Beyond Microbiota and Colorectal Cancer: A Critical Review
Received: 01-Sep-2018 / Accepted Date: 03-Sep-2018 / Published Date: 10-Sep-2018
Abstract
Introduction: Cancer is known to be a major cause of morbidity and mortality, making it vital to deepen cancer research. Colorectal cancer is one of the most common cancers and represents the fourth most common cause of death from cancer worldwide. It is a multifactorial disease associated with environmental exposure, DNA mutations, lifestyle, inflammation and recently, microbiota.
Aim: The aim of this review is to discuss the most recent scientific advances in the establishment of a causality link between intestinal microbiota and colorectal cancer, as well as explore the underlying physiopathological mechanisms and theories. It will also be discussed the clinical relevance of the subject.
Results: Most of the studies corroborate a relationship between the microbiota changes and tumorigenesis. However, the existence of a causal relation has not yet been fully clarified. More and more evidence suggests a collective and multifactorial role of the microbiome. Nevertheless, some microorganisms appear to play a prominent role, such as Fusobacterium nucleatum, Escherichia coli and Enteroxigenic Bacteroides fragilis. There has been increasing interest in the clinical potentials of this association, namely by microbiome modulation and creation of potential screening biomarkers.
Conclusions: There is solid evidence of the relationship between microbiome and colorectal cancer. However, it’s not entirely clear yet the causal nature of this relationship, as well as the mechanisms and interactions that characterize it to its fullest extent. The most consistently associated microorganism, and therefore with the greatest clinical potential is Fusobacterium nucleatum. For now, the clinical application of these findings is not a reality, but the potentialities are huge.
Keywords: Microbiome; Microbiota; Cancer; Colorectal; Dysbiosis
Background
Colorectal cancer – Impact and carcinogenesis
Colorectal cancer is defined as a carcinoma, most commonly adenocarcinoma, in the colon or rectum [1]. It is the third most common cancer diagnosed in both men and women in the United States, representing 10% of the global cancer incidence burden [2]. It is the fourth most common cause of death from cancer worldwide. The incidence and death rates for colorectal cancer increase with age and more than 65% of the new cases occur in developed countries [3]. The exact etiology of colorectal cancer is still unknown, but it has been linked with genetic, epigenetic and environmental factors [4]. Most colorectal cancer cases are sporadic (70-80%), but it can have a hereditary component (20-30%), due to some susceptibility syndromes, such as Human Non-polyposis Colon Cancer and Familiar Adenomatous Polyposis. A small part of CRC cases (1–2%) can arise as a consequence of inflammatory bowel diseases [5]. Colorectal cancer is a heterogeneous disease, both in terms of molecular and morphologic carcinogenesis pathways. Three molecular carcinogenesis pathways are generally accepted: the chromosomal instability, microsatellite instability and CpG island methylator phenotype or epigenetic instability pathway [6]. The chromosomal instability pathway is the most common in sporadic colorectal cancer (about 85%), and includes activation of oncogenes like KRAS and the inactivation of tumor suppressor genes such as APC and p53 [1,4]. Although the triggers leading to the onset of DNA mutations and molecular alterations are not fully understood, it is known that colorectal cancer is mainly a “lifestyle disease” [1]. In most cases, environmental factors such as dietary composition, lack of physical activity, overweight, cigarette smoking and heavy alcohol consumption play a vital role in an individual’s risk of colorectal cancer [7,8]. Over the past decade, there has been increasing experimental evidence linking gut microbiome and colorectal cancer [8]. Therefore, colorectal cancer is one of the major cancers for which modifiable causes may be identified and possibly prevented. A better understanding of the mechanisms involved and interactions between the environmental factors is crucial in the quest for CRC prevention and screening.
Overview of the gut microbiome
Most of the human epithelial surfaces host a variety of microorganisms, mainly bacteria, that outnumber the human somatic cells by at least a magnitude of 10. However, the greatest number of bacterial cells reside in the digestive tract. The microbiome, defined as the collective microflora genetic material, is 150 times larger than the human genome [9]. In the gastrointestinal tract, the colon contains 70% of the human microorganisms, approximately 1014 bacteria [10]. It provides a good environment for bacterial growth due to an almost neutral pH, reduction of bile salt concentration and pancreatic secretion, associated with a slow transit time [11]. There is a natural interindividual variation in the microbiota composition and each person has its own unique microbiota [12,13]. However, there are some bacterial species commonly found in human gut microbiota, mainly from the phyla Firmicutes (30%-50%), Bacteroidetes (20%- 40%), followed by Actinobacteria and Verrucomicrobia. This complex, diverse and dynamic communities of microbiota are known to play a significant role in health. The microbiota participates in digestion and extraction of nutrients, protection against infection, in the host immune response, drugs metabolism and is also involved in regulation of host metabolism [14]. We can divide the microbiota in mucosaassociated and luminal flora, whether the microbes penetrate the mucosal layer or are located in the lumen [15]. The luminal bacteria are less abundant than the mucosa-associated bacteria [16]. Furthermore, when comparing microbiome from stool and mucosal tissue samples, different populations are found [17]. The colonic mucosal communities are adherent to surface-associated polysaccharide matrices and are therefore less affected by hydrodynamic shear forces. These communities rooted to the mucosa interact with the immune system and appear to be more relevant to diseases such as CRC.
