Research Article Open Access
Bacterial Mannitol-1-Phophate Dehydrogenase (mtlD) Transgene, Confers Salt Tolerance in the Fourth Generation Transgenic Maize (Zea Mays. L) Plants
| Thang Nguyen, Hussien Alameldin and Benjamin Goheen, Mariam Sticklen* | ||
| Department of Plant, Soil and Microbial Sciences, Michigan State University, East Lansing, MI, USA | ||
| Corresponding Author : | Mariam Sticklen Department of plant, soil and microbial sciences, Michigan State University East lansing, MI, USA E-mail: stickle1@msu.edu |
|
| Received February 14, 2013; Accepted September 16, 2013; Published September 18, 2013 | ||
| Citation:Sticklen M, Nguyen T, Alameldin H, Goheen B (2013) Bacterial Mannitol- 1-Phophate Dehydrogenase (mtlD) Transgene, Confers Salt Tolerance in the Fourth Generation Transgenic Maize (Zea Mays. L) Plants. Adv Crop Sci Tech 1: 112. doi: 10.4172/2329-8863.1000112 | ||
| Copyright: © 2013 Sticklen M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | ||
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
||
Visit for more related articles at Advances in Crop Science and Technology
Abstract
About 15% of global agricultural lands are exposed to high salinity, resulting in low crop yields and reduced food supplies. Attempts to develop salinity tolerant crops via selection and breeding have not been sufficient. Bacterial mannitol-1-phophate Dehydrogenase (mtlD) has been known for its tolerance to salinity. Maize is the third cereal crop (after wheat and rice) that is severely affected by soil salinity. We have genetically engineered maize plants with the bacterial mtlD gene, confirmed the integration and expression of this transgene in upto forth progenies, and have confirmed that transgenic plants transcribing the mtlD gene have higher rate of photosynthesis and are more tolerant to different concentrations of NaCl (especially at 200 mM level) as compared with their wild-type nontransgenic control plants.
| Keywords | |
| mtlD; Corn; maize; transgenic plant; Zea mays; Salinity | |
| Abbreviations | |
| Act1: Actin Rice Promoter; BAP: Benzylaminopurine; CaMV35S: Cauliflower Mosaic Virus (CaMV) 35S Promoter; IBA: Indole-3-butyric Acid; JS101: plasmid containing bar and mtlD gene; MS: Murashige and Skoog; Nos: nopaline synthase terminator; PCR: Polymerase Chain Reaction; PinII: potato proteinase inhibitor terminator | |
| Introduction | |
| Salinity occurs due to low rain and high soil water evaporation when water brings up the soluble salts to the top soil at a level that it causes a damaging impact on crops and on economic welfare of the farmers [1]. It is believed [2] that about 15% of the global agricultural lands have high salinity, resulting in drastic effects on crop losses and on reduced global food supplies. Such effects occur when the level of soil salinity reaches above the crops salt tolerance threshold. | |
| Methods of germplasm selection and breeding for development of salinity tolerant varieties have been used for many years [3]. However, screening of crops for salinity tolerance in the field is encountered by spatial heterogeneity of soil Physico-chemical properties and the seasonal rainfall fluctuations [4]. Therefore modern methods, including crop genetic engineering are needed to supplement the crop screening and breeding for salinity tolerance. Maize is considered to be the third major cereal crop (after wheat and rice) that is severely affected by soil salinity [4], and therefore it should be a priority to develop salinity tolerant maize via genetic engineering. | |
