Astrovirus: A Challenge for Humans, Animals and the Environment
Received: 21-Nov-2023 / Manuscript No. JIDT-23-120597 / Editor assigned: 24-Nov-2023 / PreQC No. JIDT-23-120597 (PQ) / Reviewed: 07-Dec-2023 / QC No. JIDT-23-120597 / Revised: 14-Dec-2023 / Manuscript No. JIDT-23-120597 (R) / Published Date: 21-Dec-2023 DOI: 10.4172/2332-0877.575
Abstract
During the last two decades, Astroviruses (AstVs) have gained more attention most likely due to the discovery of their immense diversity, and extragastrointestinal spread. The number of living species as well as the tracks that are infected by AstVs are continuously raising which highlights the significance of this viral family. importantly, the presence of AstVs in the environment not only gives the virus an arsenal of options to spread, but also completes the cycle of the one-health approach to challenge the intertwined health of humans, animals, and the environment. Here, we review the most current knowledge on the virus from different perspectives including its presence in food, and environmental matrices
Keywords: Astrovirus; Environmental matrix; Animals; Food; Encephalitis
Introduction
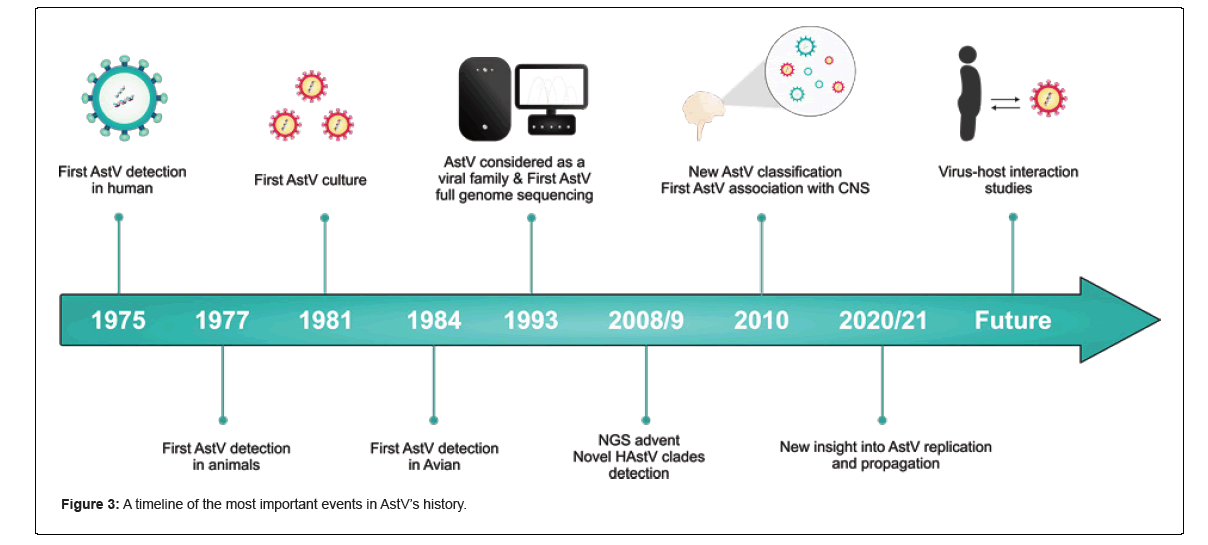
In 1975, an enteropathogenic agent was observed through electron microscopy in stool samples from children who had diarrhea. The agent was called Astrovirus (AstV) because of its appearance and shape [1]. AstV is a positive sense, single-stranded, non-enveloped, spherical RNA virus, considered the second most prevalent cause of gastroentrotites in children following rotavirus [2-4]. The Astroviridae family is highly widespread, and it has been reported that up to 90% of the pediatric population has antibodies against Human Astrovirus Type 1 (HAstV-1), making it the most prevalent HastV [5]. AstVs typically cause diarrhea and gastroenteritis in infants, the elderly, and immunocompromised people and may lead to an outbreak where these high-risk people live in close contact in places such as kindergartens, dormitories, care centers, and hospitals [6]. Importantly, recent reports indicate that the virus is associated with non-intestinal organs, including the Central Nervous System (CNS) and respiratory tract [1], highlighting their growing clinical importance. The first report on AstV in animals was published in 1977, revealing astrovirus-like microorganisms in domestic animals suffering from diarrhea [7]. Since then, the virus has been detected in a wide range of animals, from domestic to wild, including avian species and those living in aquatic environments. Thanks to accurate molecular techniques, the detection rate has significantly increased in recent decades, and so far, the virus has been identified in more than 80 species [1,8,9].
Namely, astrovirus-like genomes and/or viruses have also been described in fish, amphibians, reptiles, invertebrates, and even plants [10]. Real-time PCR using pan-astroviral primers is now considered a reliable method for detecting AstVs. Also, Next-Generation Sequencing (NGS) may be used for further investigations to obtain massive genomic information [11]. Nevertheless, AstVs have been shown not to be host-specific viruses, so interspecies transmission is often reported; for example, the MAstV-pseudo genome has been reported in a bird in Hungary and AAstV-like AstVs in farmed minks in China [11]. A comprehensive timeline of the most important events in AstV research is shown in Figure . AstVs have not been widely studied, probably because there are no highly reliable cell culture systems, suitable small animal models, and some other practical restrictions [1]. Additionally, in the past two decades, the presence of the virus in the environment, water, and food as possible transmission methods has been of interest to be researched. Here we review the virus from different perspectives, including its existence in food and the environmental matrix. We have searched popular databases such as PubMed, Google Scholar, and Web of Science, and included papers published in high-quartile and impact- factor journals to review the current state of research on AstVs (Figures 1-3).