Methods
For this review, a literature search was performed using the PubMed database. The search key terms were (“microbiome” or “microbiota” or “intestinal flora”) and (“colorectal” or “colon” or “rectal” or “rectum”) and (“cancer” or “neoplasm” or “neoplasia”). Only articles published in English and during the period of January 2013 to June 2016 were selected. The articles of the initial search have been screened for their potential eligibility according to the content of the title and/or abstract. In a second phase, the initially selected papers were full text reviewed, and a final selection was made. Articles considered off-topic, nonrelevant or redundant in their results were excluded.
Microbiota and Colorectal Cancer
Tumorigenesis and dysbiosis – Is there an association?
One third of cancers worldwide are associated with identified single microbial infections [18]. Concerning CRC, investigation linking microbiota with cancer begun in 1969 [19]. Since then, the interplay between intestinal microbiota and CRC has been a growing area of research. CRC is still one of the most prevalent and lethal cancers, and the screening methods available (colonoscopy and FOBT) are either invasive or lack high sensitivity and specificity [20]. The human gut hosts a broad and diverse community of bacteria that play key roles in the host immunity and metabolism, and possibly in disease. Several studies used next generation sequencing techniques to compare luminal (fecal samples) and/or mucosa-associated (mucosal biopsy samples or rectal swabs for mucosa-adherent specimens) microbiota of CRC patients and healthy individuals. Many of them consistently found CRC associated dysbiosis. Most colorectal carcinomas develop through an adenoma-carcinoma sequence, so several studies that include microbiome analysis of subjects with adenomas were reviewed and the results are shown in Table 1. The overall bacteria diversity in adenomas is reported to be higher than that of the normal individuals. Regarding CRC patients, studies found both lower bacterial population diversity [21-23] and an increasing diversity [24,25]. Is has been hypothesized that the lower diversity detected in diseased sites is due to a selective enrichment of a reduced number of potentially pathogenic microorganisms due to specific characteristics of the tumor microenvironment, at the expense of many other species, possibly more sensitive to environmental changes [26]. The higher bacterial diversity detected in adenoma patients and in some groups of CRC patients, could be due to the intense irrigation of tumors and polyps, providing more nutrients [27]. Another factor that might be associated with the divergent results in CRC microbiome diversity is the different stage of tumor progression in different studies. As we can conclude from Tables 1 and 2, the most consistent finding is an enrichment of Fusobacterium in CRC patients and in patients with adenomas, both in the gut lumen and mucosa. There is also an enrichment of genus Porphyromonas, Peptostreptococcus and the family Enterococcaceae in CRC patients compared to healthy controls. Regarding the phylum Proteobacteria, some papers report an enrichment in the lumen and mucosa of adenoma patients, specifically a significantly increase in some genus such as Pseudomonas, Helicobacter and Acinetobacter. On the other hand, for CRC patients, there is a decrease in Proteobacteria abundance in some studies, while other studies show that Campylobacter and Escherichia coli were significantly more abundant in CRC cases (Table 2). Lactococcus was also found to be over- represented in the mucosa of adenoma and CRC patients (Tables 1 and 2).
| Phylum | Microbiota | Change in adenomas compared to control | Type of sample | References |
|---|---|---|---|---|
| Firmicutes | Reduced | Mucosa | [19] | |
| Enterococcaceae | Increased | Feces and mucosa | [25] | |
| Lactobacillus | Increased | Mucosa | [20] | |
| Lactococcus | Increased | Mucosa | [19] | |
| Ruminococcaceae | Increased | Feces | [36] | |
| Faecalibacterium | Reduced | Mucosa and feces | [25] | |
| Solibacillus | Reduced | Mucosa | [19] | |
| Bacteroidetes | Prevotella | Increased | Mucosa and feces | [25] |
| Porphyromonadaceae | Increased | Feces | [36] | |
| Bacteroides | Increased (mucosa and feces) | Mucosa and feces | [25] | |
| B. dorei | Increased | Feces | [35] | |
| B. massiliensis | Increased | Feces | [35] | |
| Cloacibacterium | Increased | Mucosa | [20] | |
| Lachnospiracae | Reduced | Feces | [36] | |
| Actinobacteria | Bifidobacterium | Reduced | Feces and mucosa | [25] |
| Fusobacteria | Fusobacterium | Increased | Mucosa and feces | [25,43] |
| Proteobacteria | Increased | Mucosa and feces | [19,44] | |
| Enterobacteriaceae | Increased | Feces | [36] | |
| Helicobacter | Increased | Mucosa | [20] | |
| Acinetobacter | Increased | Mucosa | [20] | |
| Pseudomonas | Increased | Mucosa and feces | [19,20,36] | |
| Acidovorax | Increased | Mucosa | [20] |
Table 1: Studies that include microbiome analysis of subjects with adenomas.
