Research Article Open Access
Bioefficiency of Indigenous Microbial Rhodanese in Clean-up of Cyanide Contaminated Stream in Modakeke, Ile-Ife, Osun State, Nigeria
Oluwatosin A Adedeji1*, Omolara T Aladesanmi1, Oluwaseyi A Famakinwa2and Rapheal E Okonji2
1Institute of Ecology and Environmental Studies, Obafemi Awolowo University, Ile-Ife, Osun State, Nigeria
2Department of Biochemistry, Obafemi Awolowo University, Ile-Ife, Osun State, Nigeria
- *Corresponding Author:
- Oluwatosin A
Adedeji, Institute of Ecology and Environmental Studies
Obafemi Awolowo University
Ile-Ife, Osun State, Nigeria
Tel: 2347030262067
E-mail: oluwatosinadedeji92@gmail.com
Received Date: March 03, 2017 Accepted Date: March 28, 2017 Published Date: March 30, 2017
Citation: Oluwatosin AA, Omolara TA, Oluwaseyi AF, Rapheal EO (2017) Bioefficiency of Indigenous Microbial Rhodanese in Clean-up of Cyanide Contaminated Stream in Modakeke, Ile-Ife, Osun State, Nigeria. J Bioremediat Biodegrad 8: 390. doi: 10.4172/2155-6199.1000390
Copyright: © 2017 Oluwatosin AA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Cyanide pollution of aquatic environment has become a great concern in Nigeria because of the increase in cassava cultivation. In Nigeria, cassava processing milling plants are usually situated around streams or rivers such that the waste from each stages of processing easily find their way into these water bodies as effluents and waste waters. Extracellular rhodanese of Klebsiella edwardsii isolated from Atutulala stream, Modakeke, where cassava is being processed, was assessed for its bioremediation potential. Cyanide concentration of the stream was analysed for six months. Four bacterial isolates were screened for their ability to degrade free cyanide and the best strain was further screened for rhodanese producing ability. The enzyme was purified by 85% ammonium sulphate precipitation and diethyl aminoethyl-cellulose ion-exchange chromatography. The pure enzyme had a specific activity of 0.0473 Rhodanese Unit mg-1 with a purification fold of 4.56 and a percentage yield of 30.30%. The enzyme demonstrated a broad pH range but the optimum pH was at 6.0 while the optimum temperature was 60°C. The bioremediation potential of the enzyme was assessed under various conditions such as the field pH and temperature as well as optimum pH and temperature using the cyanide contaminated water as substrate source in a typical assay protocol. The enzyme was able to convert 1.6481 μmol of cyanide to thiocyanate in the water sample at optimum pH and temperature of the enzyme. It could be concluded from the study that at optimum pH and temperature, rhodanese exhibited remediation activity in cyanide contaminated aquatic ecosystems and thus, can be used for its restoration.
Keywords
Bioremediation; Rhodanese; Cyanide; Klebsiella edwardsii; Atutulala stream
Introduction
Cyanide, a compound containing CN– as its functional group is highly toxic and an important constituent of industrial effluents [1]. In the biosphere, plants, bacteria and fungi are the major sources of cyanide [2]. It is found naturally in cassava roots, potato like tubers grown in tropical countries in the form of cyanogenic glucosides (the precursors of HCN) in various concentrations depending on the variety and growth condition [3]. Cassava (Manihot esculenta ) is known to be the third most important source of calories in the tropics, after rice and corn [4]. Depending on the absence or presence of toxic levels of cyanogenic glucosides, cassava varieties are often categorized as either sweet or bitter. The sweet cultivars are able to produce as little as 20 milligrams of cyanide (CN) per kilogram of fresh roots, while the bitter ones may produce more than 50 times as much (1 g/kg) [5]. These cyanides which exist as cyanogenic glycosides are hydrolyzed into the toxic compound hydrogen cyanide during processing [5,6]. In South Western Nigeria, cassava processing milling plants are usually situated around running streams or rivers such that the waste from each stages of processing easily find their way into these water bodies as effluents and waste waters [6]. The cassava are even washed and rinsed in these water bodies before and after peeling. In south-western Nigeria especially, local farmers detoxify their cassava tubers/roots by steeping/retting, soaking and wetting or moistening the whole peeled or unpeeled tubers/roots in these running streams for about 3-5 days; after which the skins of the unpeeled roots peel off easily [7-9]. Subsequent washing and sieving with baskets which separate the fibres from the starch are also done in the water bodies. The cassava is now largely detoxified by the prolonged soaking in the running water. All these activities contribute to the release of cyanogenic glycosides from enzymatic degradation following maceration to produce hydrogen cyanide, which may result in acute cyanide poisoning [10]. Cyanide binds and inactivate several enzymes, particularly those with iron in the ferric (Fe3+) state and cobalt [11]. It binds to the active site of cytochrome c oxidase, the terminal protein in the electron transport chain