Review Article Open Access
Biochemical Sulfuryl Group Transfer From 3'-Phosphoadenosine 5'-Phosphosulfate (PAPS) Versus Phosphoryl Transfer From ATP: What Can Be Learnt?
| KV Venkatachalam* | |
| Department of Biochemistry, College of Medical Sciences, Nova Southeastern University, Ft. Lauderdale, FL-33328, USA | |
| Corresponding Author : | KV Venkatachalam Department of Biochemistry, College of Medical Sciences Nova Southeastern University, Ft. Lauderdale, FL-33328, United States Tel: (954)262-1870 E-mail: venk@nova.edu |
| Received: November 18, 2015; Accepted: December 21, 2015; Published: January 07, 2016 | |
| Citation: Venkatachalam KV (2016) Biochemical Sulfuryl Group Transfer From 3’-Phosphoadenosine 5’-Phosphosulfate (PAPS) Versus Phosphoryl Transfer From ATP: What Can Be Learnt? Biochem Physiol 5:192. doi:10.4172/2168- 9652.1000192 | |
| Copyright: © 2016 Venkatachalam KV. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Biochemistry & Physiology: Open Access
Abstract
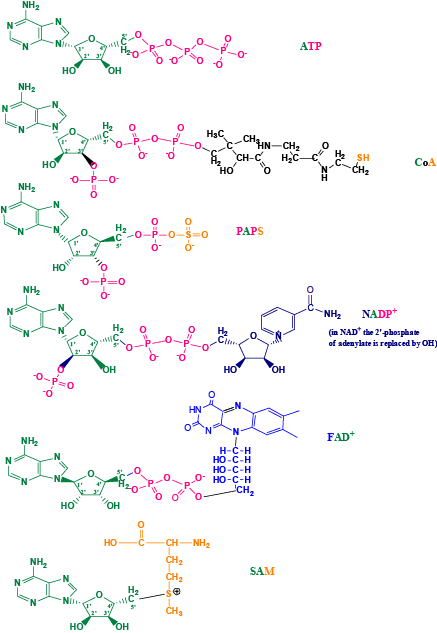
Phosphotransferases (kinases) use ATP as the universal phosphoryl donor whereas sulfotransferases use PAPS as the universal sulfuryl donor. Once phosphoryl group is transferred to a recipient molecule, it receives total of two negative charges changing its physicochemical properties. Similarly, upon sulfuryl transfer the recipient molecule receives one negative charge from the sulfonate group. Aside from this difference are there any other advantages of choosing an additional element in the biological systems? It appears that the phosphoryl transfer reactions take place during cell cycle/signaling and in primary metabolism. Whereas, sulfuryl transfer reaction happens mainly to the secondary metabolites/protein transformations? In this paper, I have compared the overall biochemical aspects of phosphate/ sulfate metabolic activation and the variety of phosphate/sulfate based cofactors. ATP, CoA, PAPS, NADP+, NAD+, FAD+ and SAM, all share the presence of adenosyl moiety. ATP, SAM and FAD+ contain only 5’ phosphate. CoA and PAPS in addition to 5’ phosphate have 3’ phosphate. CoA has the terminal pantotheine sulfur in the reduced thiol form whereas PAPS contain sulfate (the most oxidized form of sulfur). SAM has sulfur in cationic form that is attached to adenosyl group. NADP+ has 2’ phosphate in addition to the unique 5’-5’ linked phosphates. Thus, the nucleotide cofactor varieties from sulfur and phosphates are intriguing and add interesting evolutionary combinations to the biological systems.