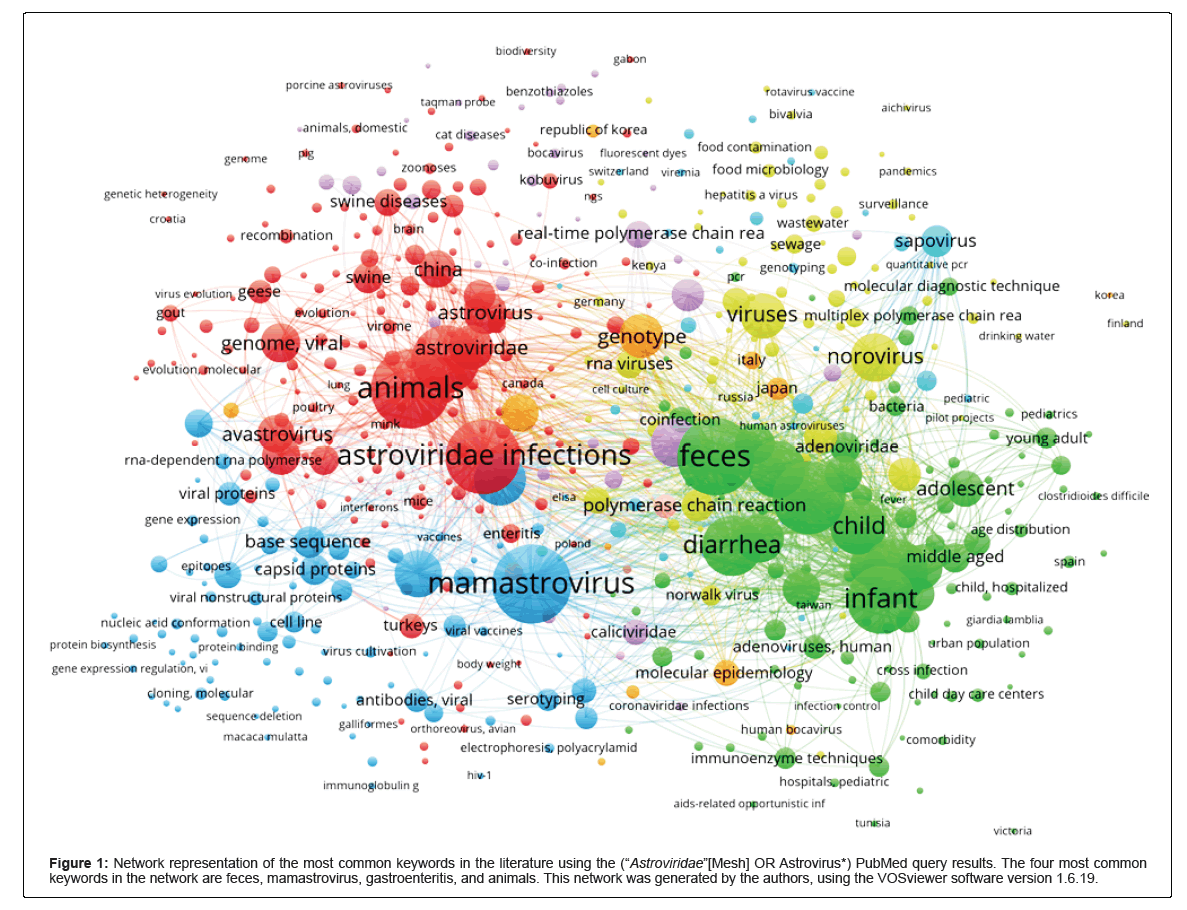
Figure 1: Network representation of the most common keywords in the literature using the (“Astroviridae”[Mesh] OR Astrovirus*) PubMed query results. The four most common keywords in the network are feces, mamastrovirus, gastroenteritis, and animals. This network was generated by the authors, using the VOSviewer software version 1.6.19.
Figure 2: An overview of the number of papers published on PubMed annually from 1977 to 2023 in this matter. The following PubMed query was used for the generation of this figure: (“Astroviridae”[Mesh] OR Astrovirus*). The plot shows that after a rise in publications in 2021, there was a steady decline in 2022 and 2023. The authors used version 4.3.2 of the R programming language to generate this plot.
Literature Review
Taxonomy
Classification: In 2009, the Astroviridae study group of the International Committee on Taxonomy of Viruses (ICTV) released the updated means of classification of AstVs based on the amino acid sequence of the ORF2 genome region instead of host species [3]. The Astroviridae family is currently classified into two genera, including Mamastroviruses (MAstVs) and Avastroviruses (AAstVs), and each one of the genera is also split into two genogroups (I, II) and several species within any genogroup [12].
HAstVs (MAstV1) include 8 genotypes classically known to infect humans (AV1-8), along with two novel clades [1]. Additionally, 14 species for MAstVs (MAstV20-33) and 4 species for AAstVs (AAstV4-7) have been proposed, and their acceptance by ICTV is now pending [11].
Novel clades of HAstV
Until 2008, HAstV was assumed to consist of eight genotypes of closely similar variants, so-called classical HAstVs. Subsequently, the development of metagenomic techniques along with other molecular tools such as RT-PCR allowed the identification of eight novel genotypes.
These novel clades were phylogenetically distinct from classical HAstVs and were named based on the place of their identification: Melbourn (MLB1-3) (MAstV6) and Virginia (VA1-5) (MAstV8-9), the latter also called HAstV-HMO because of a phylogenetic close association with human, mink, and ovine astroviruses. Phylogenetic analyses showed that these clades are closer to animal AstVs than classic human AstVs, hypothesizing that over time some AstVs might have crossed the species barrier and been transmitted from animals to humans as a zoonotic agent [11].
MLB and VA clades have been found in different countries, including Nigeria, Pakistan, India, the US, Gambia, China, Egypt, Nepal, and Kenya, showing their worldwide occurrence [13-16]. These novel clades have recently been of interest because of their non-enteric infections in humans and animals [10].
Virion structure and genome organization
Virion structure: The ~90KDa Capsid Protein (CP) of AstVs is encoded by ORF2 through subgenomic RNA and consists of several domains, including both conserved and hypervariable ones. The ~41nm in diameter CP protects the viral genome and also performs cell entry; however, it is the main target for the host immune system as it activates the immune response [17], resulting in heterogeneity in this region that might have been derived through positive selective pressure imposed by the host immune system on capsids [18].
Upon cleavage, the 180-subunit CP gets resolved. It has also been demonstrated that T=3 icosahedral symmetry has around 90 spike proteins around the defining cell tropism. AstV spike protein is different among genera, as AAstVs have a slightly different structure in their spike protein [17].
Genome organization
AstVs have a single-stranded, positive-sense RNA genome of approximately 6.8-7.9 kb, which includes two Untranslated Regions (UTRs) at both the 3 and 5´ terminals and a VPg protein covalently linked at the upstream of the genome [19]. Additionally, a conserved poly (A) tail called stem-loop II motif (s2m) containing approximately 30 adenines is found at the 3´ end of the majority of AstVs. The s2m has been suggested not only to have an evolutionary role but also may help the viral genome gain stability and is probably involved in viral replication [12, 20-23]. Of interest, this motif is not seen in the most prevalent genotype of porcine astrovirus (PAstV2) [21-23]. The AstV genome possesses three overlapping Open-Reading Frames (ORFs), including ORF1a, ORF1b, and ORF2, each of which is different in length based on strain, possibly due to insertion and deletion in the 3´ terminal of ORF1a [12].
Both ORF1a and conserved ORF1b regions are located at 5’. ORF1b encodes RNA-dependent RNA polymerase (RdRp), and ORF1a produces different proteins such as the VPg, Transmembrane Proteins (TM), Coiled-Coil (CC) domains, the Protease domain (PRO), a putative Helicase domain (HEL), a putative Death Domain (DD), a Nuclear Localization Signal (NLS), and a Hypervariable Region (HVR) [12].
