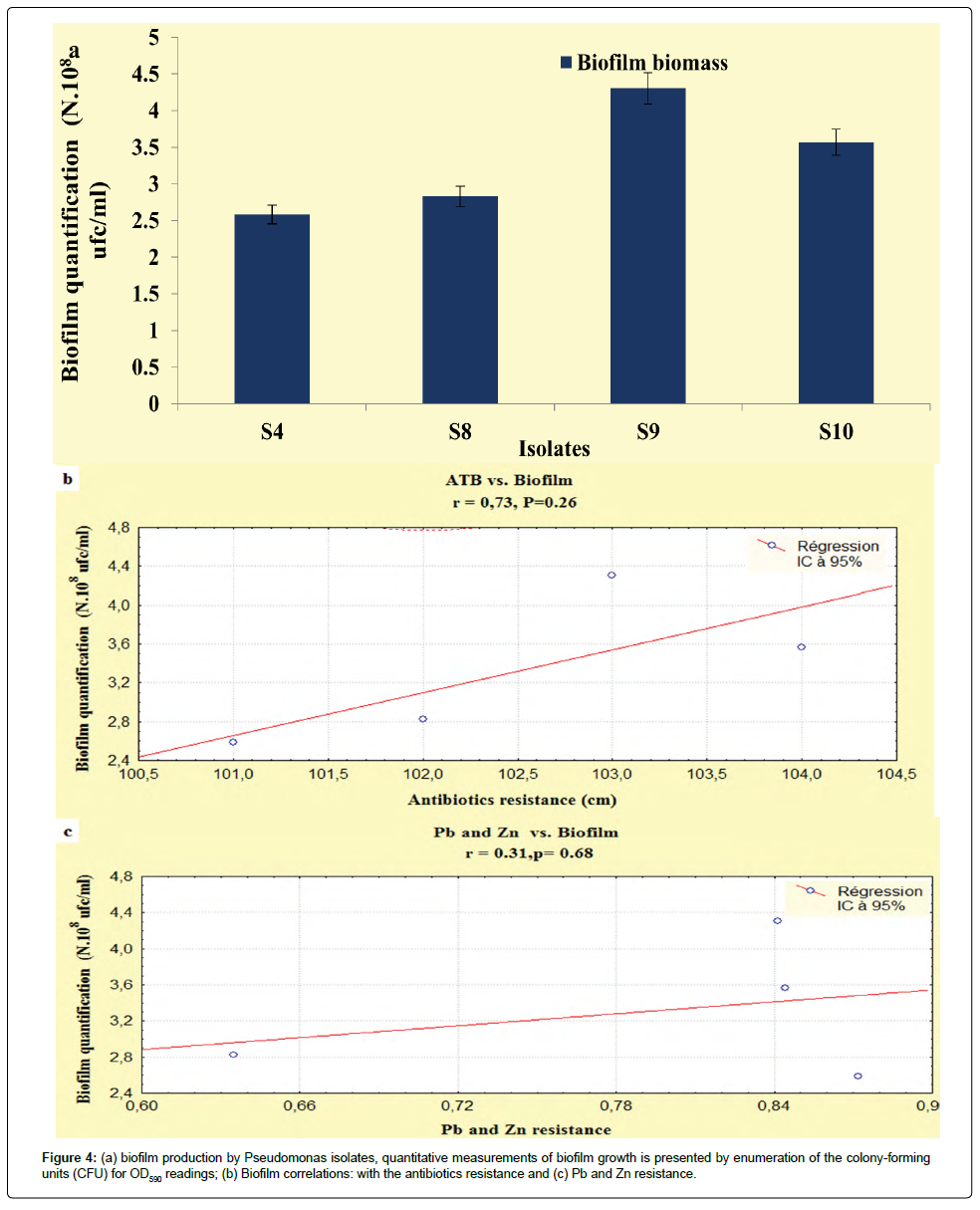
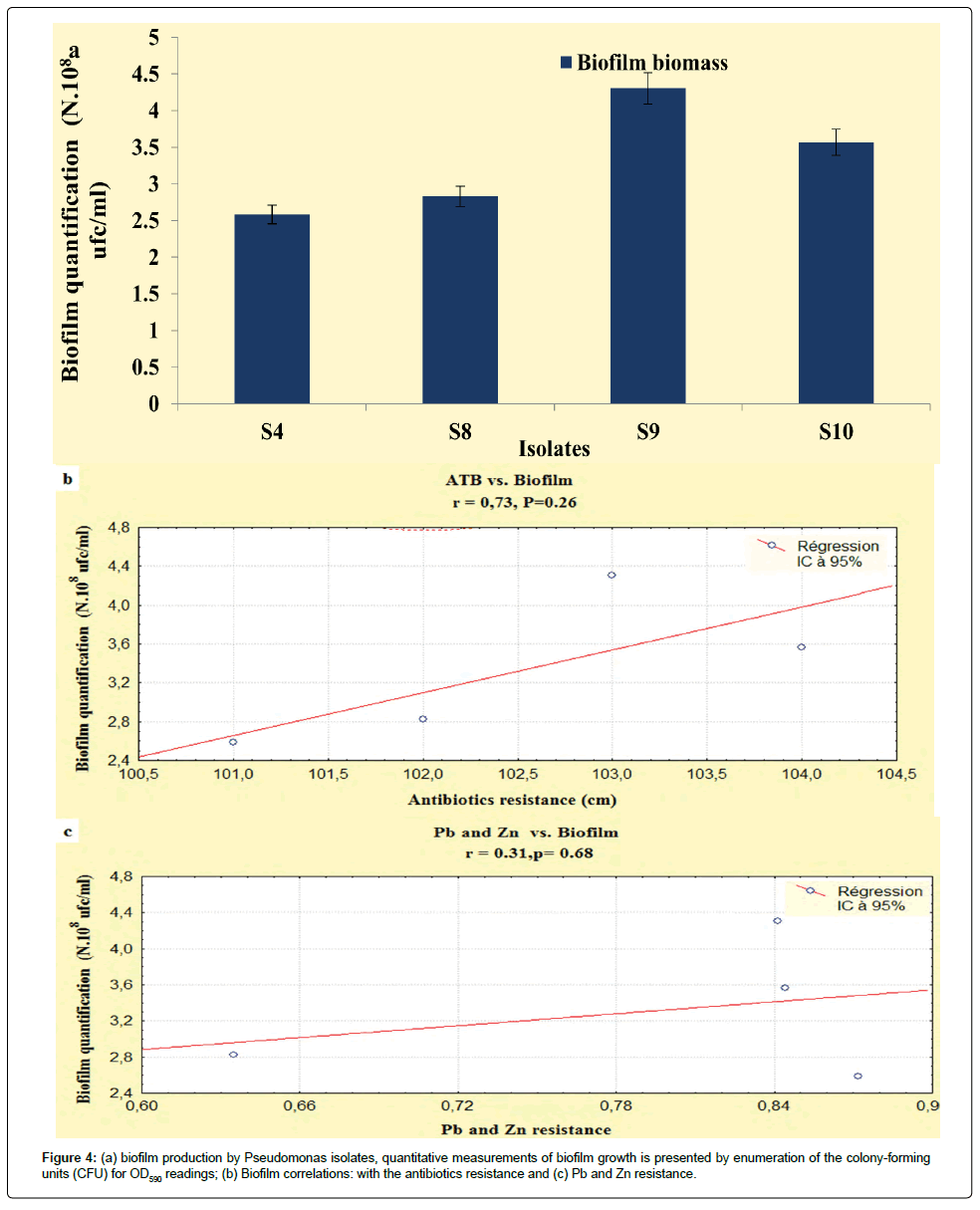
The results developed in this study, showed that our isolates exhibited an important biofilm mass, the P. fluorescens (S9) and P. aeruginosa (S8) exhibited increased biofilm formation. The Pseudomonas isolates had OD590 readings ranging from 0.29 to 0.35 (Figure 4a), according to StepanoviÄÂ? et al. [15] and Perez et al. [29] classification, these isolates were categorized as having moderate biofilm adherence properties, despite they were categorized as slimeproducers and exhibited a coordinated multicellular behavior called swarming with a crystal structure of the EPS matrix [30].