located within mitochondrial membranes where it exerts its ultimate lethal effect of histotoxic anoxia [12]. The binding of cyanide ion to cytochrome hinders the transport of electrons from cytochrome c oxidase to oxygen, a process which prevents the cells from utilizing the oxygen present in the bloodstream, resulting in cytotoxic hypoxia or cellular asphyxiation [13]. Many methods are available for the treatment of cyanide such as ferrocyanide precipitation, prussion-blue and copper catalyzed H2O2 precipitation, ozonation, incineration, alkaline chlorination and electrolysis [14]. These methods remediate cyanide contaminated wastewaters with attendant high reagent costs and may often not achieve complete breakdown of some cyanide complexes [15]. Biological method for cyanide removal from industrial wastewaters often termed bioremediation, is cost effective and environmentally friendly [16-18]. Certain microorganisms such as bacteria and fungi are capable of metabolizing cyanide to produce non-toxic end products using the cyanide as the sole nitrogen and carbon sources under aerobic and anaerobic environments [19]. Some species of Bacillus and Klebsiella are resistant to cyanide even at concentrations higher than 1 mM as the enzymes within them called rhodanese have been reported to be involved in cyanide detoxification [20-23]. Rhodanese is a ubiquitous enzyme that is known to be responsible for the biotransformation of cyanide to less toxic thiocyanate using thiosulphate as the donor substrate [24-27]. The use of enzymatic proteins may represent a good alternative for overcoming most disadvantages related to the use of microorganisms [28,29]. They are not inhibited by inhibitors of microbial metabolism such as occur in industrial wastewaters, such as cassava mill wastewater with low nitrogen (N) and high chemical oxygen demand (COD), leading to a nutritionally imbalanced wastewater [30]. Moreover, they can be used at extreme conditions limiting microbial activities and are effective at low pollutant concentrations. This study involved the isolation, partial purification and characterization of microbial rhodanese isolated from Atutulala stream with further assessment of the bioremediation potential of the enzyme.
Materials and Methods
Sampling area and water sample collections
Water samples were collected from Atutulala Stream also called Odo-Isale, Kola Village, Modakeke, Ile-Ife city in Osun State, Southwestern, Nigeria. The village is made up of predominantly farmers. Atutulala lies within Latitude 7°46'17.5''N and Longitude 4°49'54.5'' E. It is used by the villagers for domestic purposes and also for cassava processing as villagers wash and soak already peeled cassava in the stream for days before milling into fufu and gari products. Water samples were collected from three different designated points for six months from July to December 2015 in sample bottles using grab method. The samples were then brought to the laboratory for cyanide analysis (Figure 1).
Cyanide analysis
The cyanide content of the water samples was determined to ascertain the presence of cyanide in the water samples by adding 4 ml alkaline picrate to 1 ml of the sample filtrate in a corked test tube and incubated in a water bath for 5 mins. After colour development (reddish brown colour), the absorbance of the corked test tube was measured in spectrophotometer at 490 nm. The absorbance of the blank containing only 1 ml distilled water and 4 ml alkaline picrate solution was also read. The cyanide content was extrapolated from a cyanide standard curve prepared by measuring different concentrations of KCN solution containing 5 to 50 μg cyanide and 25 ml of 1 N HCl. The different concentrations were used to prepare the cyanide standard curve.
Isolation, characterization and identification of rhodanese producing bacterium
One millilitre (1 ml) of the water sample was taken from each of the sample bottle into a test tube containing 9 ml of sterile distilled water. Serial dilution was carried out to thin out the load of microorganisms to be plated. Serial dilution up to 10-6 was carried out and dilution 1, 2, 4, 6 were pour plated with a composed basal medium containing agar agar (1.5% w/v), peptone (1% w/v), yeast extract (0.5% w/v), sodium chloride (0.5% w/v), potassium cyanide (0.3% w/v) and were incubated in duplicates at 37°C for 48 h. After incubation, plates were examined and representative colonies were isolated from each plate for further identification. The representative colonies were purified by repeated streaking on the same medium and were then maintained on nutrient agar slants at 4°C for further study.
The different isolates were grown separately in composed broth medium basal broth containing peptone (1% w/v), yeast extract (0.5% w/), sodium chloride (0.5% w/v), potassium cyanide (0.3% w/v) dissolved in deionized water. The initial pH was adjusted to 9.5. MacFarland standards (0.5) of the isolates were prepared and used for inoculation of the media. The media were incubated at 37°C on a rotary shaker at 230 rpm for 48 h. Each of the culture media was then assayed for rhodanese production.