While non-enveloped +ssRNA viruses use an RNA-helicase domain to translate the RdRp gene, AstVs employ the Ribosomal Frameshifting Signal (RFS) mechanism in their overlapping, conserved AAAAAAC sequence region between ORF1a and ORF1b, by which a portion of cellular ribosomes that translate ORF1a carry out a minus one nucleotide (-1 nt) shift into ORF1b in order to encode RdRp [1,3,12,24,25]. However, an interesting exception to the RFS mechanism was found in 2011 when a Chicken Astrovirus (CAstV) genome was shown to enjoy an independent start codon for ORF1b[26]. Also, a Duck Astrovirus (DAstV) genome was shown not to have an overlapping site between ORF2b and ORF2 [27].
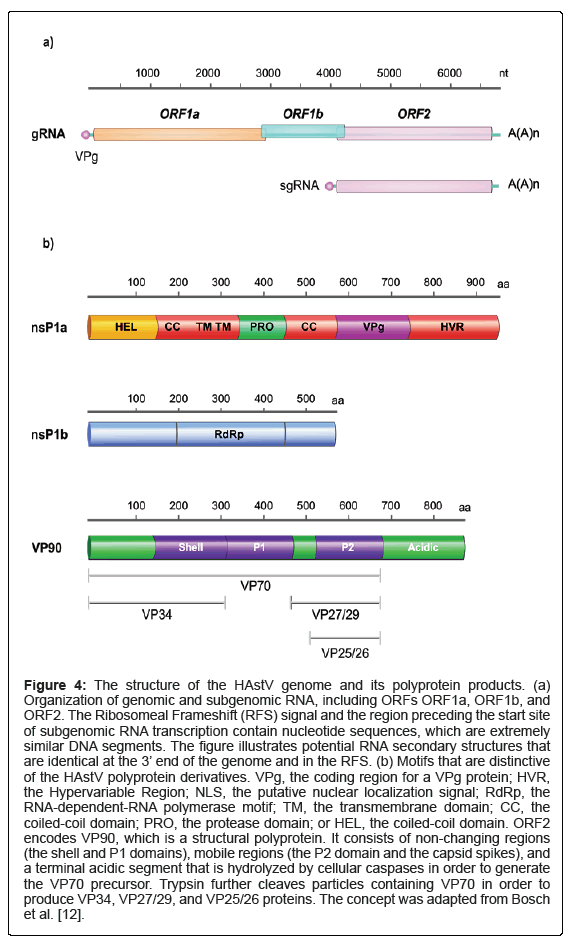
The 2400-nt-length ORF2, located at the 3´ end, encodes the viral capsid protein (VP90) and is translated through a subgenomic RNA mechanism similar to other +ssRNA viruses such as alphaviruses (Figure 4) [1,3]. The promotor of this subgenomic RNA is found in the overlapping region of ORF1b-ORF2 with a highly conserved sequence, which might be used for diagnostic assays [1,28]. Sequence analysis, however, revealed that ORF2, particularly its 3’ end, is a divergent region, so it is used for classification purposes [28]. Additionally, another ORF called ORFX with an unknown product, located at the 5´ end of the genome, which is assumed to be translated via a leaking scanning procedure, has been described in some MAstV [3, 12, 25].
Lulla et al. recently demonstrated that a 12 kDa, 112 aa, functional protein termed XP is encoded by ORFX during HAstV-1 in vitro infection. This protein plays a role in virus assembly, formation, and/ or release, probably through its viroporin-like kinetics; experimentally knocking ORFX out significantly impacts the virulence of the virus [25], providing novel insights into designing therapeutics (Figure 4).
Figure 4: The structure of the HAstV genome and its polyprotein products. (a) Organization of genomic and subgenomic RNA, including ORFs ORF1a, ORF1b, and ORF2. The Ribosomeal Frameshift (RFS) signal and the region preceding the start site of subgenomic RNA transcription contain nucleotide sequences, which are extremely similar DNA segments. The figure illustrates potential RNA secondary structures that are identical at the 3’ end of the genome and in the RFS. (b) Motifs that are distinctive of the HAstV polyprotein derivatives. VPg, the coding region for a VPg protein; HVR, the Hypervariable Region; NLS, the putative nuclear localization signal; RdRp, the RNA-dependent-RNA polymerase motif; TM, the transmembrane domain; CC, the coiled-coil domain; PRO, the protease domain; or HEL, the coiled-coil domain. ORF2 encodes VP90, which is a structural polyprotein. It consists of non-changing regions (the shell and P1 domains), mobile regions (the P2 domain and the capsid spikes), and a terminal acidic segment that is hydrolyzed by cellular caspases in order to generate the VP70 precursor. Trypsin further cleaves particles containing VP70 in order to produce VP34, VP27/29, and VP25/26 proteins. The concept was adapted from Bosch et al. [12].
Also, the Internal Ribosome Entry Site (IRES), which is a useful motif in the viral genome of some +ssRNA families such as Picornaviridae and Flaviviridae for rapidly recruiting the host cell ribosomes, is either absent in AstVs or has not been discovered yet [12, 29].
Recombination
It is assumed that the high genetic diversity of AstVs might have emerged from frequent transmission within different species [30].
Recombination events are thought to be a key factor for AstV evolution, as they allow for quick acquisition of those sequences that are more likely favorable compared with random mutations. Recombinant variants may have considerable consequences for epidemiological surveillance, phylogenetical analysis, and even pharmaceutical attempts like vaccine design [12]. Like other +ssRNA viruses, e.g., picornaviruses and caliciviruses, recombinant variants are frequent in AstVs as well, and both intra- and inter-species recombinations may ease the development of new and different variants [31].
Full genome sequence analysis has shown that these recombinations mostly occur within the ORF1b-ORF2 conserved junction [24,32], although recombinations might be seen within the ORF1a region as well [3]. In a stool sample from a kid in Kenya, an interesting recombinant AstV was recognized, where ORF1b and ORF2 have a 92% and 94% nucleotide identity to HAstV-3 and HAstV-2, respectively [33]. Another recombination in HAstV-2 types c and d was described with a recombination point in the ORF1b-ORF2 joint, in which the viruses might derive ORF1b from HAstV-3 and 1, respectively [34]. Recombination events have also been shown to occur within phylogenetically close variants, suggesting their critical role in AstVs’ evolution [35,36].