| Phylum | Microbiota | Change in adenomas compared to control | Type of sample | References |
|---|---|---|---|---|
| Firmicutes | Increased | Mucosa | [45] | |
| Lactobacillales | Increased | Mucosa | [22] | |
| Erysipelotrichaceae | Increased | Feces | [22] | |
| Peptostreptococcus | Increased | Rectal swab, feces, mucosa | [22,26,46] | |
| Mogibacterium | Increased | Rectal swab | [22] | |
| Clostridium difficile | Increased | Feces | [37] | |
| Enterococcaceae | Increased | Feces | [36,47] | |
| acidaminobacter | Increased | Feces | [48] | |
| Phascolarctobacterium | Increased | Feces | [48] | |
| Parvimonas | Increased | Mucosa | [46] | |
| Parvimonas micra | Increased | Feces | [26] | |
| Solobacterium moorei | Increased | Feces | [26] | |
| Lactococcus | Increased | Mucosa | [45] | |
| Blautia | Reduced (rectal swab)/increased (mucosa and feces) | Rectal swab, mucosa and feces | [22,25] | |
| Ruminococcus | Reduced (mucosa, feces), increased (mucosa) | Mucosa, feces | [36,46,48,49] | |
| Roseburia | Reduced (feces)/increased (mucosa) | Feces, mucosa | [24,46,47] | |
| Lactobacillus | Reduced | Feces | [47] | |
| Dorea | Reduced | Feces | [48] | |
| faecalibacterium | Reduced | Mucosa, rectal swab, feces | [22,25,47] | |
| Clostridia | Reduced | Feces | [21,50] | |
| Anoxybacillus | Reduced | Mucosa | [24] | |
| Bacteriodetes | Lachnospiraceae | Reduced | Feces | [48,50] |
| B. ovatus | Increased | Feces | [35] | |
| B. vuldatus | Increased | Feces | [35] | |
| Porphyromonas | Increased | Rectal swab, feces, mucosa | [21,22,36,46,50] | |
| Prevotella | Reduced (feces), increased (mucosa and feces) | Mucosa, feces | [25,48] | |
| Parabacteroides | Reduced | Mucosa | [49] | |
| Fusobacteria | Fusobacterium | Increased | Rectal swab, mucosa, feces | [21,22,25,26,36,37,45-47,49,50] |
| Leptotrichia | Increased | Mucosa | [49] | |
| Euryarchaeota | ||||
| Methanobacteriales | Increased | Feces and mucosa | [25] | |
| Proteobacteria | Reduced | Mucosa | [45] | |
| Citrobacter farmeri | Increased | Feces | [48] | |
| Esherichia coli | Increased | Feces, mucosa | [35,51,52] | |
| Campylobacter | Increased | Mucosa, feces | [27,33] | |
| Verrucomicrobia | ||||
| Akkermansia muciniphila | Increased | Feces | [48] | |
| Actinobacteria | ||||
| Coriobacteriaceae | Increased | Feces | [22] | |
| Microbacterium | Reduced | Mucosa | [24] | |
| Bifidobacterium | Reduced | Rectal swab, feces, mucosa | [22,25,35,51] |
Table 2: CRC associated dysbiosis.
Individual studies discovered some over-represented bacteria listed in Tables 1 and 2. It seems relevant to refer that Parvimonas micra showed strong association with CRC across five different cohorts, four of them ethnically different [28]. There were also many discording results regarding some bacteria. These differences and results that don’t replicate between studies seem to reveal that there is a diversified pattern in human CRC microbiome community. This makes sense since there is a natural inter-individual variation in normal gut microbiome, and regional differences must be taken into account. The different stage of tumor progression in different CRC individuals may also account for some part of this variation. Differences in sampling methods (stool sampling or mucosal biopsies), in the experimental approaches and the small sample size of some studies also contributes to these different results. The fact that in many studies, the microbiota of individuals with polyps was found to be significantly different to that of controls suggests a possible role of microbiota in the early stages of tumorigenesis, and that the microbiota differences in patients with cancer are not secondary to the cancer itself.