Four bacterial isolates were selected because they showed appreciable rhodanese activities and were screened for their ability to degrade free cyanide. The best strain with the highest rhodanese activity was selected for further study and characterized. The selected rhodanese-producing strain was identified based on its cell morphology, cultural and biochemical characteristics [31,32].
Enzyme production and partial purification
The time course of the enzyme production was determined and compared with growth. This was done using Zlosnik and Williams [33] growth methods with appropriate modifications by inoculating a 100 ml of the sterile basal broth with 1 ml standard inoculum of 0.5 McFarland standard in an Erlenmeyer flask and incubated at 37°C for 48 hr with agitation at 140 rpm. At intervals of 3 hr, 5 ml of samples were collected aseptically for a period of 48 hr, the optical density of each sample was checked at 630 nm using a colorimeter and recorded as the cell optical density. The samples each at the different time intervals was further centrifuged at 12,000 rpm for 30 mins to separate the cell from supernatant. The supernatant which served as the crude enzyme was checked for rhodanese activity at 460 nm according to the principles of the colorimetric determination of thiocyanate formation by Lee et al. [34], the reaction mixture consists of 0.5 ml of 50 mM Borate buffer (pH 9.4), 0.2 ml of 250 mM KCN, 0.2 ml of 250 mM Na2S2O3, and 0.1 ml of the enzyme solution in a total of volume of 1.0 ml. The mixture was incubated for 1 min at room 37°C and the reaction was stopped by adding 0.5 ml of 15% formaldehyde, followed by the addition of 1.5 ml of Sorbo reagent. The absorbance was taken at 460 nm. One Rhodanese Unit (RU) was defined as the amount of the enzyme that will convert one micro-mole (1 μmol) of cyanide to thiocyanide in one minute at 37°C [35]. Protein concentration was determined by using Bovine Serum Albumin (BSA) as the standard, where the protein absorbance was extrapolated from the standard curve [36]. The absorbance was read at 595 nm.
In order to achieve partial purification of the enzyme, the supernatant obtained from the centrifugation of the crude homogenate was brought to 85% ammonium sulphate saturation i.e., 55.6 g of ammonium sulphate (analytical grade) was added to 200 ml of crude enzyme and stirred slowly for 1 hr until all the salt had dissolved completely in the supernatant. The mixture was left overnight in the fridge, followed by centrifugation at 4,000 rpm for 30 min at 10°C. The supernatant was discarded and precipitate was collected and suspended in 0.1 M Phosphate buffer pH 7.2. It was evaluated for rhodanese activity and the protein content was determined. The ammonium sulphate precipitate was dialysed against several changes of 0.1 M solution of phosphate buffer (pH 7.2) at 4°C for 18 hr. The dialysate was centrifuged at 4,000 rpm at 10°C for 30 mins to remove insoluble materials and the supernatant was assayed for rhodanese activity and protein. The yield and fold of purifications were calculated. The enzyme was further purified using DEAE-Cellulose cation exchanger equilibrated with 0.1 M phosphate buffer (pH 7.2) followed by elution with 1.0 M NaCl in the same buffer. Fractions of 3 ml were collected from the column at a rate of 128.57 ml per hr. The fractions were assayed for rhodanese activity and protein profile. The active fractions from the column were pooled and dialyzed against 50% glycerol in 0.1 M phosphate buffer, pH 7.2. The yield and fold of purifications were calculated [37].
Characterization of the enzyme
The kinetic parameters (Km and Vmax) of the enzyme were determined by varying concentrations of KCN between 0.05 M and 0.005 M at fixed concentration of 0.25 M Na2S2O3. Also, the concentration of Na2S2O3 was varied between 0.05 M and 0.005 M at fixed concentration of 0.25 M KCN. Plots of the reciprocal of initial reaction velocity (1/V) versus reciprocal of the varied substrates 1/[S] at each fixed concentrations of the other substrate were made [38]. Lines through the points were drawn by using the method of regression.
To determine the effect of pH on the activity of the enzyme, the enzyme was assayed using 50 mM of citrate buffer (pH 3-6); 50mM phosphate buffer (7-8) and 50mM borate buffer (pH 9-11) in place of the reaction buffer of the rhodanese assay [37].
The optimum temperature of the enzyme was investigated by assaying the enzyme at temperatures between 30°C and 80°C at an interval of 10°C. The assay mixture was first incubated at the indicated temperature for 10 min before addition of an aliquot of the enzyme which had been equilibrated at the same temperature.