Also, it is hypothesized that HAstV-8 likely originated when HAstV-4 and 5 recombined [37]. Recombinations can occur among novel human clades of the virus as well, as it has been reported that HAstV-MLB3 can be recombined with HAstV-MLB1 or 2 [34]. Although recombination events happen between human and animal AstVs such as California sea lion [18] and turkey, or recombination of an astrovirus-like pathogen with Hepatitis E [10], strangely enough, no recombination between the novel and classic clades of HAstV has been reported yet, possibly because of insufficient studies and sampling [1].
In the environment, the marine matrix may have a key role in the virus ecology as different AstVs from different sources, such as animals and humans, can shed the virus through feces, farming runoff, and sewage in the same matrix, like a waterhole where it is regularly drunk from by different beasts, and it may provide a possibility of co-infection and recombination.
As mentioned, it should be taken into account that recombinations among human and animal strains are possible, most likely because the virus is environmentally durable, and marine matrices may have a key role in viral ecology, as different harbors can shed the virus through feces, farming runoff, and sewage in the same matrix like a waterhole where is regularly drunk from by different beasts, as a result of which, if either an animal or even human gets co-infected, we may face a possible new variant because the virus shows a high propensity for recombination, indicating how the virus challenges one-health approach, as it can simultaneously infect humans, animals, and the environment.
Astrovirus disease
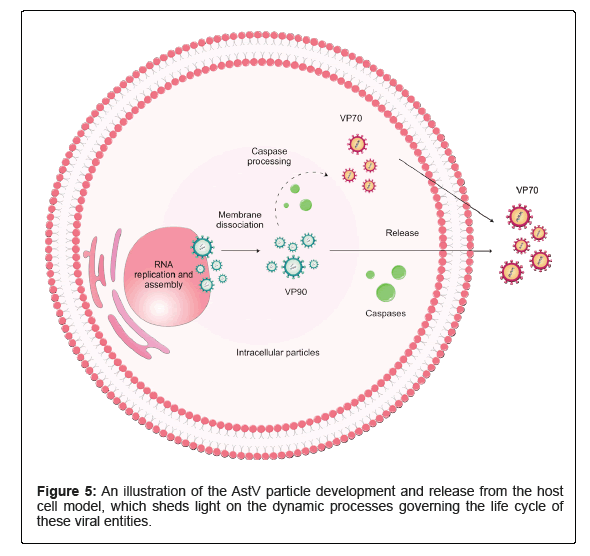
AstVs are considered enteropathogenic agents that are transmitted through both person-to-person and fecal-oral routes, like the consumption of contaminated food and water [11,38]. Vectors seem not to play a role in AstV transmission, although an arthropod is thought to be a vector for Chicken Astrovirus (CAstV) [39]. Figure 5 illustrates a model of AstV maturation and release from cells. Coinfection is common among AstVs, and it has been reported that AstV clinical cases can be accompanied by other enteric viruses, e.g., Rotaviruses (RVs) and/or Noroviruses (NVs) [40]. Also, possible correlations between acute AstV and hepatitis [41] as well as AstV and Immune Thrombocytopenia (ITP) have been hypothesized [10]. Although some novel research shows Nitazoxanide (NTZ) has positive impacts on HAstVs, including both classic and non-classic clades, through in vitro impeding their replication kinetics, leading to a decrease in the shedding viral load in the animal model, this is partially similar to other in vitro studies on the antiviral activity of ribavirin and favipiravir against human astroviruses [42,43]. So far, no approved anti-astrovirus vaccine or specific medicine is available on the market (Figure 5) [44,45].
Nevertheless, there have been numerous studies in this area that will further help the development of therapeutic agents [43,46]. For instance, Hargest et al. found that the FDA-approved nitazoxanide drug can effectively inhibit HAstV replication with a 50% effective concentration of approximately 1.47 μM in vitro [6]. A study done by Ren et al. suggests that the ORF2 protein of the novel goose AstV, GAstV-GD, along with the OASL gene, can serve as potential antiviral targets for AstV. The research revealed that GAstV-GD and its ORF2 protein trigger a robust innate immune response, leading to increased OASL expression, which, in turn, restricts the replication of the virus [47]. Ongoing research in this field aims to gather further data on potential targets within AstVs, such as their replication, with the ultimate goal of uncovering avenues for therapeutic development [48].
Astrovirus disease in humans
HAstVs are mostly observed in infants, the elderly, and/or immunosuppressed people, and classic human astroviruses (MAstV-1) usually involve typical organs such as the alimentary tract, estimated to account for approximately 3% to 5% of severe pediatric diarrhea cases [49]. Also, AstVs in children may cause long-term excretion of the virus [50].
The exact immune response and its mechanism of action against AstVs are little known, but some studies suggest that the remarkable impact of virus-specific IgA and interferons against gastrointestinal viral infections, including AstVs, cannot be ruled out [12,51]. However, maternal-fetal anti-AstV antibodies significantly drop at approximately 7 months of age, making newborns vulnerable to the infection [52].
The exact cell receptor for viral entry is currently undefined [1]. However, it is suggested that C1q receptors located on the surface of the gut epithelial cells are used by the viral capsid protein for cell entry, possibly through the Receptor-Mediated Endocytosis (RME) process [12,53]. It is suggested that the virus disorders the process whereby the intestine track absorbs water and minerals, and as a result of that, diarrhea happens [54].
In some cases, AstV infection in the gastrointestinal tract might be asymptomatic after a 3- or 4-day incubation period; however, clinical symptoms including watery diarrhea, headache, abdominal pains, and fever are displayed, and vomiting might be less frequent compared with other enteric pathogens such as rotaviruses [12]. Also, it is suggested that AstVs induce diarrhea in a way different from directly destroying intestinal epithelium; therefore, AstV-diarrhoea causes less cell death [55].
AstVs in immunosuppressed people can have life-threatening consequences as they can go beyond their typical organs to cause upper respiratory infection [12], liver disease [10], fatal brain encephalitis, meningitis [56-62], and lethal viremia [62].
AstVs beyond the gastrointestinal tract
Non-classic HAstVs are highly suspected to go beyond the gastrointestinal tract, and recent studies have associated HAstVs including HAstV-4, HAstV-VA1 (HMO-C), and MLB-2 with encephalitis in immunocompromised patients [56,59,60]. Accordingly, HAstV-VA1 (MAstV-9) is thought to be the most common cause of human encephalitis [63]. Furthermore, a recent study has indicated that extracellular vesicles may serve as a potential mechanism for AstVs to spread beyond the confines of the gastrointestinal system [64].
A possible explanation for these multi-tissue tropisms could be the expression of trypsin, as this enzyme acts in both viral capsid cleavage and virion assembly processes [12]. It is hypothesized that since classic HAstVs’ pathogenicity mostly depends on the presence of trypsin, it may significantly restrict viral replication to those organs where the enzyme is abundant, such as the gastroenteritic track. Novel HAstVs, on the other hand, may not have such limitations and be able to replicate even where trypsin is not as high as in the gastroenteritic track, e.g., the brain [10,63]. Also, most novels in vitro evidence shows non- classic clades, in particular VA-1, which shows a different replication procedure in vitro as it replicates slower than classic clades and induces a considerably different cell reaction [42].