Microbiome Role in Tumorigenesis Promotion
Bacterial driver-passenger model and alpha-bug hypothesis
One of the main questions investigators have asked is how bacteria play a role in oncogenesis. Is it driven by multiple species acting collectively? Are there protagonists, or even single microbes capable of promoting cancer by themselves?
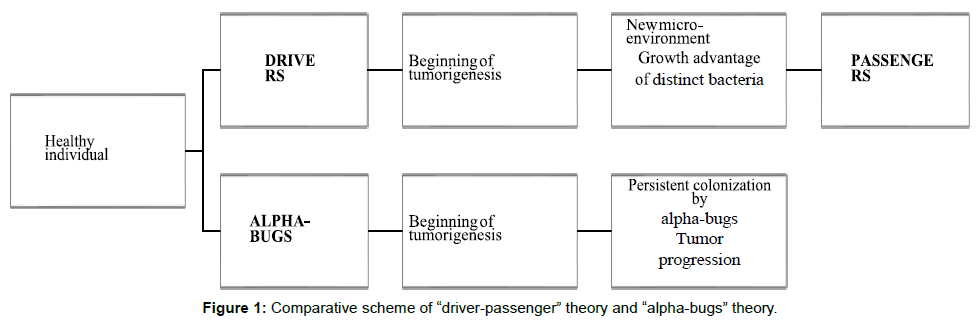
Harnold et al proposed a bacterial counterpart of the genetic “adenoma-carcinoma sequence” model formulated by Fearon and Vogelstein, the driver–passenger model. According to this model, the colonic mucosa of individuals more prone to develop CRC is colonized with certain pathogenic intestinal bacteria. These bacteria drive epithelial DNA damage by persistent inflammation, increasing cell proliferation and/or production of genotoxic substances. This would trigger the subset of pre•malignant lesions and the subsequent accumulation of mutations, leading to the initiation of carcinogenesis. There are designed “bacterial drivers” [29].
Then tumorigenesis, with rupturing and bleeding of the cancerous tissue, induces local intestinal alterations like changes in colonic barrier permeability and cellular metabolism. These changes would favor the proliferation of not only opportunistic bacteria, but maybe also commensal or probiotic bacteria, being these bacteria designated the “bacterial passengers”. Bacterial drivers may disappear from cancerous tissue as they are outcompeted by passenger bacteria with a growth advantage in the new tumor microenvironment [30]. So, according to this theory, bacterial drivers and passengers have distinct temporal associations and probably distinct roles in tumorigenesis [31]. Another model, the “alphabug” hypothesis, suggests that some microorganisms, designated “alpha•bugs” are directly pro•oncogenic, by their ability to modify the mucosal immune response and alter the colonic bacterial community. By these mechanisms, the alpha bugs together with helper bacteria, possibly “crowd out” some beneficial species. This “alpha bugs” and the helper bacteria correspond to the driver bacteria, in the driverpassenger model [32]. However, according to this hypothesis, drivers persistently colonize the developing tumor and are not outcompeted by other bacteria. This theory arose from their studies of enteroxigenic Bacteroides fragilis [33] and this microorganism is the protagonist of this model. The authors of the driver-passenger model propose as possible main bacterial-drivers pathogenic members of Bacteroides, such as enteroxigenic Bacteroides fragilis or the family Enterobacteriaceae, highlighting the uncertainty of this hypothesis. For main bacterial passengers are proposed the pathogens Fusobacterium or Streptococcus spp. and commensals like members of the Coriobacteriaceae family in Figure 1. After these proposals many studies have emerged. Different studies found an enrichment of Fusobacterium in CRC but there’s also a study reporting over-representation in adenomas, noting that there was higher abundance in tumor that in adenoma samples [34]. This suggests a possible active involvement also in early CRC development. According to the alpha-bug theory, Fusobacterium could possibly be considered an alpha-bug. This information isn’t also incompatible with the driver-passenger theory since, as the authors highlight, this model does not exclude the involvement of some of the bacterial passengers in the beginning of pathogenic alterations. If the new tumoral microenvironment provides preferable conditions for these bacteria, then they will continue to grow and participate in CRC progression [35]. Therefore, the bacterial driver-passenger model is apparently more complete, including the alpha-bug definition, but also proposing new hypothesis.
In Tables 1 and 2 we can see that many of the bacteria reported as over and underrepresented in adenomas were not reported in CRC, and vice versa. For example, Porphyromonas, and Peptostreptococcus were not mentioned in any of the studies with adenomas but were reported as over-represented in some of the studies with CRC samples. According to the driver-passenger theory, these bacteria could be possible bacteria-passengers. Mira-Pascual et al hypothysed that the family Enterococcaceae could be a possible driver, since besides being present in all samples, its proportion was higher in the adenoma group, comparing to healthy controls and adenocarcinoma.