The substrate specificity of the enzyme was determined by using different sulphur compounds such as sodium sulphite, 2- mercaptoethanol, ammonium persulphate, ammonium sulphate and sodium metabisulphite in a typical rhodanese assay mixture. The different sulphur compounds were substituted in place of sodium thiosulphate. The activity was taken and expressed as a percentage activity of the enzyme using sodium thiosulphate as the control [37].
Assessing the enzyme for bioremediation potential
The cyanide content of the water sample from Atutulala stream was determined to ascertain the presence of cyanide in it. The pH and temperature of the water sample were also determined. The assay protocol of the enzyme was set up at varying conditions such as pH of the water sample (pH=6.5), the optimum pH of the enzyme (pH=6.0), temperature of the room (37°C), the optimum temperature of the enzyme (60°C), presence or absence of buffer (50mM Citrate Buffer pH=6.0) in the protocol, introduction of sulphur compounds (250mM sodium thiosulphate) and the use of the water sample as the sulphur source with the water samples as the cyanide source to be remediated by the enzyme. Eight different conditions were set up in the assay protocol assessment of the remediation potential of the rhodanese enzyme as stated below:
1. pH=6.5; Temperature=37°C; Water sample as Sulphur source; No Buffer
2. pH=6.5; Temperature=60°C; 250mM of Na2S2O3; No Buffer
3. pH=6.0; Temperature=60°C; Water sample as Sulphur source; No Buffer
4. pH=6.0; Temperature=60°C; 250mM of Na2S2O3; No Buffer
5. 50mM Citrate Buffer (pH=6.0); Temperature=37°C; Water sample as Sulphur source
6. 50mM Citrate Buffer (pH=6.0); Temperature=37°C; 250mM of Na2S2O3
7. 50mM Citrate Buffer (pH=6.0); Temperature=60°C; 250mM of Na2S2O33
8. 50mM Citrate Buffer (pH=6.0); Temperature=60°C; Water sample as Sulphur source
Each of the assay protocols have control which follows the same protocol as the test except that the 0.5 ml of the purified enzyme solution was added after all the other reagents have been added and incubated for 1 min at the appropriate temperatures. The amount of cyanide converted to thiocyanide was calculated by finding the difference between the test and the control.
Results and Discussion
The cyanide concentrations for the six months (July-December) were all above World Health Organisation (WHO) and Nigeria International Standard (NIS) limits of 0.07 mg/l and 0.01 mg/l for cyanide in drinking water respectively (Table 1). This study agrees with Akinpelu et al. [39] who reported high levels of cyanide from cassava production effluent and further stated that intake of much of hydrogen cyanide could damage body cells.
| July | Aug | Sept | � Oct� | Nov� | Dec | WHO (2003) | NIS (2007) | |
|---|---|---|---|---|---|---|---|---|
| Cyanide | 1.44 | 3.84 | 2.67 | 0.71 | 1.6 | 2.11 | 0.07 | 0.01 |
| (mg/l) | �± 0.95 | �± 2.13 | �± 0.91� � � � � � � � � � � � | � �± 0.36 | �± 1.14 | �± 1.25 |
Table 1: Cyanide Concentration (Mean �± SD) of Atutulala Stream from July-Dec, 2015.
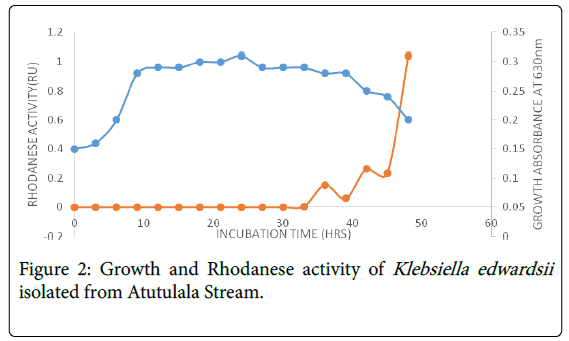
A total of four bacterial strains were isolated from Atutulala stream, out of which one was selected for further study based on its appreciable rhodanese production. Cultural, morphological, biochemical and physiological characteristics of the isolate was examined and they were identified as a strain of Klebsiella edwardsii. Klebsiella edwardsii rhodanese production was maximum at 48 h in the death phase and this may be due to the inability of the bacteria to adapt as early as possible to the environmental conditions such as pH, temperature (Figure 2). This has been noted by Goyal et al. [40] and Prakash et al. [41], who stated that inhibition of enzyme production may occur as a result of catabolic repression by metabolizable monosaccharides such as glucose, increase in concentration of protease and rapid change in pH with the aim of conserving energy by bacteria.