| Introduction-Elements of Fundamental Biology |
| My inquiry on this subject came about when one of my scientific colleague asked me the question, why sulfonation as opposed to phosphorylation? This was back in 1993 while not much was known about the mammalian PAPS relative to ATP. I felt this question would be relevant at a point when more information is gathered on the mammalian, especially when human PAPS biosynthetic enzymes and the sulfur metabolism genes (cDNA) are cloned, expressed and well characterized. Now with the availability of human genome sequence, crystallographic information on sulfo and phospho transferase enzyme(s) and with the knowledge on the phosphate and sulfate metabolism one could ponder and try to understand the biological and evolutionary significance of these two related biochemical processes. |
| Comparison of Phosphate and Sulfate Metabolism |
| Phosphate transport |
| The element phosphorus (P) belongs to group number 15 (atomic weight 30) and in the oxygenated state it is referred to as phosphate (PO4 2-) and its acidic version is referred to as phosphoric acid (H3PO4). In higher organisms phosphorus element is obtained in the form of inorganic phosphate [for e.g., Ca(PO4)2, trisodium phosphate (Na3PO4)], etc. There are many mechanisms by which inorganic phosphate can be transported from the extracellular milieu to the intracellular regions via specific carrier proteins. One of the mechanisms involve either a sodium or proton dependent transport [1]. Regardless of their cation dependency these transporters have consensus amino acid sequence of “GANDVANA” on the amino and carboxy end. The aspartic residues of the human phosphate transporter are very crucial in binding to inorganic phosphate [1]. |
| Phosphate activation |
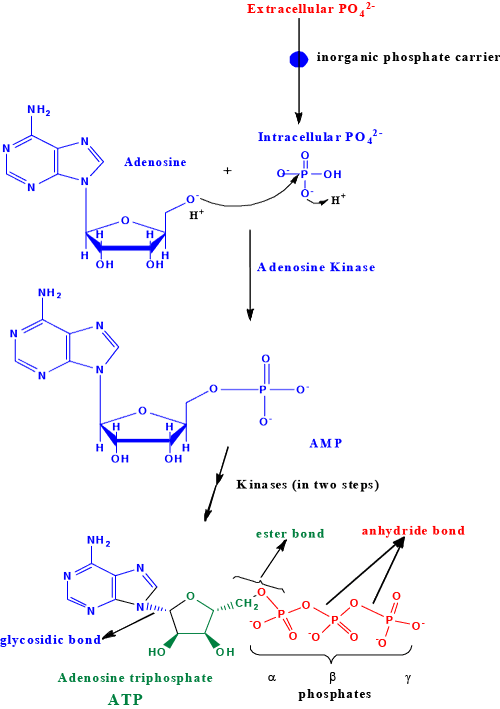
| Inorganic phosphate referred to as ortho phosphate (Pi) becomes organo phosphate by combining with glucose in glycolysis to form glucose 6-phosphate that eventually forms ribose 5-phosphate through pentose phosphate pathway. Ribose 5-phosphate is converted to PRPP. PRPP is the phosphoribosyl donor for salvage as well as de novo synthesis of AMP nucleotide. Once AMP is formed the only route to make ADP is through adenylate kinases that are dependent on ATP. This raises a serious question as to how ATP would be formed from ADP in a tissue that lacks preexisting ATP? Tissues those are rich in mitochondria forms ATP from ADP and Pi through ATP synthase (Fo/F1 ATPase). However, a tissue that lacks mitochondria must rely on ATP independent AMP kinase/adenylate kinase. Regardless of mitochondrial or non/a-mitochondrial cells: preexisting intracellular ADP and ATP are expected to be present for interconversions. However, here I hypothesize that in primordial cell/differentiating cells once adenosine and phosphate are taken up by the cell the adenosine kinase would have to make AMP using Pi and adenosine. This adenosine kinase could make ADP and ATP using Pi which would then form the fundamental pools of AMP, ADP and ATP. Once the basic/fundamental pools of mono, di and tri phosphates of adenosine are established the rest of the evolutionary sophistications can be established (Venkatachalam). This proposed area must be investigated further (Venkatachalam). |