If all of these scenarios are the case, not only may it show a different strategy of novel HAstVs to capsid cleavage, replication, and in general distinct mechanisms of disease inducement, but it may also mean that some specific tissues, like those with low-trypsin or any other advantageous features for the virus, might be highly susceptible to getting infected by non-classic HAstVs, particularly immunocompromised patients. Furthermore, some seroprevalence surveys showed that 65% of adults in the US were exposed to VA-1, a substantially high rate [42,65]. Yet, the possible correlation between trypsin, proteases, or any other suspected factors and the infectivity of HAstVs or their tissue tropisms needs to be studied.
AstV and encephalitis
Encephalitis may occur through the involvement of some antiviral factors, such as CXCL10, a 10 kDa inflammatory chemokine protein that intervenes in immune responses through both the activation and recruitment of immune cells and the actuation of apoptosis [66]. Abortive AstV viral particles have been shown in vitro to still cause inflammatory reactions through CXCL10 level elevation [66]. Cases of encephalitis related with astrovirus have been documented in Asia, North America, and Europe. The initial instance of encephalitis related with bovine astrovirus was detected in the country of Uruguay. The phylogeographic research indicates that the virus was initially transmitted to South America from Europe, and subsequently propagated to North America and Japan [67]. Koukou et al. reported the first instance of AstV causing encephalitis in an immunocompetent person. The case involved a 16-month-old infant with a history of watery diarrhea and subsequent neurological symptoms. Despite initially appearing as fever-associated seizures, further examination revealed altered mental status, leading to a diagnosis of encephalopathy [68].
Given the fact that up to two-thirds of encephalitis cases might be etiologically unexplained, AstVs have just been recently revealed to be a neurotropic pathogen, and the virus used to be commonly considered an enteropathogen, it might be argued that the etiological role of AstVs in neurological illness might have been unintentionally underestimated [66], highlighting the crucial need for advanced and accurate techniques for detecting, typing, and sequencing novel strains of AstVs.
AstV disease in animals
AstVs induce disease in several animals, including mammalian and avian species [3]. While the virus is mostly expected to cause enteric illnesses, other organs may be involved as well; for example, kidney lesions, lethal hepatitis, nephritis in avians, or neurological manifestations in mink and cattle [3,12]. Also examined are AstV- induced gene expression changes. A study examined the immunological response to Goose Nephritic Astrovirus (GNAstV). It was demonstrated that GNAstV damaged splenic and renal cells, stimulating innate and adaptive immune responses. The virus altered immune activation gene expression, suggesting immunological evasion [69]. A study conducted by Wu et al. examined the effects of GAstV on goslings and concluded that intramuscular infection can result in characteristic gout and substantial mortality. Virus concentrations were discovered to be elevated in the kidneys and liver of infected goslings, in addition to kidney and liver abnormalities. An association was identified between increased uric acid concentrations in the plasma and hepatic alterations that caused uric acid-related enzymes to become overexpressed [70].
Moreover, in a study dealing with 97 etiologically unknown cattle encephalitis cases, it was estimated that in slightly more than one-third of samples, Bovine Astrovirus (BoAstV) proteins and RNAs have been detected [53]. Zaccaria et al. also identified BoAstV in cows exhibiting neurological symptoms in Italy, providing further evidence of the role of AstV as a neurotropic agent [71]. In that context, another study focused on calf diarrhea, and identified four viruses, including BoAstV, Bovine Coronavirus (BCoV), Bovine Kobuviru (BKoV), and Bovine Norovirus (BNoV) in fecal samples [72].
Furthermore, some reports documented the detection of AstVs in both sheep and pigs suffering from encephalitis [66]. Kauer et al. provided the first identification of AstV in goats as well as in small ruminants [73]. Farmed avians are frequently infected with different strains of AAstVs such as TAstV (turkey), DAstV (duck), CAstV (chicken), GFAstV (guineafowl), PiAstV (pigeon), and GAstV (goose) [3,74,75]. For example, a recent study has found CAstV to be the major cause of hatchery condemnation in commercial turkeys in Nigeria [76]. Additionally, another recent study has demonstrated that the GAstV-1 strain is a contributing factor to the ongoing goose gout disease in China [77]. Furthermore, Pakbin et al. discovered that among 492 samples of raw cow milk in Qazvin, Iran, astrovirus predominated, being present in 66.86%, and that it was frequently co-present with norovirus GI [78].
The viral interference phenomenon has been recently described for murine AstVs in vivo, as Ingle et al. showed chronic murine AstV infection in highly immunocompromised mice could induce resistance against other Enterovirus (Ev) infections through IFN-lambda signaling in gut epithelial cells. Interestingly, the viral interference immunity could be successfully transferred to other in vivo murine models [79].
Propagation and in vivo investigation
Having an efficient cell culture system for AstVs has always been a challenge, hindering the study of the pathogenicity of the virus. It is assumed different cell lines might be needed based on the target strain; also, the presence of trypsin to get HAstVs cultivated seems crucial [12]. Although at least HAstV-VA1 could be successfully propagated without additional trypsin [63], a possible justification might be the trypsin-independent biology of VA1 to interact with the target cell.
Efforts to cultivate AstVs failed until 1981, when for the first time Lee and Kurtz could successfully grow HAstVs in Human Embryonic Kidney (HEK) cells, but significant progress was made in 1990 when MAstV1 was grown in colonic Carcinoma Cells (CaCo-2) straight from feces [80]. CaCo-2 cells simulate some important functional and constructional features of gastrointestinal epithelial cells, making them a gold cell line for investigating AstVs over the last decades, especially when the virus is pretreated with trypsin [12,81,82].
Even though the adenocarcinoma and hepatoma cell lines PLC/ PRF/5 are used to grow HAstV, CaCo-2 is mainly considered a permissive cell line to culture MAstV1 [63]. Tassoni et al. could also culture porcine AstV in Newborn Swine Kidney (NSK) and Porcine Kidney-15 (PK15) cell lines [83]. Furthermore, it has been suggested that some AstVs can be cultured in embryonated eggs, which may facilitate studying AstVs [28]. Canine AstV has been isolated in the Madin-Darby Canine Kidney (MDCK) by Martella et al. [84] as well as an emerging novel goose AstV that could be isolated in LMH cells in China, providing a possible growth system for AastVs [44].