As described in the literature microbial EPS contribute to protect cells from hostile environments and can bind significant amounts of heavy metals. Forming biofilms by these strains is considered a natural strategy to maintain a favorable niche in stressful environments with increased metals concentrations. As reported by Workentine et al. [31] biofilms may reduce metal toxicity by altering their physiology to protect the sensitive chemical targets of the reactive metal species. Upon metal binding, the concentration of the free toxic ions in the cytoplasm is minimized. Biosorption of toxic metals is known from cell membranes, cell walls and extracellular polymeric substance (EPS) of biofilms [16]. For example, the EPS matrix and the contained polysaccharides were reported to bind heavy metals [20]. Thus, the metal tolerance of the bacteria belonging to that biofilm was enhanced.
Discussion
In the current study, three PGPR strains of P. fluorescens and P. aeruginosa were tested for biofilms, biosurfactans production and were evaluated for their resistance toward Zinc and lead.
In the drop collapsing test a flat drop was observed around the colonies, which indicates a biosurfactant activity. According to Ron and Rosenberg [32] several Pseudomonas are able to synthesize biosurfactants of diverse chemical nature. The most studied of these compounds are the rhamnolipids produced by P. aeruginosa. Microbial compounds that exhibit particularly high surface activity and emulsifying activity are classified as biosurfactants. These are structurally diverse surface active compounds capable of reducing surface and interfacial tension at the interfaces between liquids, solids, and gases, thereby allowing them to mix or disperse readily as emulsions in water or other liquids [2].
Recently biofilms have become a focus of interests for the researchers in the field of bioremediation of xenobiotic compounds. Microbial biofilms, natural or engineered, could be used to remediate heavy metal pollution by biochemical modification and/or the accumulation of toxic metal ions [33,34]. Biofilms are clusters of microbial cells that are attached to a number of different surfaces such as natural aquatic and soil environments, living tissues, medical devices or industrial or portable water piping systems [6]. Biofilms give support to the high density of microbial biomass which facilitates the mineralization processes by maintaining optimal pH conditions, localized solute concentration and redox potential in the vicinity of the cells [27,35].
Heavy metal remediation can be achieved by immobilization, concentration and partitioning to an environmental compartment, thereby minimizing the anticipated hazards [27]. There are reports on the application of biofilms for the removal of heavy metals [34] reported that biofilm process with simultaneous nitrogen and phosphorus removal. Zinc, cadmium and nickel were rapidly adsorbed in 20 min by the harvested sludge from a continuous flow pilot plant. Nilanjana et al. [36] reported the zinc sorption by bacterial biofilm and the implication of extracellular polymeric substances (EPS).
Costley and Wallis [37] investigated the efficiency of biofilm of a rotating biological contactor (RBC) for the treatment of wastewaters contaminated with cadmium, copper and zinc in multiple sorption desorption cycles. The removal pattern observed in the initial cycle was Cu>Zn>Cd which was repeated in the subsequent cycles. The system successfully removed the metals in the order Cu>Zn>Cd with removal capacities of approximately 73, 42 and 33% respectively.
It is interesting to point out that statistically, biofilms production was correlated with the antibiotics resistance (r=0.73; P<0.05), unlike, this relationship the heavy metals had not recorded any relationship (r=0.31; P<0.05) (Figures 4b and 4c). On the basis of these observations the correlations obtained in the present study are not statistically significant, this disparity can be attributed to the regulatory processes that indirectly activate the genetic and biochemical pathways that are shared in the response of microorganisms to antibiotic and metal exposure [38].
Taken together, our data are consistent with Harrison et al. [16] reports, they had signaled that toxic metals present problems to biofilm communities that are distinct from those of antibiotics because the different metal species have distinct chemistries, and function through diverse biochemical routes of toxicity.