| The enzyme adenosine kinase (EC 2.7.1.20) [2-7] reported earlier is catalytically dependent on ATP and therefore it is possible that this particular enzyme could indeed be a salvage pathway enzyme. I feel being a primordial Pi activation reaction there must be a catalytically ATP independent adenosine kinase/and or AMP kinase that must be present in eukaryotic cell that is involved in the de novo synthesis, that is yet to be isolated. The adenosine kinase (EC 2.7.1.20) that is described by Kornberg et al. [8] perhaps uses ATP as an allosteric regulator as opposed to a source of phosphoryl donor to make AMP from adenosine. This aspect is not clear from their assay systems. Nonetheless, the inhibition of adenosine kinase by bisubstrate analogs could mean direct transfer of phosphoryl from ATP to adenosine in the active site to form AMP and ADP [9]. Careful inspection of the amino acid sequences of the adenosine kinase [9] reveals the characteristic ATP binding motif (GxxxxGK), which could mean this enzyme, could use ATP as a phosphoryl donor or could use ATP as an allosteric regulator. From the crystal structure of human adenosine kinase it is proposed that an aspartic residue is involved in the deprotonation of the ribosyl hydroxyl during phosphoryl transfer [7]. The salvage pathway enzyme adenine phosphoribosyl transferase (APRT, E.C. 2.4.2.7) that transfer adenine onto C-1 of PRPP to make AMP and PPi had been well described [9- 11]. The formation of PRPP also relies on preexisting intracellular ATP. |
| Here I have proposed a reaction mechanism for a possible catalytically ATP independent de novo adenosine/AMP kinase reaction (Figure 1). Such an enzyme is warranted in the intracellular milieu since it excludes preexisting ADP or ATP in the primordial cell. Therefore, isolation of this putative catalytically ATP independent adenosine/ AMP kinase is very crucial in understanding the fundamentals of biology and the evolution of phosphate activation in organisms (work in progress: Venkatachalam et al.). AMP can then be activated to 2ADP by combining with ATP catalyzed by adenylate kinase (EC 2.7.4.3) [12-18]. The formation of ATP itself can occur by de novo mechanisms through oxidative phosphorylation by combining ADP with inorganic phosphate catalyzed by the mitochondrial proton coupled ATP synthase/ATPase system (EC 3.6.3.14) [19-22]. In cells that lack mitochondria for example (erythrocytes) ATP could be formed by ADP kinase that would use soluble NDP kinase to make ATP (poorly identified). Very little light has been shed on this over all context of ATP formation between these cell types. |
| Phosphoryl transfer |
| ADP has one phosphoanhydride bond that could in theory serve as a high-energy reservoir for driving the thermodynamically unfavorable reaction to the direction of product formation. However, the phosphotransferase enzymes that we know thus far use only ATP and not ADP as a phosphoryl donor. In other words the scission that happens between β−γ phosphates of ATP forms the γ-phosphoryl that is eventually transferred to a recipient moiety by the phosphotransferases to form phosphorylated product [23]. Therefore, we observe ADP only as an intermediate during the biosynthesis of ATP or as a bi product of ATP hydrolyzed reactions [24]. In biochemical systems, ATP cleavage at the