The increasing importance of AstVs regarding CNS-associated problems requires new approaches to AstV propagation to evaluate human CNS-derived cell lines’ interaction with the virus. Recently, the first successful HAstV cultivation in CNS cells has been reported, in which MAstV9, the most common causer of human encephalitis [57,61,66,85], could be cultivated in both primary SK-N-SH and astrocyte cell lines [66]. These cell lines could bear the full life cycle of the virus, documented through a meaningful increase of viral gRNA, sgRNA, and intracellular viral capsid over time [66]. MAstV9 was also attempted to be propagated in other CNS-derived cell lines such as U87MG and SW-1088, where both virus entry and replication happened, but the virus could not complete its life cycle, possibly because of the expression of antiviral inhibitors like IL-6 and IL-8, resulting in the generation of abortive viral particles [66]. On the other hand, MAstV1 (HAstV4), another human encephalitis-related agent [86], could be incubated in the same CNS-derived cell lines that only yielded abortive viral particles similar to MAstV9 in U87MG and SW- 1088 [66].
Recently, Vu et al. successfully cultured two genotypes of the MLB clade in human liver-derived cell lines, Huh7 and A549, without adding exogenous trypsin and showed that the viruses induce persistent infection in vitro [87].
For those viruses that do not have a highly reliable cell culture system, the Human Intestinal Enteroid (HIE) model is often used. The culture contains multiple intestinal epithelial cell types and is obtained through expanding stem cells in 3D propagation [88,89]. Recently, Kolawole et al. could successfully culture all three clades of HAstV in HIE [88], providing new insights into the AstV-cell interaction investigation.
In all, different cell lines such as Cos-1, HEK293, CaCo-2, SK-CO-1, HT-29, MA-104, and Vero cell lines, as well as human-derived cell lines including HeLa, HEp-2, and A549, along with CNS-derived cell lines such as primary SK-N-SH and astrocytes, are considered to support HAstVs propagation [63], and different cultivation methods may be used based on the clade and/or the in vitro system [82,90]. Yet, the lack of a suitable animal model is considered a roadblock to broadening our knowledge of AstV biology, but recently, turkey has been used as an animal model for studying gastroenteric infections like AstVs [10]. However, in another study, attempts at incubation of HAstVs in murine models were unsuccessful since the virus could not cause any disease in the animal [45].
Occurrence in the food and environmental matrix
Food and environmental matrices have always been one of the main origins for enteric viruses like rotavirus, norovirus, and astrovirus to spread, as they have been frequently linked to food- or waterborne gastroenteritis [91]. It is estimated that the absence of the lipid envelope in the structure of these viruses makes them more stable in the environment and marine matrices [1]. Ready-to-eat foods and vegetables, as well as both treated and untreated water and drinking sources, may get contaminated with these viruses, providing the opportunity for the virus to reach its hosts (Table 1).
| Country | Year | Matrix | Prevalence | Outcome | Refs. |
|---|---|---|---|---|---|
| China | 2006 | water samples | 6% | The seasonal trend was hypothesized | [92] |
| China | 2013 | raw sewage | 40% | HAstV-1, 2, and 4 were characterized | [93] |
| Hungary | 2004 | sewage | 46% | HAstV-1 and 2 were characterized | [94] |
| - | - | raw sewage | 58.30% | - | - |
| - | - | treated sewage | 33.30% | - | - |
| Egypt | 2017-18 | sludge | 66.60% | - | [95] |
| - | - | river water | 25% | - | - |
| - | - | river sediment | 16.60% | - | - |
| Argentina | 2012 | dam water | 50% | The seasonal trend was hypothesized | [96] |
| - | - | beach water | 32% | - | - |
| Brazil | 2005 | raw and treated sewage | 17% | HAstV was detected | [97] |
| Uruguay | 2011-13 | sewage | 20% | MLB1 | [98] |
| - | - | - | 70% | HAstV-1, 2, 5 | |
| France | 2013-14 | water samples | 83% | HAstV-1, 2, 5, 6 were characterized | [99] |
| France | 2013-14 | river water and wastewater treatment plant | 36% | AstVs were the second most prevalent virus. | [100] |
| U.S. and Nepal | 2018 | wastewater | 73% | classic and novel HAstV, as well as animal AstVs, were characterized | [101] |
| Japan | 2007-8 | wastewater | 100% | novel HAstVs were predominant | [102] |
| Haiti | 2018 | wastewater | 50% | - | [103] |
| - | - | seawater | 20% | - | |
| Türkiye | 2017 | mussel samples | 63.46% | AstVs were the most prevalent virus. | [104] |
| Italy | 2018 | fecal samples from a raw seafood-associated outbreak | 73.30% | HAstV-1, 3, and VA1 were characterized | [105] |
| Italy | 2014-15 | shellfish samples | 28.70% | AstVs were the most prevalent virus. | [106] |
| - | - | - | - | HAstV-1 and 2 were characterized | |
| Italy | 2015-17 | mussels and clams | 20.80% | - | [107] |
| Tunisia | 2000-1 | shellfish samples | 61% | - | [108] |
| Japan | 2001-12 | fecal samples from oyster-related epidemics | 2.80% | - | [109] |
| Japan | 2002-6 | fecal samples from oyster-related epidemics | - | HAstV-4 was characterized | [110] |
| Vietnam | 2015-16 | pacific oysters and white hard clams | 12.40% | HAstV-1 was characterized | [111] |
| Vietnam | 2015-16 | pacific oysters and white hard clams | 12.40% | HAstV-1 was characterized | [111] |
| Vietnam | 2015-16 | pacific oysters and white hard clams | 12.40% | HAstV-1 was characterized | [111] |
Table 1. Summary of the most important surveillance of AstVs in food and environmental matrices over the last two decades.
AstVs in the environment
Water treatment methods are not always perfectly successful; therefore, sewage that often carries a high volume of microorganisms can still remain a potential infection source even after being treated, as enteric viruses are highly resistant. For example, AstVs have stayed infective at 60˚C for 10 minutes and could resist a pH of three. They also stay infectious in the underground water sources [91,94,112]. Some research, however, indicates the significant impact of methacrylate monolith chromatography implemented for the disinfection of waterborne viruses [113].
The presence of AstVs in natural sources has been a neglected area of research, and not many surveillance studies have been carried out. Clearly, environmental virology’s limitations and difficulties helped the area stay less studied. Viral detection in aquatic samples needs efficacious and cost-effective concentration methods; on the other hand, a high volume of bacterial contamination makes the work more complicated. Yet, researchers have tried to tackle these problems.
In 2006, a study group in China performed a one-year monthly monitoring of enteric viruses in three sewage treatment plants in Beijing and showed that in almost one-third of their samples, at least one of the main human enteric viruses is detectable, and HAstVs consisted of more than 6% of all samples. A seasonal trend was hypothesized as all astrovirus-positive samples were taken during either winter or spring [92]. A few years later, in 2013, another study in China showed a relatively higher rate of AstV infection in raw sewage samples, with nearly 40% of the specimens being positive for one of the genotypes HAstV1, 2, 4, and 5 [93]. In 2004, HAstV genotypes I and II were detected in 43% of sewage samples in Hungary. It was the first report on AstV in water sources in the country and showed how the virus circulates in the Hungarian population [94].