Mechanisms of interaction with the host
The studies that have been presented so far have focused on identifying the taxonomic classes associated with CRC. Furthermore, it’s important to understand if and what bacterial products may have an impact in health and disease and explore the potential bacterial interactions.
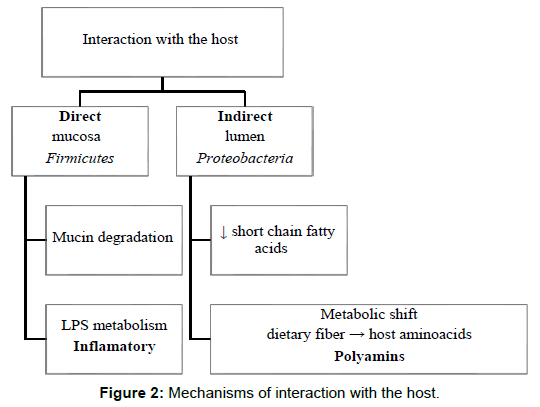
Chen et al postulate that bacteria and its components function by directly interacting with the host or in an indirect way, through cometabolism or metabolic exchange with the host. According to their results, the predominant phylotypes in lumen of CRC patients have all been associated with metabolic disorders or energy metabolism and linked with obesity and high-fat diets. They also compared fecal and mucosal samples and found higher Firmicutes in gut lumen and higher Proteobacteria in mucosa. Firmicutes has been demonstrated to enhance energy harvest from diet and Proteobacteria potentially exhibits direct interaction with intestinal cells.
Zeller et al. found a global metabolic shift between CRC cases and controls, from utilization mainly of dietary fiber in the healthy participants, to predominant host-derived energy sources in CRC. Host cell-derived metabolites like amino acids were more abundant in the tumor environment, and data suggested an increased capacity of amino acids uptake and metabolism by the microbiota, via the putrefaction pathway. The metabolites from this pathway include polyamins, which if accumulated intracellularly can promote tumorigenesis. This finding raises the question of whether this increased putrefaction is a tumoral consequence or has a causal role in tumorigenesis. They also observed an enrichment in LPS metabolism, which has pro- inflammatory potential, triggering an inflammatory signaling cascade [36].
Ohigashi et al detected lower concentrations of short chain fatty acids in feces of CRC individuals, and an associated increase in pH. More precisely, three types of organic acids (acetic acid, propionic acid and butyric acid), usually the most abundant in the gut, were reduced [37]. Short chain fatty acids are important final products of bacterial carbohydrate fermentation in the gut. Butyrate in particular is thought to be important in maintenance of a healthy intestinal environment. It’s considered to be the preferred energy substrate for the colonocytes, and apparently stimulates a physiologic pattern of cell proliferation and suppresses tumor cells proliferation in the colonic crypts. It also participates in the maintenance of intestinal acidity, prevention of toxin absorption and promotion of cancer apoptosis [38]. According to Hold et al, some of the main butyrate-producing bacteria are Roseburia intestinalis, Faecalibacterium prausnitzii and Eubacterium hallii [39]. In Tables 1 and 2 we can see that Faecalibacterium and Roseburia were found diminished in CRC/adenoma cases in some studies. In adenoma cases, fecal short chain fatty acids and pH were intermediate between normal individuals and CRC cases, and there were no differences detected between different CRC stages. This suggests that these variations are not consequent to the cancer itself [40] Baxter et al. found a negative correlation between the number of tumors and butyrate production capacity. It was also found a positive correlation between tumor count and mucin degradation. Disruption of the mucosal barrier integrity by mucin degradation could possibly lead to increased inflammation in Figure 2. These are some of the main mechanisms that are thought to take part in promotion of carcinogenesis by bacterial populations. However, there’s a much wider range of possible interactions and mechanisms studied and a lot of questions to answer.
Beneficial roles of bacteria
Many authors hypothesized that certain bacteria may have a role in protection against pathogens and possibly prevent the progression of cancer. Feng et al observed that some of the control-enriched species were lactic acid-producing bacteria Bifidobacterium animalis, Streptococcus mutans and S. thermophilus. Lactic acid participates in gut acidification and inhibits intestinal amino acid degradation. It was also reported to accelerate colon epithelial cell turnover in mice. There is evidence that advanced colorectal adenoma or carcinoma patients are deficient in lactic acid-producing commensals such as Bifidobacterium, that could potentiate daily epithelial renewal and inhibit potential pathogens [41]. Lactococcus also a lactic acid-producing bacteria, were over-represented in CRC patients besides playing a probiotic role in colon. Short chain fatty acids are important microbial metabolites and butyrate has been shown to have substantial anti-tumorigenic properties [42]. Butyrate is thought to be important in the maintenance of a healthy intestinal environment, participating in several benefic and antitumoral processes. Some of the main butyrate-producing bacteria (Roseburia intestinalis, Faecalibacterium prausnitzii) were found diminished in CRC/adenoma cases in some studies. This loss of short chain fatty acids producing bacteria populations is likely to play a synergistic role in potentiating tumorigenesis [43]. Lactobacillus spp. interacts with the host by binding to human mucus and they are currently used as probiotics. It is not yet understood if the effect is direct (through immune modulation, for example) or indirect (via alteration of the intestinal microbiota) [44].