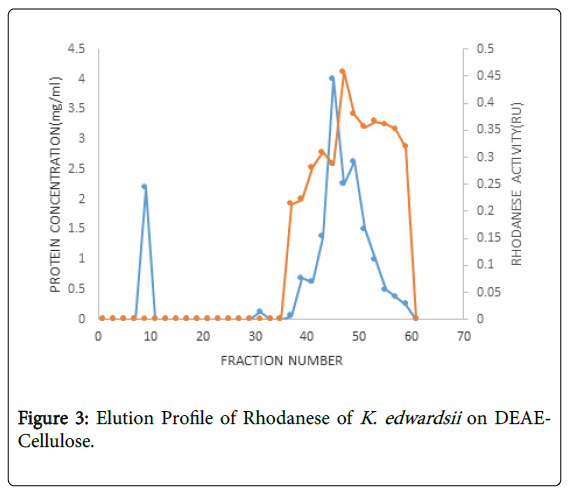
The enzyme from K. edwardsii was purified 2.26 folds using ammonium sulphate precipitation at 85% saturation and 4.56 folds through DEAE-Cellulose with a yield of 30.3% (Table 2). The chromatography result showed a specific activity of 0.047 Rhodanese Unit/mg with a percentage recovery of 30.3, the activity increased dramatically because the bounded proteins were released and increased the volume of the enzyme during ion-exchange chromatography.
| Purification Steps | Total Activity (RU) | Total Protein (mg/ml) | Specific Activity (RU/mg) | Fold | Yield |
|---|---|---|---|---|---|
| Crude Extract | 58.34 | 562.5 | 0.01037 | 1 | 100 |
| Ammonium Sulphate Precipitation | 4.802 | 205.625 | 0.0234 | 2.26 | 36.56 |
| Ion Exchange on DEAE Cellulose | 8.07 | 170.625 | 0.0473 | 4.56 | 30.3 |
Table 2: Summary of Partial Purification Process of Crude Rhodanese Obtained from K. edwardsii
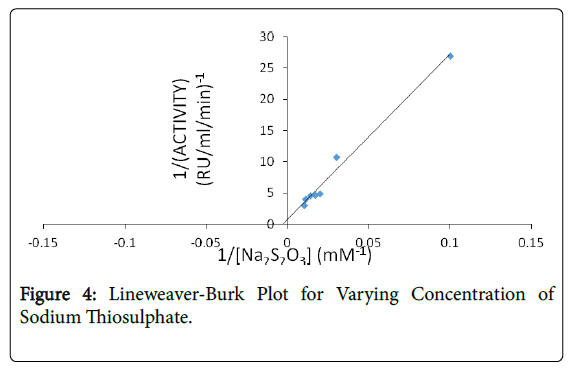
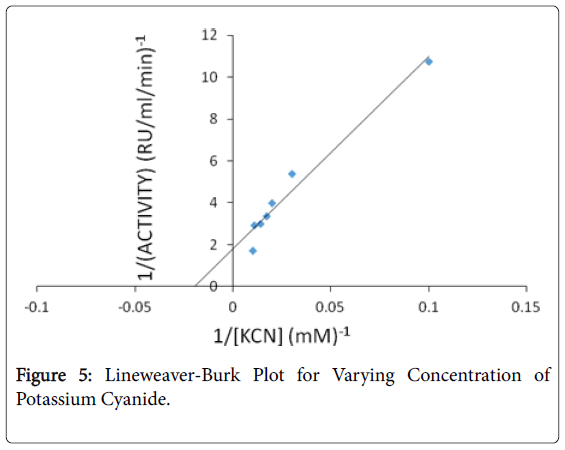
The kinetic parameters (Km and Vmax) of Klebsiella edwardsii rhodanese for potassium cyanide are 50 mM, 0.56 RU/mg and for sodium thiosulphate are 200 mM, 1 RU/mg respectively (Figures 4 and 5). This indicate great affinity of the Klebsiella edwardsii rhodanese for these substrates and that it would catalyze the detoxification reaction with great efficiency.