In 2017, a report from Egypt showed a high prevalence of enteric viruses, in particular AstV. They examined raw sewage, treated sewage, sludge, river water, and river sediment, and the figures for AstV were 58.3%, 33.3%, 66.6%, 25%, and 16.6%, respectively [95], comparable with similar studies in the country during 2007 and 2014 in which AstV was detected [114,115]. More recently, environmental monitoring in Egypt showed AstVs circulate in both wastewater and river water samples and highlighted the negative role of a local drain canal in the quality of the aquatic environment [116].
AstVs have also been detected in water samples in South America, where Masachessi et al. conducted a 2-year study to screen both dam and beach water plants, so they could detect HAstV at 50% and almost 32%, respectively, showing the circulation of the virus in recreational waters in the area [96]. These results were relatively higher than what was already reported from South America, in which almost 17% of raw and treated sewage samples were positive for HAstV [97]. Still in South America, there is a report on the detection of new genotypes of MLB1 in sewage samples in Uruguay [98]. A one-year wastewater treatment plant assessment in 2015 showed HAstVs, including HAstV-1, 2, 5, and 6, are circulating in Paris. These findings were consistent with the evaluation of clinical samples while at the same time highlighting the possible relationship between the presence of infection in humans and the environment [99]. Also, another survey at the same time, sampling the Saint-Royal in Paris, could detect different human enteric viruses, including AstVs [100].
In 2012, Hata et al. could detect diverse AstVs, including both classic and non-classic HAstVs, as well as other animal AstVs, in water samples, and interestingly, next-generation sequencing revealed possible recombination between human and feline AstVs in their positive samples [101]. This result is almost similar to another report that reveals how a wide range of HAstVs are circulating in the Japanese population, where the MLB clade was suggested to be dominated [102].
Kaas et al. reported that 50% and 20% of their total 11 wastewater and seawater samples, respectively, were contaminated with AstV [103].
AstVs in food
In the various preparation steps, AstVs can also contaminate food. Seafood, particularly shellfish, is thought to be the most common way of transmitting the virus to humans [1,12]. It is probably due to their nature that shellfish filter the water they live in, so they may absorb viruses [117].
A study in Istanbul reported that approximately two-thirds of their mussel samples were positive for AstV, showing a high rate of contamination in a city where mussels are the second most consumed seafood [104], highlighting the possibility of a local outbreak, similar to what has been reported from Bari, a port city in the south of Italy, where Lanave et al. could detect HAstV-1, 3, and VA1 in a local acute gastroenteritis outbreak associated with raw seafood consumption [105]. Another study from the Campania region in southern Italy carried out between January 2014 and December 2015 reported AstV as the most frequent virus in the shellfish samples, as more than 28% of the samples were AstV-contaminated [106]. Also, Fusco et al. monitored 289 shellfish specimens, including mussels (Mytilus galloprovincialis) and clams (Ruditapes philippinarum), and sampled different production places in the southwestern seashore of the Campania region of southern Italy between 2015 and 2017 and reported AstV as the second most prevalent virus (20.8%) following NoVGII [107]. The high rate of infection in shellfish samples has also been demonstrated in another coastal area, where almost two-thirds of the samples were positive for AstV in two sites in Tunisia [108], highlighting the possibility that in raw or poorly cooked food destined for human consumption, there may be astroviruses that go undetected.
In Japan, Iritani et al. studied 286 fecal samples from 88 oyster- related epidemics over an 11-year period and could detect AstVs in 5 epidemics [109]. Similarly, another study from Japan highlighted the detection of HAstV-4 in one gastroenteritis outbreak in Japan [110].
According to Suffredini et al., AstV was the second most common virus they found in Vietnam. They screened 121 shellfish samples from wet markets and stores, including Pacific oysters and white hard clams, and found that 12.5% of them were positive for AstV [111]. Various inactivation methods such as high-pressure processing (HPP) have been employed against food-borne viruses such as HAstV, Human Adeno Virus (HuAdV), Aichi Virus (AiV), Sapo Virus (SaV), and Ev. HPP operates by subjecting solid or liquid foods to isostatic pressure at temperatures ranging from 0 to 100 degrees celsius for a few seconds to over 20 minutes [118].
Conclusion
The growing number of living species and their tracks that are reported to be infected by AstVs, as well as frequent reports associating AstVs with water- and foodborne agents, make the Astroviridae family a serious concern to public health and a heavy financial burden to the food industry and animal husbandry. The novel developments in viral metagenomic practices have significantly increased our knowledge of the diversity and molecular virology of AstVs. There are, however, more questions to be addressed, particularly in the field of virus- host interaction. Recent progress to propagate AstVs in different cell lines has provided us with more insights into studying AstV biology, developing an effective medicine, or designing a safe vaccine against AstVs. The limited understanding of the fundamental replication cycle, transmission dynamics, and propagation of AstV demands focused attention. To this day, there are no specific antiviral drugs or vaccines available for treating or preventing AstV infections. Developing antiviral therapies is a significant challenge and requires a detailed understanding of the virus’s replication cycle and host interactions. To bridge these gaps, researchers should advocate for collaborative efforts, increased funding, and the development of innovative research tools. Additionally, integrating recent advances in viral metagenomics and cell line propagation into the discussion can provide a more cohesive narrative on how these developments contribute to advancing the field. The host range of AstV and their zoonotic potential (ability to transmit between animals and humans) remain unknown. Addressing existing gaps in our knowledge is crucial. Ultimately, due to the tremendous progress in metagenomic and NGS technologies, it is probable that numerous host AstVs will be identified and their sequences determined, which will reveal valuable information for us to be able to discover antivirals and vaccines and understand the propagation and evolution of AstV.
Declarations
Ethical approval
Not applicable.
Conflict of interests
The authors declare that they have no conflict of interest.
Funding
Not applicable.
Data availability statement
All data generated or analyzed during this study are available as supplementary material.
Acknowledgments
None.
References
- Wohlgemuth N, Honce R, Schultz-Cherry S (2019) Astrovirus evolution and emergence. Infect Genet Evol 69:30-37.
- Matsui SM, Kim J, Greenberg H, Young L, Smith L, et al. (1993) Cloning and characterization of human astrovirus immunoreactive epitopes. J Virol 67: 1712-1715.
- Donato C, Vijaykrishna D (2017) The broad host range and genetic diversity of mammalian and avian astroviruses. Viruses 9: 102.