α−β position results in adenylate (AMP) and PPi formation [25,26]. This process happens during adenylylation, PAPS synthesis, etc. [25-27]. Here we see that the whole AMP (adenylate) portion of ATP is further used in the biotransformations [27] and PPi is only a bi product that is cleaved into two inorganic phosphates by the ubiquitous pyrophosphatase enzyme to push the thermodynamically unfavorable reaction in the forward direction. Thus we see that the molecule ATP can be utilized in two independent ways: a) Upon β−γ cleavage, the resulting γ−phosphate can be transferred to a recipient and the resulting ADP itself is a side product [23] b) Upon α−β cleavage of ATP the resulting adenylate (AMP) is the useful biotransformed unit and PPi is just a waste product [25-27]. On rare occasion during the course of s-adenosyl methionine (SAM) synthesis, methionine combines with ATP and the adenosyl moiety is transferred to the sulfur of the methionine to form SAM catalyzed by methionine adenosyltransferase (EC 2.5.1.6) [28-30]. The three phosphates in this reaction of (EC 2.5.1.6) are released as Pi and PPi [28-30]. This is only one enzyme that we know of that splits the bond of ATP between ribosyl unit and the α-phosphate. Thus we see that by using ATP in different modes, an additional variety to the biological processes had formed through evolution. |
| ATP being a primordial compound various enzymes had evolved based on the principles of the variety that are available for binding ATP. For example, the overall binding of ATP could be anti (i.e., in two dimensions adenine facing westward direction and ribosyl triphosphate(s) facing eastward) or syn (both adenine moiety and ribosyl-phosphate(s) facing the same direction). Although not absolute, it seems like many prokaryotic enzymes bind ATP in syn configuration whereas eukaryotic ATP utilizing enzymes that contains P-loop bind ATP in the anti configuration. By tweaking the molecule into shapes some more variations in binding modalities are achieved. |
| Sulfate transport |
| In contrast to the element P, sulfur belongs to group 16 (atomic weight 32) and in the oxygenated state it is referred to as sulfate (SO4 2-) and its acidic version is referred to as sulfuric acid (H2SO4). In higher organisms sulfur element is obtained in the form of inorganic sulfate [for e.g., CaSO4, MgSO4, etc.]. From the extracellular milieu sulfate is transported to the intracellular regions via specific carrier proteins [31]. The human sulfate anion transporter family (sulfate/anion exchanger) is called SLC26. SLC26A1 has 12 transmembrane domains and a cytoplasmic STAS (sulfate transporters and antisigma factor antagonists) domain at the carboxy end [32]. The STAS domain has been predicted to bind NTP based on the β-sheet scaffold that could accommodate NTP [32]. The proteins of the three human diseases diastrophic dysplasia/achondrogenesis type IB (gene DTD that encodes sulfate transporter) [33], Pendred’s syndrome (PDS gene that encodes iodide-chloride transporter) [34] and congenital chloride diarrhea (CLD gene that encodes chloride-NaHCO3 - exchanger) [35] contains STAS domain similar to bacterial antisigma factor and has once again been predicted to be regulated by NTP (ATP) hydrolysis [32]. In addition a sodium independent sulfate transporter (SLC26A11) from high endothelial venules has been reported to contain the prosite SLC26 sulfate transporter signature (PS01130), the Pfam sulfate transporter domain (Pf00916) and the STAS domain (pf01740) [36]. |