| Mannitol-1-phophate dehydrogenase or mtlD gene is encoded by an enzyme that can improve morphological and physiological characteristics that are associated with salinity tolerance in crops. For example, Abebe et al. [5] demonstrated that the expression of the E. Coli mtlD gene in third generation transgenic (T2) wheat (Triticum aestivum L. cv Bobwhite) resulted in an improved tolerance to salinity, enhanced fresh weight, dry weight and plant height as compared with the wild-type non-transgenic wheat plants. Another report, [6], indicates that salinity reduced the growth of non-transgenic tobacco plants by 40%, whereas it had no negative effect on growth of the mtlD transgenic tobacco plants. | |
| Another group of researchers reported that mtlD transgenic poplar trees (Populus tomentosa) survived up to 40 days in a hydroponic culture containing 75 mM NaCl solution, while their non-transgenic plant counterparts only survived 25 mM NaCl [7]. In this experiment, the stomatal conductance and photosynthesis of transgenic poplar were also higher than those of the wild-type control plants under the same salinity condition [7]. | |
| Prabhavathi et al. [8] reported that the expression of mtlD transgene in the first generation transgenic (T0) eggplant (Solanum melongena L.) seedlings grown in vitro under 200mM NaCl resulted in good growth, whereas the wild-type control seeds did not germinate to produce seedlings under the same NaCl condition. Also, canola (Brassica napus L.) transgenic seeds, expressing the mtlD gene, germinated and their seedlings survived up to 24 days under in vitro conditions on Murashige and Skoog medium [9] containing 350 mM of NaCl, whereas their wild-type control seeds failed to germinate under the same NaCl concentration [10]. | |
| The expression of the bacterial mtlD gene in transgenic potato plants resulted in an increased salt tolerance under both in vitro and hydroponic stress conditions. The effect of 100 mM NaCl on potato shoot fresh weight under hydroponic condition was reduced by only 17.3% as compared to 76.5% reduced fresh weight of control nontransgenic plants [11]. Maheswari et al. [12] also reported 1.7 to 2.8- fold shoot and root growth enhancement in the sorghum plants that were expressing the mtlD gene. A summary of mtlD transgenic crops with enhanced salinity tolerance is shown on Table 1[13-17]. | |
| To our knowledge, there has been no report on genetic transformation of maize solely with the bacterial mtlD gene. In the research presented here, we have explored the possibility of developing salinity (NaCl) tolerant maize plants via transfer of the bacterial mtlD gene into the maize genome, testing of transgenic plants for the integration and expression of this heterologous gene, and testing of the T3 plants for photosynthesis rate, stomatal conductance and NaCl tolerance at greenhouse level. | |
| Materials and Methods | |
| Transgene construct | |
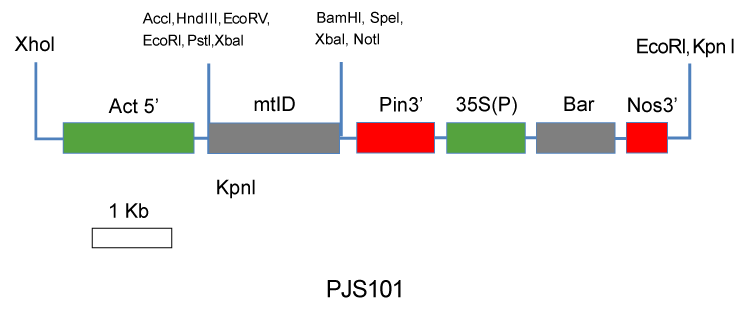
| The JS101 plasmid (Figure 1) construct was used in this research. This construct has a cassette containing the bacterial mtlD gene regulated by rice actin promoter (Act1) and the potato protease inhibitor II terminator. This cassette is linked to a second cassette containing the bar herbicide resistance selectable marker gene regulated by the Cauliflower Mosaic Virus (35S) promoter and nopaline synthase (Nos) terminator. | |
| Genetic transformation of maize immature embryo-derived cell lines | |
| Highly proliferating, immature-embryo-derived maize calli were co-bombarded via the Biolistic™ gun with the pJS101construct containing the mtlD and the herbicide resistance selectable marker genes. The bombarded calli were kept on the same conditioning medium for 24 h, transferred to callus proliferation medium [18] for 5 d, and then placed on a selection medium containing 2.0 mg/L bialaphos, and maintained for 6-8 wk, with 2-wk subcultures into fresh medium. All cultures were maintained in the dark up to this point. | |