Similar to studies of biofilm antibiotic susceptibility [19], simple time and dose dependent killing assays have shown that there are subpopulations of cells in biofilms, and that these cells die at different rates after the exposure of the entire population to metal ions. The concentration-dependent killing of microbial populations is exemplified by a plateau in the activity of the antimicrobial. Examples include Pseudomonas aeruginosa ATCC 27853 killing by most antibiotics, Ni or Zn cations for exposure times of up to 24 hours. These results are also typical of the exposure of biofilms to most toxic metal compounds for short exposure times (only a few hours). Under certain in vitro conditions, and with sufficient exposure times, other highly toxic metal species can eradicate 100% of bacterial biofilms populations [16]. Examples include P. aeruginosa biofilms that are exposed to Cu or Ag cations for 24 hours. As was pointed out by Harrison et al. [17] many metal ions can exert toxicity on biological systems by multiple biochemical pathways simultaneously. The susceptibility of microorganisms to toxic metal species has been linked to several metal ion specific physicochemical parameters. As the correlations are different for biofilms and planktonic cells, these trends indicate that the chemical mechanisms of toxicity are different between each modes of growth as signaled by many researchers.
Variant formation by Pseudomonas spp. might be an important contributor to biofilm metal susceptibility, as the switch to the SCV (small colony variant) and other variant phenotypes is correlated with the emergence of multidrug and multi metal resistance [39]. The response of P. fluorescens and P. chlororaphis biofilms to metal ion exposure is reasonably anticipated to be highly complex, as it is linked to the formation of multiple colony morphotypes, such as the wrinkly spreader (WS) phenotype in the case of P. fluorescens [16].
It is also interesting to note that the structure dependent metabolic heterogeneity may also explain, in part, the tolerance of bacterial biofilms to metal ions. In P. aeruginosa biofilms that are less than 100 μm thick, the cells that are nearest to the substratum are in anoxic zones and are slow growing, which leads to an intrinsic tolerance to killing by antibiotics relative to the aerobic fast growers in the outer biofilm layers 50.
Consistent with these observations, it is obvious that extracellular signaling events affecting biofilm physiology. One prominent signal system that probably has a role in biofilm susceptibility to metal toxicity is extracellular signaling by quorum sensing (QS) systems. Depending on the growth environment of the biofilm, QS systems have a conditional role in biofilm formation and/or development [16]. One of the other ways that signaling events contribute to this resistance is by regulating the synthesis of the extracellular matrix (ECM) components that facilitate biosorption. Through experiment and mathematical analysis, researchers have shown that the extracellular matrix (ECM), a mesh of proteins and sugars that can form outside bacterial cells, creates osmotic pressure that forces biofilms to swell and spread.
Consistent with these observations, Hu et al. [40] found that Zn was evenly equilibrated across thin (approximately 12 μm) biofilms, but penetrated less than 20 μm into thick (approximately 350 μm) biofilms. Previous studies of biofilm and heavy metal interactions have mainly focused on the sorption of heavy metals. Several researchers have reported that biofilms are capable of removing heavy metal ions from bulk liquid [41], and the use of biofilms to remove heavy metals from wastewater has been investigated [42]. Electron microscopy revealed that a P. aeruginosa biofilm was capable of sequestering heavy metals [43], while mercury-reducing Pseudomonas putida biofilms were found to accumulate elemental mercury on the exterior of the biofilms [44]. Burkholderia cepacia biofilms on hematite and alumina surfaces were found to preferentially accumulate Pb2+ at concentrations higher than 1 μM, implying that the chemical nature of the attachment surface affects metal sequestration. Within a biofilm it has been found that EPS, specifically the polysaccharide components, binds heavy metals [20].
Conclusion
During the last few decades, extensive attention has been paid on the management of environmental pollution caused by hazardous heavy metals. However, understanding the mechanisms by which metals are toxic to Pseudomonas biofilms has remained elusive. Here, we emphasize that biofilms formation seems to be more correlated to the antibiotics resistance than the heavy metals resistance, in which the stationary phase may be responsible more for this resistance. It is important to note that future endeavors are needed to elucidate mechanisms that reduce the susceptibility of biofilms to antibiotics since many of biological mechanisms that function in metal resistance might also explain the recalcitrance of biofilms towards multiple, structurally unrelated antibiotics.