| Sulfate activation |
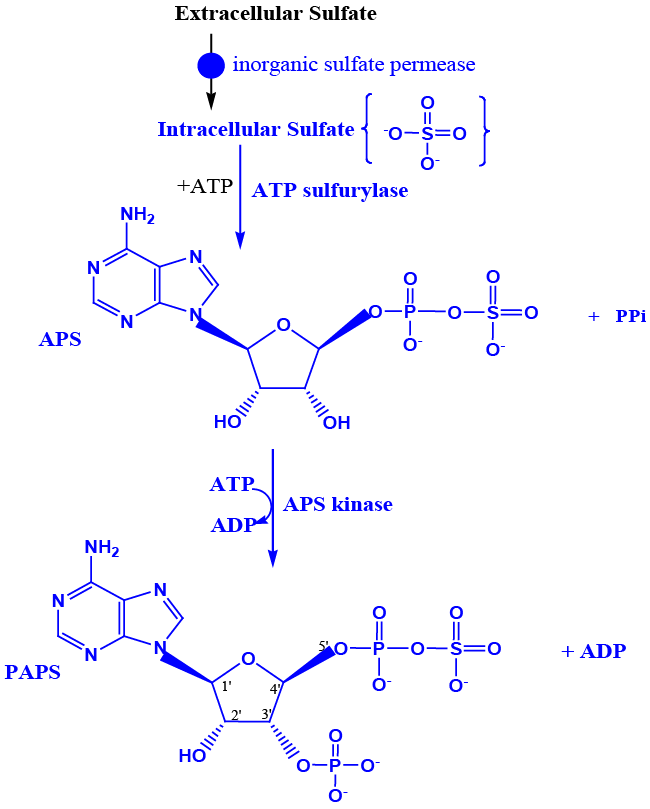
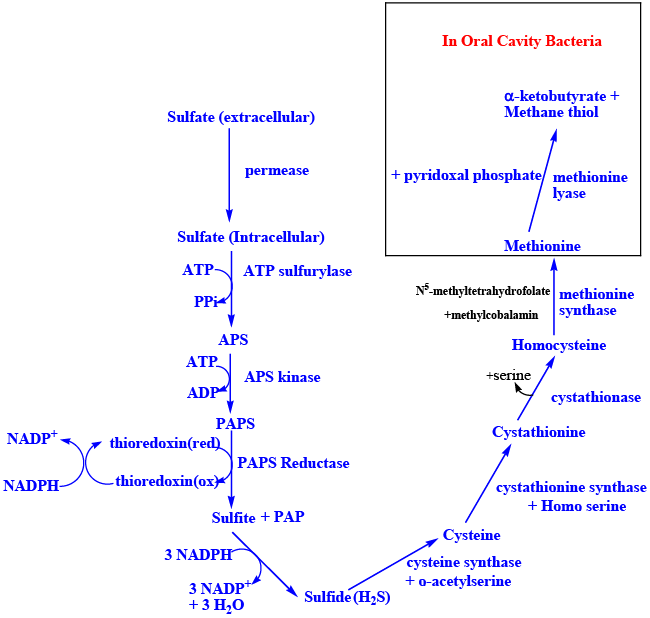
| Inorganic sulfate becomes organo sulfate by combining with ATP to form adenosine phosphosulfate (APS) catalyzed by ATP sulfurylase (EC 2.7.7.4), inside the cell [25,26,37,38,39]. APS combines with another molecule of ATP to form PAPS catalyzed by the enzyme APS kinase (EC 2.7.7.4). PAPS, is synthesized by the fused gene product referred to as PAPS synthase in humans (EC 2.7.1.25) [26,37,39] (Figure 2A). Once PAPS is formed intracellularly it cannot permeate to the extracellular regions similar to ATP. PAPS serve as the universal sulfuryl donor for many sulfotransferase reactions [23]. In addition, in lower organisms APS/PAPS also serves as the activated oxygenated carrier of sulfur to be reduced to sulfide [40]. Sulfide is then used for methionine synthesis in bacteria, plants etc., [41,42] (Figure 2B). De novo synthesis of methionine doesn’t happen in humans making it an essential amino acid that would have to come from the diet. Methionine is converted into odoriferous compound(s) methylthiol in the human oral cavity, catalyzed by the methionine lyase of the gingival bacteria; Porphyromonas gingivalis (Venkatachalam et al., accepted) (Figure 2B). The compounds methyl thiol, dimethyldisulfide, dimethylsulfide etc., causes the characteristic morning breath. |
| Sulfuryl Transfer |
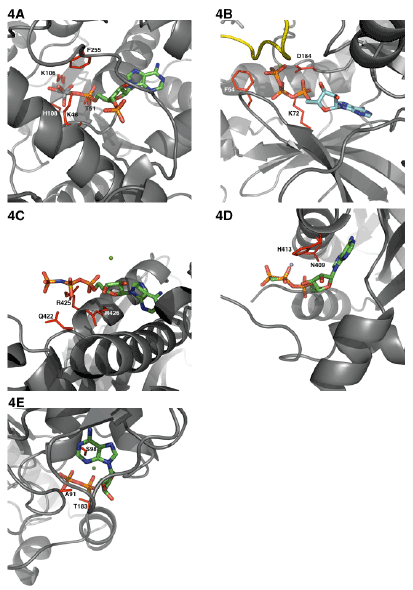
| Comparison of active site of phospho and sulfo transferases binding: Among the various elements from the periodic table only the oxyanions of group 15 element (P) and group (16) element (S), that is (PO4 2-) and (SO4 2-) respectively, had been selected during the course of evolution to be incorporated into the biological systems. What if any evolutionary advantages that these two oxyanions have that the organisms were selective, is poorly understood? Or it could simply mean that both P and S are available in equal amounts from earth. As discussed earlier the molecule ATP can bind to the enzyme either in the anti or syn mode. In Figure 3, I have depicted the syn geometry of ATP. In the anti configuration the adenine portion would assume 180o horizontal flip, upon which the ribose cyclic oxygen unit is mostly masked by the indole (nitrogen 9) of adenine. This rule applies to all cofactors that contain the adenosine/adenylate unit as part of the cofactor moiety. Thus, PAPS can also assume either anti or syn configuration. It appears that from all the known crystal structures of sulfotransferases, PAPS seems to bind in the anti orientation similar to the bacterial ATP binding to its respective active sites and similar to the various P-loop containing eukaryotic kinases [43] (Figure 4A). |