| The bialaphos-resistant surviving callus clones were placed on our maize regeneration medium [18] and exposed to 60 µmol quanta/ m2/s light for 4-6 wk. Plantlets were transferred to a rooting medium containing the MS salts and vitamins and 1 mg/L Indole butyric acid (IBA). In vitro cultured bialaphos resistant callus lines were selected and regenerated as per our previous report [18]. Eight to ten centimeters long plantlets were transferred to small pots containing soil mix, and pots were covered with plastic bags and kept under light to mimic their former in vitro culture conditions. Small holes were made daily in each plastic bag to acclimate the plants to the greenhouse conditions before they were transplanted into 7.6 L pots, and transferred and grown in a long-day (16 h/d light) greenhouse until maturity. | |
| Leaves of fertile transgenic plants were tested for the integration and expression of the mtlD, and mature transgenic plants were selfpollinated towards homozygosity. Seeds were harvested 35-45 days after pollination, when they were dry. | |
| Conformation of the bacterial mtlD integration and expression in plants | |
| Polymerase chain reaction (PCR) analyses were performed on the leaves of different putatively transgenic plant progenies to confirm the presence of the mtlD transgene in plants. The sequences of primers used for each PCR analysis to confirm the mtlD transgene integration included the 5’ ATC GGT CGT GGC TTT ATC GG 3’ (forward primer) and the 5’ TCG ACA AAG CCA ACG TGT TC 3’ (reverse primer). The PCR program was set at 94°C for 3 min for one cycle; the following 35 cycles of 30 s at 94°C, 30 s at 55.5°C, 45 s at 72°C; one cycle at 72°C for 10 min; and the final cycle at 40C. The amplified 431bp segment of the mtlD gene was also used as DNA probes for Northern blot hybridizations. The PCR products were analyzed by electrophoresis in 0.8% agarose gels containing ethidium bromide, and visualized under ultraviolet light. | |
| Northern blotting was performed to confirm the transcription of the mtlD transgene in transgenic plants. Total RNA was isolated from the PCR-positive and from the wild-type control untransformed plants using Trizol reagent following the manufacturer instructions (Invitrogen, CA). RNA gel blot analysis was carried out following modifications of our previous procedure [18]. The Northern blots were exposed to X-ray film and developed in an X-OMAT Processor (Hyblot CL, Denville, Scientific INC, E3018). | |
| Net photosynthesis and stomatal conductance test | |
| The net photosynthetic rate (µmol CO2 m-2 leaf area s-1) and stomatal conductance (mmol H20 m-2 leaf area s-1) of the plants 2nd uppermost leaves were determined using the portable photosynthesis LI-6400XT device as recommended by the manufacturer (LICOR, Lincoln NB). | |
| Salinity tolerance test | |
| A total of 40 seeds of one of the T3 generation lines and a nontransgenic line were sown in small square plastic pots containing BACCTO High Porosity Professional Planting Mix (Michigan Peat Company, Houston, TX). The seedlings were watered daily with normal tap water for two weeks before being salt treated. Salt treatments were performed for 10 days on the seedlings that were at their four-leaf stage of growth. Plant height and the distance from ground level to the tip of the longest leaf were measured in cm for the absolute growth rate (AGR). The formula: AGR = (h2-h1)/ (t2-t1) was used, where h2 and h1 represented the final and the initial height of plant; and t2 and t1 represented the final and the initial days [19]. This experiment was replicated in two locations of the same greenhouse for accuracy. | |
| Seedlings were daily treated with equal volume of four different concentrations (0, 100, 200 and 300 mM) of NaCl, while gradually increasing 50 mM per day NaCl to reach their final concentrations within the ultimate 10 days. The commercial 20-20-20 fertilizer (Peter, Salem, OR) was weekly supplemented into the salinity solution for nutritional needs. After 10 days of salt water applications, observations were made on shoot fresh weight, and shoot and root dry weight. Plants were then watered daily for one week in order to allow them to recover from salinity stress injuries. | |