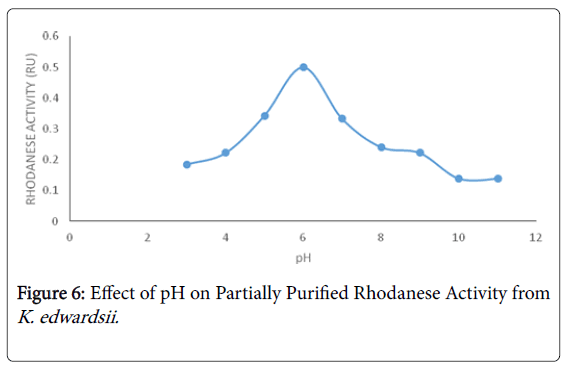
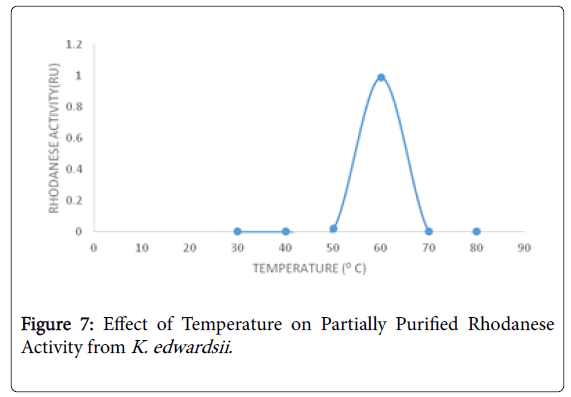
The optimum pH obtained for Klebsiella edwardsii rhodanese was 6 (Figure 6) which is similar to the work of Oyedeji et al. [19] who reported an optimum pH of 6 for rhodanese from Pseudomonas aeruginosa isolated from the soil of cassava processing site. The optimum temperature for Klebsiella edwardsii rhodanese is 60°C in this study (Figure 7) as reported by Ehigie et al. [42] for rhodanese isolated from the leaves of Mormodica charantia .
The substrate specificity study of rhodanese isolated from Klebsiella edwardsii has preference for sodium thiosulphate but also showed higher activity with ammonium sulphate and ammonium persulphate (Table 3). The higher activities shown by the enzyme for ammonium sulphate (122.89%) and ammonium persulphate (178.31%) could be as a result of their concentrations in the native environment. The substrate specificity result also showed that sodium sulphite, sodium metabissulphite, and 2-mercaptoethanol can also be used for its catalytic activities.
| Substrate | % Residual Activity |
|---|---|
| Sodium Thiosulphate | 100 |
| Sodium Sulphite | 78.31 |
| Ammonium Sulphate | 178.31 (?100) |
| Ammonium per Sulphate | 122.89 (?100) |
| 2-Mercaptoethanol | 33.7 |
| Sodium Metabisulphite | 67.5 |
Table 3: Effect of Different substrates on Partially Purified Rhodanese from K. edwardsii.
The bioremediation potential of the purified rhodanese on Atutulala water sample showed that the enzyme had highest activity of 1.6481 Rhodanese Unit (C) when the water sample was used as the sulphur source at the optimum pH and temperature of the enzyme (Table 4). This value corresponds to the amount of cyanide in μmol that was reduced to a less toxic thiocyanate in the water sample tested. This lends credence to the effect of temperature and pH of the enzyme studied. The enzyme also showed higher activity of 1.0741 Rhodanese Unit (A) as compared to the remaining experiments, when the water sample (pH 6.5) was used as the sulphur source at 37°C. This may be due to the presence of sulphur in the water sample which was just sufficient for the enzyme to act. The activities of the enzyme in the presence of citrate buffer (pH 6.0) when water sample was used as sulphur source at 60°C (G); when sodium thiosulphate was used as sulphur source at 60°C (H); and when sodium thiosulphate was used as sulphur source at 37°C are 0.8148 Rhodanese Unit, 0.5000 Rhodanese Unit and 0.4259 Rhodanese Unit respectively (Table 4). Other conditions (B, D, E and F) also resulted in low activity of the enzyme in converting cyanide to thiocyanate in the water sample probably because of the presence of sulphur compound just sufficient enough in the sampled water for the enzyme to act; and the addition of more sulphur from sodium thiosulphate might have reduced the activity as the enzyme might have reached its maximum velocity irrespective of the optimum temperature and pH.