| I have mostly eliminated the crystal structures that would contain the bound recipient cosubstrate to avoid the conformational change induced NTP interactions Figures 4A-4E. In the estrogen sulfotransferase (EST) active site that is bound to PAP and vanadate, a closer look of the ribose C-3 to the 5’-β-phosphate (the residues that are within 5Aº) distance are Lys48, Thr51, Lys106, His108 and Phe255 [43]. No metal ions were found to be associated with EST around the putative PAP(s) binding region [43]. In protein kinase A (PKA) that is bound to MgATP within 5Aº distance residues Phe54, Lys72 and Asp184 were found [44]. Between EST and PKA there seems to be at least two residues (Phe and Lys) that are common in the active site. Human hexokinase type I bound with ATP analog (AMP-PNP) along with its cosubstrate(s) interacts closely with Gln422, Arg425 and Arg426 [45]. In histidine kinase CheA domain P4, that is complexed with ADPCP and manganese, the closest residues of interactions were Asn409 and His413 [46]. |
| In contrast the homoserine kinase, a prototype of GHMP kinase superfamily, seems to bind ADP in syn mode [47]. Homoserine kinase complexed with ADP shows Ala91, Ser98 and Thr183 residues in close proximity [47]. It seems that upon cosubstrate binding the active site interaction would create a dynamic motion of the protein that would result in residues that are poised for catalysis. Histidine kinase seems to share one exact residue His and one similar residue Asn with that of EST and PKA. On the other hand homoserine that is bound to the ADP in the syn conformation seems to be quite different in terms of amino acid residue selection. |
| In ATP binding proteins the adenine moiety of ATP uses peptide backbone polar interactions that are independent of protein amino acid sequences to interact with the N-1 and N-6 of adenine ring. Instead they seem to use nonspecific hydrophobic interactions [48]. If the adenine binding loop has the same backbone direction “direct” (NH2- COOH) then the N-1 is hydrogen bonded to the backbone amide of the third residue of the loop and the amino group of the adenine at the C-6 is hydrogen bonded to the backbone carbonyl of the first residue of the loop (e.g. cAMP protein kinase). In the “reverse” orientation of the protein, the amino group of the adenine at the C-6 interacts with backbone carbonyl of residue 1 and the N-1 is hydrogen bonded to the backbone amide of the second residue of the loop. The geometry of the adenine binding thru hydrogen bonding with the respective amino acid backbone motifs exactly mimics the adenine base pairing with thymine in DNA and adenine base pairing with uracil in RNA. Thus, the adenine binding motif which includes three-residue loop and an additional hydrophobic residue probably evolved independently on different time as opposed to a more prosaic divergent mechanism [48]. The adenine rings of PAPS as expected seems to be recognized by entirely different sets of amino acid residues, however overall it possesses hydrophobic pi stacking interactions (Trp53) as seen with other proteins using residues like Trp, Tyr and His (Figure not shown). |
| Next, I have compared the ribosyl unit and the 5’ phosphate/sulfate and the 3’-phosphate and the associated interactions of the amino acid residues. The phosphates (P-P) at the 5’ carbon of the ribosyl unit are present in ATP, CoA, PAPS (phospho-sulfate), NAD+, NADP+, and FAD+. Whereas, the phosphate at the 3’ end of the ribose is present only in CoA, and PAPS. In NADP+ instead of 3’-phosphate it is phosphate at the 2’C of ribose Figure 3. Using the adenylate moiety containing coenzymes, I have earlier compared the 5’-P-P interactions versus 5’-PS interactions with its respective active site residues. |