| Results and Discussions | |
| Integration of mtlD transgene was confirmed by PCR, and confirmation of the mtlD transcription was confirmed via Northern blotting. | |
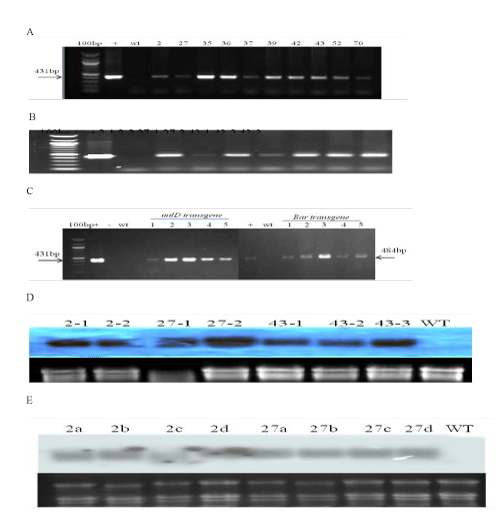
| PCR analyses of different transgenic progenies confirmed the integration of the bacterial mtlD transgene in maize plants (Figures 2A-2C). The PCR amplification products showed the expected band size of 431 bp in all transformants and in the positive control (JS101 plasmid) as expected. No band was observed in the wild-type control plants. | |
| Northern blot hybridization confirmed the transcription of the mtlD transgene in plant progenies (Figure 2D and 2E). The specific PCR amplification product of mtlD sequence was used as a probe for Northern blot hybridization. | |
| Salinity tolerance test of fourth generation transgenic plants | |
| The fourth progeny of transgenic (T3) plants were tested for salinity tolerance. Under salinity stress conditions, there was a significant reduction in plant height growth rate (cm growth per day) as salinity levels were increased in both T3 and in the wild-type control plants. However, the wild-type control plants were more retarded due to salinity stress than transgenic plants under the same salinity treatments. The average plant growth rate of the wild-type control plant under 200 mM and under 300 mM NaCl were respectively 1.5 cm per day and 1.1 cm per day, whereas the corresponding data for transgenic plants under the same NaCl concentrations were 1.6 and 1.4 cm per day respectively (Table 2). | |
| Note: in Turkey test, the mean values with the same letter (such as, a and a /b and b, etc) are not significantly different from each other, but mean values with different letters (such as, a and b / b and c) are significantly different (P<0.05).WT: Wild-type control plant. | |
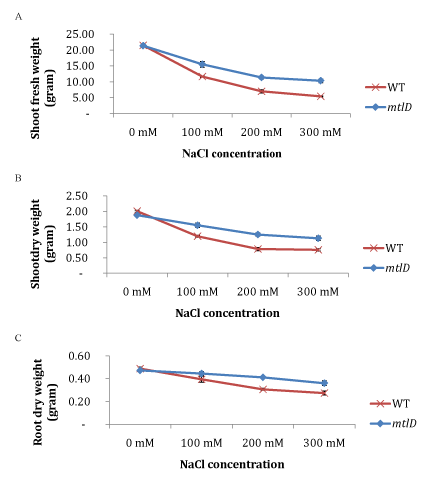
| Figures 3 represents the shoot fresh and dry weight and the root dry weight 10 days after being treated with different concentration of NaCl. | |
| Table 3 confirms that the NaCl treatment had affected the wildtype control seedling growth, resulting in reduced shoot fresh weight, and shoots and root dry weights. However, the mtlD transgenic plants were less affected by the NaCl treatments as compared to the wild-type control plants. At 100 mM NaCl treatment, the shoot fresh weight of transgenic plants was reduced by 27%, shoot dry weight by 17%, and root dry weight by 6% as compared to the no salt treated control plants. However, the reduction of shoot fresh weight, shoot and root dry weights in wild-type control plants were 45%, 40% and 19% respectively. | |
| When plants were exposed to 300 mM NaCl, the effect of salinity stress was more severe on wild-type control plants, reducing up to 74% of shoot fresh weight, 62% of shoot dry weight, and 44% of root dry weight. Table 3 also shows that T3 plants exhibited greater salt tolerance at the same salt stress condition than the wild-type plants, as they reduced shoot fresh weight, shoot dry weight and root dry weight by 52%, 40%, and 24% respectively. The effect of salinity stress also reduced the absolute plant height rate of both transgenic and wild-type plants. The plant height rates of transgenic and wild-type plants were not significantly different under no-salt stress condition. Overall, the increase in salt concentration had more negative effects on the wildtype control plants as compared to the T3 plants. | |
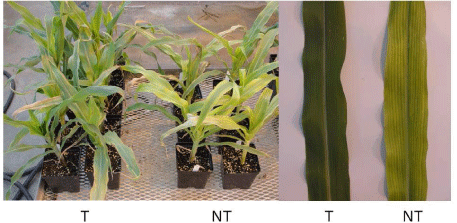
| Figure 4 represents samples of the T3 mtlD versus the wild-type control plants after 10 days of 200 mM NaCl treatment followed by seven days of water treatment. | |