Clinical Relevance
Zackular et al. identified a panel of bacterial populations that could indicate both the progression from healthy tissue to adenoma and the progression from adenoma to carcinoma, and created a screening model combining BMI, FOBT, and the microbiome data. This model provided excellent discriminatory ability. They also compared the microbiome test with the FOBT, and assessed that the likelihood ratio of a positive FOBT was lower than the likelihood ratio of a positive microbiome test. For better understanding, they explained that for a 65 years old person with a positive FOBT, there was a 1 in 15 chance of having an adenoma. This contrasts with 1 in 9 chances using a positive microbiome test in the same 65-year old. It was concluded that the sensitivity of the microbiome test was greater than the sensitivity of the FOBT [45].
George Zeller et al. used metagenomics to explore microbiota potential for CRC detection, hypothesizing that a combination of marker species could be used to improve screening. They selected the four most discriminative species, (two Fusobacterium species, Porphyromonas asaccharolytica and Peptostreptococcus stomatis) enriched in CRC patients. This metagenomic classifier proved to be slightly better than FOBT. They also combined the two tests and obtained sensitivity 45% higher than FOBT alone. The authors then assessed for external validation, applying the classifier in cohorts from different countries. They concluded that high accuracy detection was still possible even with cohort differences. It was also concluded this classifier has potential for early detection, since the sensitivity was similar for early-stage and late-stage CRC. These markers were also tested in IBD patients, and the most discriminative markers were all significantly higher in CRC, proving its specificity for CRC (29). The future application of these markers in population screening relies on the development of cost-effective methods. With this in mind, Zeller et al tested an alternative 16S sequencing classifier for CRC, and it accomplished almost as good an accuracy as the metagenomic model [46]. A recent study tested the effect of probiotic Lactobacillus salivarius REN1 in a 1,2-dimethyl hydrazine (DMH - induces colon carcinogenesis) induced mice model. Injection with DMH significantly altered the gut microbiota, while Lactobacillus salivarius REN promoted some reversion of these alterations. With carcinogenesis induction, the amount of Ruminococcus and Clostridiales increased, while Prevotella sp decreased. The probiotic bacteria reduced the amount of Ruminococcus, Clostridiales and Bacteroides dorei, and increased the amount of Prevotella.
These results suggest that some bacteria are capable of modulating other bacteria populations, and could possibly be used as a way to prevent carcinogenesis [47].
Baxter et al found a positive correlation between tumor count and mucin degradation, which would be interesting to confirm with further experiments since blockage of mucin degradation could be used as a future therapeutic target in tumorigenesis prevention or delay [48].
A recent study concluded that probiotics Clostridium butyricum and Bacillus subtilis inhibited the growth of CRC cells in mice both in vitro and in vivo, by promoting apoptosis and triggering cell cycle arrest. The anticancer effects seem to relate with the production of antiproliferative compounds (butyrate, bacitracin), suppression of inflammation and immune modulation [49]. The next step is to investigate if these findings replicate in human studies. Researchers are also exploring the possibility that the negative effects of Fusobacterium might be regulated by therapeutic interventions recently [50] Kumar et al. identified a set of target proteins suggested to be crucial for survival and pathogenicity of the bacteria. This finding can lead to future development of drugs to target these proteins and therefore diminish Fusobacterium effect on cancer progression [51]. A recent Chinese study demonstrated that Berberine, a component of the Chinese herb Coptis chinensis, reversed the F. nucleatum-mediated increase in opportunistic pathogens and also inhibited some tumorigenesisrelated pathways [52]. It’s also plausible to speculate that short chain fatty acids producing bacteria, as other beneficial bacteria, could integrate possible health biomarkers in CRC prevention.