| Conditions | Rhodanese Activity ( Rhodanese Unit ) |
|---|---|
| A) pH=6.5; Temperature=37�°C; Water sample as Sulphur source | 1.0741 |
| B) pH=6.5; Temperature=37�°C; 250mM of Na2S2O3 | -0.6482 |
| C) pH=6.0; Temperature=60�°C; Water sample as Sulphur source | 1.6481 |
| D) pH=6.0; Temperature=60�°C; 250mM of Na2S2O3 | -0.9259 |
| E) 50mM Citrate Buffer (pH=6.0); Temperature=37�°C; Water sample as Sulphur source | -0.7592 |
| F) 50mM Citrate Buffer (pH=6.0); Temperature=37�°C; 250mM of Na2S2O3 | 0.4259 |
| G) 50mM Citrate Buffer (pH=6.0); Temperature=60�°C; 250mM of Na2S2O3 | 0.8148 |
| H) 50mM Citrate Buffer (pH=6.0); Temperature=60�°C; Water sample as Sulphur source | 0.5 |
Table 4: Bioremediation Potential of K. edwardsii Rhodanese
Conclusion
The presence of rhodanese in Klebsiella edwardsii suggested that the enzyme may possess functional cyanide detoxification mechanism necessary for the survival of the organism in Atutulala stream in which cassava tubers are soaked during processing. The water sample was confirmed to contain cyanide beyond the permissible limits of WHO and NIS. The isolated enzyme was also effective at a high temperature, a very unique and important property for detoxification of cyanide. It can then be concluded from this study that the rhodanese enzyme assessed under various conditions was found to be suitable to be adopted for clean-up of cyanide polluted water bodies at the optimum pH and temperature recorded by this study. Further studies of the biochemical parameters of the enzyme is therefore recommended to understand the appropriate applications of the enzyme in the bioremediation procedure.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- Tandishabo K, Takahashi A, Nakamura K, Takamizawa K (2007) Characterization of the Rhodanese Enzyme in Coprothermobacter Strains. Proceedings of International Symposium on EcoTopia Science, ISETS07: 1202-1203.
- Ubalua AO (2010) Cyanogenic Glycosides and the Fate of Cyanide in Soil. Australian Journal of Crop Science 4: 223-237.
- Sankaranarayanan A, Gowthami M (2015) Cyanide Degradation by Consortium of Bacterial Species Isolated from Sago Industry Effluent. Journal of Environmental Treatment and Technology 3: 41-46.
- Truong QT, Naoyuki M, Keisuke I (2003) Applicability of Filamentous Fungi to Treatment of Cassava Starch Processing Wastewater Containing Cyanide. Osaka University Knowledge Archive (OUKA)
- Oti EE (2002) Acute Toxicity of Cassava Mill Effluent to the African Catfish Fingerlings. Journal of Aquatic Science 17: 31-34.
- Abiona O, Sanni L, Bamgbose O (2005) An Evaluation of Microbial Load, Heavy Metals and Cyanide Contents of Water Sources, Effluents and Peels from Three Cassava Processing Locations. Journal of Food, Agriculture and Environment 3: 207-208.
- Okeke GC, Obioha FC, Udogu AE (1985) Comparison of Detoxification of Cassava-Borne Cyanide. Nutrition Reports International 32: 139-147.
- Emmanuel OE, Ramakrishna BB (1985) Traditional Preparation and Uses of Cassava in Nigeria. Economic Botany 39: 157-164.
- Badbury JH (2004) Wetting Method to Reduce Cyanide Content of Cassava Flour. Cassava Cyanide and Diseases. Network News 4: 3-4.
- Peter C, Darren S, Janet G (2013) Cyanogenic Glycosides in Plant-Based Foods Available in New Zealand. Food Additives and Contaminants 30: 1946-1953.
- Alexander K, Volini M (1987) Properties of an Escherichia coli Rhodanese. The Journal of Biological Chemistry 262: 6595-6604.
- Agency for Toxic Substances and Disease Registry (ATSDR) (2006). Toxicological Profile for Cyanide (draft for public comment) Atlanta, Georgia, USA.
- Nelson DL, Cox MM (2000) Lehniger Principles of Biochemistry. 3rd edn, Worth Publishers, New York, USA Pp: 668-676.
- Dubey SK, Holmes DS (1995) Biological Cyanide Destruction by Microorganisms. World Journal of Microbiology and Biotechnology 11: 257-267.
- Yanase H, Sakamoto A, Okamoto K, Kita K, Sato Y (2000) Degradation of the Metal-Cyano Complex Tetracyanonickelate (ii) by Fusarium oxysporium N-10. Applied Microbiology and Biotechnology 53: 328-334.
- Watanabe A, Yano K, Ikebukuro K, Karube I (1998) Cyanide Hydrolysis in Cyanide-Degrading Bacterium, Pseudomonas stutzeri by Cyanidase. Microbiology 144: 1677-1682.
- Akcil A, Mudder T (2003) Microbial Destruction of Cyanide Wastes in Gold Mining: Process Review. Biotechnology Letter 25: 445-450.
- Siriantapiboon S, Chuamkaew C (2007) Packaged Cage Rotating Biological Contactor System for Treatment of Cyanide Wastewater. Bioresources Technology 98: 266-272.
- Oyedeji OO, Awojobi KO, Okonji RE, Olusola OO (2012) Characterization of Rhodanese Produced by Pseudomonas aeruginosa and Bacillus brevis Isolated from Soil of Cassava Processing Site. African Journal of Biotechnology, 12: 1104-1114.