Acknowledgements
The research was supported by the national research project (PNR project) of the Direction for Research Programming, Evaluation and Prospective Study (DGRSDT), Government of Algeria. The anonymous reviewers are sincerely thanked for their beneficial suggestions to improve the manuscript.
References
- Paul D, Pandey G, Pandey J, Jain RK (2005) Accessing microbial diversity for bioremediation and environmental restoration. Trends Biotechnol 23: 135-142.
- Singh P, Cameotra SS (2004) Enhancement of metal bioremediation by use of microbial surfactants. Biochem Biophys Res Commun 319: 291-297.
- Barkay T, Schaefer J (2001) Metal and radionuclide bioremediation: issues, considerations and potentials. Curr Opin Microbio l4: 318-323.
- Lloyd JR (2003) Microbial reduction of metals and radionuclides. FEMS Microbiol Rev 27: 411-425.
- White C, Gadd GM (2000) Copper accumulation by sulfate-reducing bacterial biofilms. FEMS Microbiol Lett 183: 313-318
- Flemming HC (1995) Sorption sites in biofilms. Water Sci Technol 32: 27-33.
- Glick BR, Karaturovic DM, Newell PC (1995) A novel procedure for rapid isolation of plant growth promoting pseudomonads. Can J Microbiol 41: 533-536.
- Glick BR (2001) Phytoremediation: synergistic use of plants and bacteria to clean up the environment. Biotechnol Adv 21: 383-393.
- Raj SN, Deepak SA, Basavaraju P, Shetty HS, Reddy MS, et al. (2003) Comparative performance of formulations of plant growth promoting rhizobacteria in growth promotion and suppression of downy mildew in pearl millet. Crop Prot 22: 579-588.
- Çakmakçi R, Dönmez F, Aydm A, ÅÂ?ahin F (2006) Growth promotion of plants by plant growth-promoting rhizobacteria under greenhouse and two different field soil conditions. Soil Biol Biochem 38: 1482-1487.
- Krieg NR, Holt JG (1989) Bergey’s Manual of Systematic Bacteriology. 1st edn. Williams and Wilkins Co., Baltimore, Springer, Volume: 1-4.
- Flemming HC, Wingender J (2001) Relevance of microbial extracellular polymeric substances (EPSs) - Part I: Structural and ecological aspects. Water Sci Technol 43: 1-8.
- Sheng XF, Xia JJ, Jiang CY, He LY, Qian M (2008) Characterization of heavy metal-resistant endophytic bacteria from rape (Brassica napus) roots and their potential in promoting the growth and lead accumulation of rape. Environmental Pollution 156: 1164-1170.
- O’Toole GA, Kolter R (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple convergent signaling pathways: a genetic analysis. Mol Microbiol 28: 449-461.
- StepanoviÄÂ? S, VukoviÄÂ? D, DakiÄÂ? I, SaviÄÂ? B, ŠvabiÄÂ?-VlahoviÄÂ? M (2000) A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods 40: 175-179.
- Harrison JJ, Turner RJ, Ceri H (2007) Multimetal resistance and tolerance in microbial biofilms. Nat Rev Microbiol 5: 928-938.
- Harrison JJ, Turner RJ, Ceri H (2005) Persister cells, the biofilm matrix and tolerance to metal cations in biofilm and planktonic Pseudomonas aeruginosa. Environ Microbiol 7: 981-994.
- Gilbert PS, Collier PJ, Brown MR (1990) Influence of growth rate on susceptibility to antimicrobial agents: biofilms, cell cycle, dormancy, and stringent response. Antimicrob Agents Chemother 34: 1865-1868.
- Spoering AL, Lewis K (2001) Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol 183: 6746-6751.
- Teitzel GM, Parsek MR (2003) Heavy Metal Resistance of Biofilm and Planktonic Pseudomonas aeruginosa. Appl Environ Microbiol 69: 2313-2320.
- Chapman JS (2003) Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int Biodeterior Biodegradation 51: 271-276.
- Martinez JL, Sánchez MB, Martínez-Solano L, Hernandez A, Garmendia L, et al. (2009) Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol Rev 33: 430-449.
- Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV (2006) Co-selection of antibiotic and metal resistance. Trends Microbiol 14: 176-182.