Conclusion and Future Directions
Overall, the relation between microbiome and colorectal cancer is a growing area of research, complex, broad and continually developing. There is sustained evidence that microbiome is indeed related to colorectal cancer, and there has been a lot of research exploring and comparing the composition of clinically different populations. However, it’s not fully understood if there is a causal relation, if the changes are a cause or consequence of cancer itself, or a bit of both. Further research is needed to better understand the vast and complex mechanisms and interactions involved. Another challenge is to find a microbiome signature associated with colorectal cancer, given the great interindividual variety of microbiome populations. The most consistently associated microorganism so far, and therefore the one showing most applicability potential is Fusobacterium nucleatum. It’s also important to highlight and further explore the fact that besides the potential pathogenic role, some bacterial species may also play a protective role in colorectal cancer. This area of research has a promising potential in improving prevention, diagnosis and even treatment of colorectal cancer. Modulation of the microbial flora could possibly be used in cancer prevention. The actual screening and diagnostic methods are far from ideal: FOBT lacks sensitivity and specificity and colonoscopy is invasive and expensive. Microbiota can possibly be used to create screening biomarkers and therefore improve screening and diagnosis. It may also contribute to more directed and personalized treatments. The possibilities are immeasurable, but further prospective human studies in large populations are needed to answer the unknown questions and provide better knowledge about this complex but ultimately fascinating area.
References
- Stewart BW, Wild CP (2014) World Cancer Report 2014. Lyon: International Agency for Research on Cancer/World Health Organization 17-21: 72.
- Keku TO, Dulal S, Deveaux A, Jovov B, Han X (2015). The gastrointestinal microbiota and colorectal cancer. American Journal of Physiology - Gastrointestinal and Liver Physiology 308: G351-G63.
- Muller MF, Ibrahim AE, Arends MJ (2016) Molecular pathological classification of colorectal cancer. Virchows Arch 469: 125-134.
- Bae JM, Kim JH, Kang GH (2016) Molecular Subtypes of Colorectal Cancer and Their Clinicopathologic Features, With an Emphasis on the Serrated Neoplasia Pathway. Arch Pathol Lab Med 140: 406-412.
- Haggar F, Boushey R (2009) Colorectal Cancer Epidemiology: Incidence, Mortality, Survival, and Risk Factors. Clinics in Colon and Rectal Surgery 22: 191-197.
- Tuan J, Chen Y-X (2016) Dietary and Lifestyle Factors Associated with Colorectal Cancer Risk and Interactions with Microbiota: Fiber, Red or Processed Meat and Alcoholic Drinks. Gastrointestinal Tumors 3: 17-24.
- Tlaskalová-Hogenová H, Štěpánková R, Kozáková H, Hudcovic T, Vannucci L, et al. (2011) The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cellular and Molecular Immunology 8: 110-120.
- Gagnière J (2016) Gut microbiota imbalance and colorectal cancer. World Journal of Gastroenterology 22: 501.
- Nistal E, Fernández-Fernández N, Vivas S, Olcoz JL (2015) Factors Determining Colorectal Cancer: The Role of the Intestinal Microbiota Frontiers in Oncology 5.
- Hold GL (2016) Gastrointestinal Microbiota and Colon Cancer Digestive Diseases 34: 244-250.
- Tuddenham S, Sears CL (2015) the intestinal microbiome and health. Curr Opin Infect Dis 28: 464-470.
- Marchesi JR (2011) Human distal gut microbiome Environ Microbiol 13: 3088-3102.
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, et al. (2005) Diversity of the human intestinal microbial flora. Science 308: 1635-1638.
- Sears CL, Pardoll DM (2011) Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis 203: 306-311.
- Aries V, Crowther JS, Drasar BS, Hill MJ, Williams RE (1969). Bacteria and the aetiology of cancer of the large bowel. Gut 10: 334-335.
- Quintero E (2009) Chemical or immunological tests for the detection of fecal occult blood in colorectal cancer screening? Gastroenterol Hepatol 32: 565-576.
- Cerman AA, Aktas E, Altunay IK, Arici JE, Tulunay A, et al. (2016) Dietary glycemic factors, insulin resistance, and adiponectin levels in acne vulgaris. Journal of the American Academy of Dermatology 75: 155-162.
- Sanapareddy N, Legge RM, Jovov B, McCoy A, Burcal L, et al. (2012) Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J 6: 1858-1868.
- Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, et al. (2013) Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst 105: 1907-1911.
- Chen W, Liu F, Ling Z, Tong X, Xiang C (2012). Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer PLoS One 7: e39743.
- Huipeng W, Lifeng G, Chuang G, Jiaying Z, Yuankun C (2014). The differences in colonic mucosal microbiota between normal individual and colon cancer patients by polymerase chain reaction-denaturing gradient gel electrophoresis. J Clin Gastroenterol 48: 138-144.
- Geng J, Fan H, Tang X, Zhai H, Zhang Z (2013). Diversified pattern of the human colorectal cancer microbiome Gut Pathog 5: 2.
- Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, et al. (2015) Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol 50: 167-179.
- Yu J, Feng Q, Wong SH, Zhang D, Liang QY, et al. (2015) Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer Gut 66: 70-78.
- Zhu Q, Jin Z, Wu W, Gao R, Guo B, et al. (2014) Analysis of the intestinal lumen microbiota in an animal model of colorectal cancer. PLoS One 9: e90849.