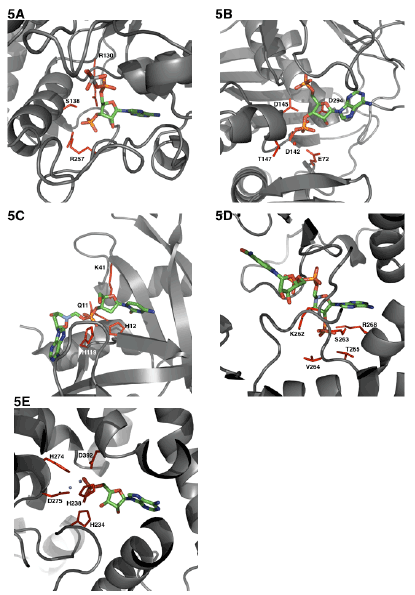
| Similarly, here below I have compared the interactions of the 3’-phosphate of various coenzymes and its interactions with its active site residues within 5Aº distance (Figures 5A-5F). While ATP phosphates are almost always complexed with magnesium the PAPS binding doesn’t seem to require metal ions for either binding of the nucleotide substrate or for catalysis. EST at the 3’-phosphate end seems to interact with Arg130, Ser138 and Arg257. The enzyme 3’-phosphonucleotidase also called PAPase (Hal2p) complexed with calcium, magnesium, and reaction substrate PAP interacts with Glu72, Asp142, Asp145, Thr147 and Asp294 [49]. PAPAse is an exonuclease that cleaves the 3’-phosphate to form inorganic phosphate (Pi) and AMP. The acidic residues in this case may interact with metal cation and the metal cation can then interact with the negative 3’ phosphate group of PAP. The indirect interaction of acidic residues with the 3’-phosphate is very similar to the phosphate transporter protein interactions with phosphate anion that I had described earlier [1]. It is very interesting that in EST since there is no metal binding, the phosphate negative charges are interacted electrostatically by the basic arginine residues (Figure 4A). In RNase A bound to dCpA mimicking the semi transition state shows the catalytic residues His12, His119 and few closely interacting residues Gln11 and Lys41 [50]. In this case the enzyme RNase A is an endonuclease that cleaves the phosphodiesterase bond at the 3’-end. Therefore one would expect very different residues. Aldose reductase that binds to NADP+ has a 2’-phosphate. This 2’-phosphate seems to closely interact with Lys262, Ser263, Val264, Thr265 and Arg268 [51]. There are at least two basic residues (Arg and Lys) that are perhaps involved in charge neutralizations. The enzyme cAMP 3’-5’ phosphodiesterase is very interesting that the phosphate is covalently bonded to 3’C and 5’C. The residues that are involved in this case are His234, His238, His274, Asp275 and Asp392 [52]. Here it could be speculated that the acidic Asp residues interacts with cation magnesium and the magnesium can then balance the charges of the 3’- phosphate. However the His residues in the 3’-5’-phosphodiesterase could be catalytically involved in a manner that is mechanistically very similar to RNase A. In Coenzyme A (CoA) [53] moiety it contains a 3’-phosphate and what exact residues interact if any at 5Aº distance among the CoA binding proteins is not clear at this point although one would be tempted to think it would be similar to PAP(S) interactions. |
| Conclusions and Future Directions |
| The molecule ATP seems to be used in a variety of mechanisms like anti or syn mode of binding. Catalytically, cleavage can happen at the β−γ bond and the phosphoryl group can be transferred to a small molecule or a macromolecule such as proteins. The phosphorylated compound would contain two negative charges. Energetically dianions are less reactive (more stable) and upon active site protonation it can be cleaved by specific phosphatases. |