- Hölzel CS, Müller C, Harms KS, Mikolajewski S, Schäfer S, et al. (2012) Heavy metals in liquid pig manure in light of bacterial antimicrobial resistance. Environ Res113: 21-27.
- de Vincente A, Avile`s M, Codina JC, Borrego JJ, Romero P (1990) Resistance to antibiotics and heavy metals of Pseudomonas aeruginosa isolated from natural waters. J Appl Bacteriol 68: 625-632.
- Bender CL, Cooksey DA (1986) Indigenous plasmids in Pseudomonas syringae pv. tomato: conjugative transfer and role in copper resistance. J Bacteriol 165: 534-541.
- Schooling SR, Beveridge TJ (2006) Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol 188: 5945-5957.
- Lewis K (2001) Riddle of biofilm resistance. Antimicrob Agents Chemother 45: 999-1007.
- Perez LRR, Costa MCN, Freitas ALP, Barth AL (2011) Evaluation of biofilm poduction by Pseudomonas aeruginosa isolates recovered from cystic fibrosis and non-cystic fibrosis patients. Brazilian Journal of Microbiology 42: 476-479.
- Meliani A, Bensoltane A (2014) Enhancement of Hydrocarbons Degradation by Use of Pseudomonas Biosurfactants and Biofilms. J Pet Environ Biotechnol 5: 68.
- Workentine ML, Harrison JJ, Stenroos PU, Ceri H, Turner RJ (2008) Pseudomonas fluorescens’ view of the periodic table. Environ. Microbiol 10: 238-250.
- Ron EZ, Rosenberg E (2002) Biosurfactants and oil bioremediation. Curr Opin Biotechnol 13: 249-252.
- Munoz R, Alvarez MT, Munoz A, Terrazas E, Guieysse B, et al. (2006) Sequential removal of heavy metal ions and organic pollutants using an algal-bacterial consortium. Chemosphere 63: 903-911.
- Chang WC, Hsu GS, Chiang SM, Su MC (2006) Heavy metal removal from aqueous solution by wasted biomass from a combined AS-biofilm process. Bioresour Technol 97: 1503-1508.
- Horn H, Morgenroth E (2006) Transport of oxygen, sodium chloride, and sodium nitrate in biofilms. Chem Eng Sci 61: 1347-1356.
- Das N, Geetanjali B, Lakshmi V, Salam JA, Evy Alice AM (2012) Application of Biofilms on Remediation of Pollutants - An Overview. Microbiol Biotech Res 2: 783-790
- Costley SC, Wallis FM (1999) Effect of disk rotational speed on heavy metal accumulation by rotating biological contactor (RBC) biofilms. Lett Appli Microbiol 29: 401-405.
- Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV (2006) Co-selection of antibiotic and metal resistance. Trends Microbiol 14: 176-182.
- Drenkard E, Ausubel FM (2002) Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416: 740-743.
- Hu Z, Hidalgo G, Houston PL, Lion LW, Abruna HD, et al. (2005) Determination of spatial distributions of zinc and active biomass in microbial biofilms by two-photon laser scanning microscopy. Appl Environ Microbiol 71: 4014-4021.
- Labrenz M, Druschel GK, Gilbert B, Welch SA, Kemner KM, et al. (2000) Formation of sphalerite (ZnS) deposits in natural biofilms of sulfate-reducing bacteria. Science 290: 1744-1747.
- von Canstein H, Li Y, Timmis KN, Deckwer WD, Wagner-Do¨bler I (1999) Removal of mercury from chloralkali electrolysis wastewater by a mercury-resistant Pseudomonas putida strain. Appl Environ Microbiol 65: 5279-5284.
- Langley S, Beveridge TJ (1999) Metal binding by Pseudomonas aeruginosa PAO1 is influenced by growth of the cells as a biofilm. Can J Microbiol 45: 616-622.
- Wagner-Döbler I, Lünsdorf H, Lübbehüsen T, Von Canstein HF, Li Y (2000) Structure and species composition of mercury-reducing biofilms. Appl Environ Microbiol 66: 4559-4563.
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 15380
- [From(publication date):
September-2016 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 13904
- PDF downloads : 1476