- Tjalsma H, Boleij A, Marchesi JR, Dutilh BE (2012) A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol 10: 575-582.
- Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, et al. (2014) Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol 10: 766.
- Ohigashi S, Sudo K, Kobayashi D, Takahashi O, Takahashi T, et al. (2013) Changes of the intestinal microbiota, short chain fatty acids, and fecal pH in patients with colorectal cancer. Dig Dis Sci 58: 1717-1726.
- Scheppach W, Weiler F (2004) the butyrate story: old wine in new bottles? Curr Opin Clin Nutr Metab Care 7: 563-567.
- Scheppach W (1994) Effects of short chain fatty acids on gut morphology and function Gut 35: S35-S38.
- Hold GL, Schwiertz A, Aminov RI, Blaut M, Flint HJ (2003) Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl Environ Microbiol 69: 4320-4324.
- Baxter NT, Zackular JP, Chen GY, Schloss PD (2014) Structure of the gut microbiome following colonization with human feces determines colonic tumor burden Microbiome 2: 20.
- Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, et al. (2015) Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun 6: 6528.
- Zackular JP, Rogers MA, Ruffin MTT, Schloss PD (2014) the human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila) 7: 1112-1121.
- Fukugaiti MH, Ignacio A, Fernandes MR, Ribeiro Junior U, Nakano V, et al. (2015) High occurrence of Fusobacterium nucleatum and Clostridium difficile in the intestinal microbiota of colorectal carcinoma patients. Braz J Microbiol 46: 1135-1140.
- Wang R, Jiang L, Zhang M, Zhao L, Hao Y, et al. (2017) The Adhesion of Lactobacillus salivarius REN to a Human Intestinal Epithelial Cell Line Requires S-layer Proteins. Sci Rep 7: 44029.
- Zhang M, Fan X, Fang B, Zhu C, Zhu J, et al. (2015) Effects of Lactobacillus salivarius Ren on cancer prevention and intestinal microbiota in 1, 2-dimethylhydrazine-induced rat model. J Microbiol 53: 398-405.
- Chen ZF, Ai LY, Wang JL, Ren LL, Yu YN, et al. (2015) Probiotics Clostridium butyricum and Bacillus subtilis ameliorate intestinal tumorigenesis. Future Microbiol 10: 1433-1445.
- Kumar A, Thotakura PL, Tiwary BK, Krishna R (2016) Target identification in Fusobacterium nucleatum by subtractive genomics approach and enrichment analysis of host-pathogen protein- protein interactions. BMC Microbiol 16: 84.
- Yu YN, Yu TC, Zhao HJ, Sun TT, Chen HM, et al. (2015) Berberine may rescue Fusobacterium nucleatum-induced colorectal tumorigenesis by modulating the tumor microenvironment. Oncotarget 6: 32013-3226.
- McCoy AN, Araujo-Perez F, Azcarate-Peril A, Yeh JJ, Sandler RS, et al. (2013) Fusobacterium is associated with colorectal adenomas. PLoS One 8: e53653.
- Goedert JJ, Gong Y, Hua X, Zhong H, He Y, et al. (2015) Fecal Microbiota Characteristics of Patients with Colorectal Adenoma Detected by Screening: A Population-based Study. EBioMedicine 2: 597-603.
- Gao Z, Guo B, Gao R, Zhu Q, Qin H (2015) Microbiota disbiosis is associated with colorectal cancer. Front Microbiol 6: 20.
- Flemer B, Lynch DB, Brown JM, Jeffery IB, Ryan FJ, et al. (2016) Tumour- associated and non-tumour-associated microbiota in colorectal cancer Gut.
- Wu N, Yang X, Zhang R, Li J, Xiao X, et al. (2013) Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol 66: 462-470.
- Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, et al. (2013) Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults PLoS One 8: e70803.
- Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, et al. (2013) Co- occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 1: 16.
- Sinha R, Ahn J, Sampson JN, Shi J, Yu G, et al. (2016) Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One 11: e0152126.
- Li Y, Zhang X, Wang L, Zhou Y, Hassan JS, et al. (2015) Distribution and gene mutation of enteric flora carrying beta-glucuronidase among patients with colorectal cancer. Int J Clin Exp Med 8: 5310-5316.
- Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, et al. (2014) Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res 20: 859-867.
Citation: Lopes V, Faria G (2018) Beyond Microbiota and Colorectal Cancer: A Critical Review. J Oncol Res Treat 3: 120.
Copyright: © 2018 Lopes V, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Open Access Journals
Article Usage
- Total views: 3342
- [From(publication date): 0-2018 - Jul 02, 2025]
- Breakdown by view type
- HTML page views: 2502
- PDF downloads: 840