- Raybuck SA (1992) Microbes and Microbial Enzymes for Cyanide Degradation. Biodegradation 3: 3-18.
- Colnaghi R, Pagani S, Kennedy C, Drummond M (1996) Cloning, Sequence Analysis and Overexpression of Azotobacter vinelandii. European Journal of Biochemistry 236: 240-248.
- Chena SC, Liu JK (1999) The response to cyanide of cyanide-resistant Klebsiella oxytoca Bacteria strain. FEMS Microbiological Ecology 175: 37-43.
- Kao CM, Liu JK, Lou HR, Lin CS, Chen SC (2003) Biotransformation of Cyanide to Methane and Ammonia Klebsiella oxytoca. Chemosphere 50: 1055-1061.
- Sorbo BH (1953) Crystalline Rhodanese—I. Purification and Physicochemical Examination. Acta Chemica Scandinavica 7: 1129-1136.
- Saidu Y (2004) Physicochemical Features of Rhodanese: A Review. African Journal of Biotechnology 3: 370-374.
- Cipollone R, Ascenzi P, Tomao P, Imperi F, Visca P (2008) Enzymatic Detoxification of Cyanide: Clues from Pseudomonas Aeruginosa Rhodanese. Journal of Molecular Microbiology and Biotechnology 15: 199-211.
- Okonji RE, Popoola MO, Kuku A, Aladesanmi OT (2010) The distribution of Cyanide Detoxifying Enzymes (Rhodanese and 3-Mercaptopyruvate Sulphurtransferase) in Different Species of Family Cichlidae (Tilapia zillii, Sarotherodon galilaeus and Oreochromis niloticus). African Journal of Biochemical Research 4: 163-166.
- Gianfreda L, Pao M (2004) Potential of Extra Cellular Enzymes in Remediation of Polluted Soils: A Review. Enzyme and Microbial Technology 35: 339-354.
- Gianfreda L, Rao MA, Scelza R, Scott R (2010) Role of Enzymes in the Remediation of Polluted Environments. Journal and Soil Science and Plant Nutrition 10: 333-353.
- Metcalf and Eddy Incorporated (1991) Wastewater Engineering: Treatment, Disposal and Reuse, 3rd edn, McGraw Hill, USA.
- Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST (1994) Bergey’s Manual of Determinative Bacteriology. Williams & Wilkins, Baltimore, Baltimore, Maryland, USA.
- Olutiola PO, Famurewa O, Sotang HG (1991) An Introduction to General Microbiology. A Practical Approach. Cab.
- Zlosnik JEA, Williams HD (2004) Methods for Assaying Cyanide in Bacterial Culture Supernatant. Letters in Applied Microbiology 38: 360-365
- Lee CH, Hwang JH, Lee YS, Cho KS (1995). Purification and Characterization of Mouse Liver Rhodanese. Journal of Biochemistry and Molecular Biology 28: 170-176.
- Sorbo BH (1951) On the Properties of Rhodanese: Partial Purification, Inhibitors and Intracellular distribution. Acta Chemical Scandinavia 5: 724-726.
- Bradford KM (1976) A Rapid and Sensitive Method for the Quantitation of Micrograme Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Analytical Biochemistry 72: 248-254.
- Agboola FK, Okonji RE (2004) Presence of Rhodanese in the Cytosolic Fraction of the Fruit Bat (Eidolon Helvum) Liver. Journal of Biochemistry and Molecular Biology 37: 275-281.
- Lineweaver H, Burk D (1934) The Determination of Enzyme Dissociating Constants. Journal of American Chemical Society 56: 658-666.
- Akinpelu AO, Amambgo EF, Olojede AO, Oyekale AS (2011) Health Implications of Cassava Production and Consumption. Journal of Agriculture and Social Research 11: 118
- Goyal K, Walton LJ, Tunnacliffe A (2005) LEA Proteins Prevent Protein Aggregation Due to Water Stress. Biochemical Journal 388: 151-157.
- Prakash JS, Sinetova M, Zorina A, Kupriyanova E, Suzuki I, et al. (2009) DNA Supercoiling Regulates the Stress Inducible Expression of Genes in the Cyanobacterium synechocystis. Molecular BioSystems 5: 1904-1912.
- Ehigie LO, Okonji RE, Ehigie AF (2015) Purification and Characterization of Bitter Melon (Monordicacharantia). International Journal Research in Applied Natural Social Science 3: 47-58.
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 4455
- [From(publication date):
May-2017 - Apr 04, 2025] - Breakdown by view type
- HTML page views : 3508
- PDF downloads : 947