| The work presented here agrees with the work of other scientists indicating that the expression of mtlD gene conferred salinity tolerance, and enhanced fresh and dry biomass of transgenic plants in sorghum [12] and potato [11]. | |
| In our studies, we did not find any abnormal phenotypes such as dwarfed or stunted growth of transgenic plants that expressed the bacterial mtlD transgene as compared to their wild-type control plants under no-salt condition. The same result was reported for the mtlD transgenic eggplant [8]. | |
| In the research presented here, salt stress experiments demonstrated significant differences in shoot fresh and dry weight, and root dry weight of transgenic as well as the wild-type control plants under high salinity concentrations. Also the mtlD transgene plants showed greater plant height growth rate and less shoot and root reduced growth under salt stress than the wild-type control plants. Similar results were reported in mtlD transgenic wheat [5], eggplant [8] and tobacco [19]. | |
| The effects of salinity stress on photosynthesis and stomatal conductance of T3 mtlD and wild-type control plants are shown in Table 4 and Figure 5. Table 4 shows that the photosynthesis rate and stomatal conductance declined with increased salinity concentrations in the wild-type non-transgenic plants, and were significantly lower as compared to transgenic plants at 100 mM NaCl. However, the photosynthesis rate and stomatal conductance were not significantly different in transgenic plants that were treated with 100 mM and 200 mM of NaCl followed by seven days of water treatment recovery. | |
| Figure 5 shows that the effect of salinity on photosynthesis in the wild-type control plants was more severe than on those of transgenic plants that were treated with 100 mM of NaCl for 10 days, followed by seven days of water treatment recovery. All wild-type control plants were dead with shrunken and dried leaves after 7 days of water treatment, not being able to recover from 10 days of 200 mM of NaCl treatment. The data presented in table 4 confirm that the mtlD transgenic plants exhibited higher photosynthesis and stomatal conductance than the wild-type control plants that were treated with 100 mM NaCl for 10 days, agreeing with the report from Hu et al. [7] indicating that there were higher stomatal conduction and photosynthetic rates in poplar transgenic plants as compared with their wild-type control plants treated with 50 mM of NaCl for 21 days. | |
| The authors hope that the research presented here will be one step closer to developing of a maize genotype that can tolerate the salinity in parts of the world that farmers suffer from losses of this abiotic stress factor. | |
| Acknowledgements | |
| The pJS101 was obtained from the late Professor Ray Wu of Cornel University and the maize immature embryos were purchased from the Transformation Center of the Department of Agronomy at Iowa State University. The authors wish to thank Professor Wayne Loescher of the Department of Horticulture at Michigan State University for his advice on the photosynthesis and stomatal conductance studies of this research. | |
| References | |
References
|
|
Tables and Figures at a glance
| Table 1 | Table 2 |
Figures at a glance
 |
 |
 |
 |
 |
||||
| Figure 1 | Figure 2 | Figure 3 | Figure 4 | Figure 5 |
Relevant Topics
- Agricultural science
- Agronomy
- Climate impact on crops
- Crop Productivity
- Crop Sciences
- Crop Technology
- Field Crops Research
- Hybrid Seed Technology
- Irrigation Technology
- Organic Cover Crops
- Organic Crops
- Pest Management
- Plant Genetics
- Plant Breeding
- Plant Nutrition
- Seed Production
- Seed Science and Technology
- Soil Fertility
- Weed Control
Recommended Journals
Article Tools
Article Usage
- Total views: 15604
- [From(publication date):
September-2013 - Apr 04, 2025] - Breakdown by view type
- HTML page views : 10621
- PDF downloads : 4983