| The molecule ATP seems to be used in a variety of mechanisms like anti or syn mode of binding. Catalytically, cleavage can happen at the β−γ bond and the phosphoryl group can be transferred to a small molecule or a macromolecule such as proteins. The phosphorylated compound would contain two negative charges. Energetically dianions are less reactive (more stable) and upon active site protonation it can be cleaved by specific phosphatases. |
| From the reports that are available it appears that PAPS bind in an anti mode to sulfotransferases (e.g. EST) much like ATP binding to PKA. The sulfotransferases [54] transfer sulfuryl group from PAPS to the recipient molecule [55,56]. The recipient molecule receives one negative charge and as a monoanion it is less stable (more reactive) and the sulfuryl group is removed by the sulfatases that would require less energy relative to phosphate. The process sulfurylation/desulfurylation mechanism occurs universally in organisms. Comparison of putative phospho-sulfo bond interaction at the 5’ end seems to have very little global consensus. Although one would be tempted to say that there is at least one basic residue that could be present in most of them (Lys/Arg/ His). Interestingly the phosphate at the 3’-C has Arg residues to interact with the negatively charged phosphates if there are no metal binding requirements, for e.g. EST. If there is metal binding involved then the bound 3’-phosphate is coordinated to metal cation (magnesium) and the positively charged magnesium interacts with negatively charged Asp residues (many phosphotransferases/kinases). If the 3’-phosphate is meant to be catalytically removed, then it seems like there are at least one or two basic (His) residues (e.g. RNAase, 3’-5’-phosphodiesterase). |
| In future, I propose to isolate, molecularly clone the cDNA/gene, express and characterize the primordial adenosine kinase in relation to the de novo inorganic phosphate activation in mammals. |
| Acknowledgements |
| I thank Dr. Arun Malhotra Ph.D., of University of Miami Miller School of Medicine, Department of Biochemistry and Molecular Biology for the immense help in performing the computer aided protein structure and ligand binding analysis from the PDB deposited protein X-ray crystallographic structures. |
References
|
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2a | Figure 2b |
 |
 |
 |
| Figure 3 | Figure 4 | Figure 5 |
Relevant Topics
- Analytical Biochemistry
- Applied Biochemistry
- Carbohydrate Biochemistry
- Cellular Biochemistry
- Clinical_Biochemistry
- Comparative Biochemistry
- Environmental Biochemistry
- Forensic Biochemistry
- Lipid Biochemistry
- Medical_Biochemistry
- Metabolomics
- Nutritional Biochemistry
- Pesticide Biochemistry
- Process Biochemistry
- Protein_Biochemistry
- Single-Cell Biochemistry
- Soil_Biochemistry
Recommended Journals
- Biosensor Journals
- Cellular Biology Journal
- Journal of Biochemistry and Microbial Toxicology
- Journal of Biochemistry and Cell Biology
- Journal of Biological and Medical Sciences
- Journal of Cell Biology & Immunology
- Journal of Cellular and Molecular Pharmacology
- Journal of Chemical Biology & Therapeutics
- Journal of Phytochemicistry And Biochemistry
Article Tools
Article Usage
- Total views: 12211
- [From(publication date):
March-2016 - Jul 06, 2025] - Breakdown by view type
- HTML page views : 11241
- PDF downloads : 970