- Capozza P, Martella V, Lanave G, Catella C, Diakoudi G, et al. (2021) An outbreak of neonatal enteritis in buffalo calves associated with astrovirus. J Vet Sci 22: e84.
- Kriston S, Willcocks M, Carter M, Cubitt W (1996) Seroprevalence of astrovirus types 1 and 6 in London, determined using recombinant virus antigen. Epidemiol Infect 117: 159-164.
- Hargest V, Sharp B, Livingston B, Cortez V, Schultz-Cherry S (2020) Astrovirus replication is inhibited by nitazoxanide in vitro and in vivo. J Virol 94:e01706- e017019.
- Snodgrass D, Gray E (1977) Detection and transmission of 30 nm virus particles (astroviruses) in faeces of lambs with diarrhoea. Arch Virol 55:287-291.
- Zhu Q, Li B, Sun D (2022) Bovine Astrovirus-A comprehensive review Viruses 14: 1217.
- Cortez V, Margolis E, Schultz-Cherry S (2019) Astrovirus and the microbiome. Curr Opin Virol 37: 10-15.
- Janowski AB (2021) Beyond the gastrointestinal tract: The emerging and diverse tissue tropisms of astroviruses. Viruses 13:732.
- Boujon CL, Koch MC, Seuberlich T (2017) The expanding field of mammalian astroviruses, opportunities and challenges in clinical virology. Adv Virus Res 99: 109-137.
- Bosch A, Pintó R M, Guix S (2014) Human astroviruses. Clin Microbiol Rev 27: 1048-1074.
- Meyer C T, Bauer I K, Antonio M, Adeyemi M and Saha D Oundo, et al. (2015) Prevalence of classic MLB-clade and VA-clade astroviruses in kenya and the gambia. Virol J 12:1-7.
- Wang Y, Li Y, Jin Y, Li D, Li X, et al. (2013) Recently identified novel human astroviruses in children with diarrhea, China. Emerg Infect Dis 19: 1333.
- Ahmed SF, Sebeny PJ, Klena JD, Pimentel G, Mansour A, et al. (2011) Novel astroviruses in children, Egypt. Emerging Infectious Diseases 17: 2391.
- El Taweel A, Kandeil A, Barakat A, Alfaroq Rabiee O, Kayali G, et al. (2020) Diversity of astroviruses circulating in humans, bats and wild birds in Egypt. Viruses 12: 485.
- Toh Y, Harper J, Dryden K A, Yeager M, Arias C F, et al. (2016) Crystal structure of the human astrovirus capsid protein. J Virol 90: 9008-9017.
- Rivera R, Nollens HH, Venn-Watson S, Guland FM, Wellehan Jr JF, et al. (2010). Characterization of phylogenetically diverse astroviruses of marine mammals. J Gen Virol 91: 166-173.
- Fuentes C, Bosch A, Pintó R M, Guix S (2012) Identification of human astrovirus genome-linked protein (VPg) essential for virus infectivity. Virol J 86: 10070-10078.
- Jonassen C M, Jonassen T, Grinde B (1998) A common RNA motif in the 3’end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. J Gen Virol 79: 715-718.
- Reuter G, Pankovics P, Boros Á (2011) Identification of a novel astrovirus in a domestic pig in Hungary. Arch Virol 156:125-128.
- Lan D, Ji W, Shan T, Cui Land Yang Z, et al. (2011) Molecular characterization of a porcine astrovirus strain in China . Arch Virol 156: 1869-1875.
- Luo Z, Roi S, Dastor M, Gallice E, Laurin MA, et al. (2011) Multiple novel and prevalent astroviruses in pigs. Veterinary Microbiology 149: 316-323.
- Babkin IV, Tikunov AY, Zhirakovskaia EV, Netesov SV, Tikunova NV (2012) High evolutionary rate of human astrovirus. Infect Genet Evol 12: 435-442.
- Lulla V, Firth AE (2020) A hidden gene in astroviruses encodes a viroporin. Nat Commun 11: 4070.
- Kang K, Icard AH, Linnemann E, Sellers H S and Mundt E (2012) Determination of the full length sequence of a chicken astrovirus suggests a different replication mechanism. Virus Genes, 44: 45-50.
- Fu Y, Pan M, Wang X, Xu Y, Xie X, et al. (2009). Complete sequence of a duck astrovirus associated with fatal hepatitis in ducklings. J Gen Virol 90: 1104-1108.
- Pantin-Jackwood M, Todd D, Koci MD (2012). Avian astroviruses. Astrovirus research 151-180.
- Mailliot J, Martin F (2018) Viral internal ribosomal entry sites: Four classes for one goal. Wiley Interdiscip Rev RNA 9: e1458.
- Karlsson E A, Small C T, Freiden P, Feeroz M, Matsen IV F A, et al. (2015) Non-human primates harbor diverse mammalian and avian astroviruses including those associated with human infections. PLoS Pathog 11: e1005225.
- Simmonds P (2006) Recombination and selection in the evolution of picornaviruses and other mammalian positive-stranded RNA viruses. J Virol 80: 11124-11140.
- Belliot G, Laveran H, Monroe S (1997) Detection and genetic differentiation of human astroviruses: Phylogenetic grouping varies by coding region. J Virol 142:1323-1334.
- Wolfaardt M, Kiulia NM, Mwenda JM, Taylor MB (2011) Evidence of a recombinant wild-type human astrovirus strain from a Kenyan child with gastroenteritis. J Clin Microbiol 49: 728-731.
- de Grazia S, Medici M, Pinto P, Moschidou P, Tummolo F, et al. (2012) Genetic heterogeneity and recombination in human type 2 astroviruses. J Clin Microbiol 50: 3760-3764.
- Martella V, Medici MC, Terio V, Catella C, Bozzo G, et al. (2013) Lineage diversification and recombination in type-4 human astroviruses. Infect Genet Evol 20: 330-335.
- Medici MC, Tummolo F, Martella V, Banyai K, Bonerba E, et al. (2015). Genetic heterogeneity and recombination in type-3 human astroviruses. Infect Genet Evol 32:156-160.
- Taylor MB, Walter J, Berke T, Cubitt WD, Mitchell, et al. (2001) Characterisation of a South African human astrovirus as type 8 by antigenic and genetic analyses. J Med Virol: 256-261.
- Beikpour F, Pellegrini F, Lanave G, Camero M, Catella C, et al. (2023) Exploring the astrovirome of shellfish matrices using nanopore sequencing. Vet Sci 10: 175.
- Smyth VJ (2017) A review of the strain diversity and pathogenesis of chicken astrovirus. Viruses 9: 29.
- Bon F, Fascia P, Dauvergne M, Tenenbaum D, Planson H, et al. (1999) Prevalence of group A rotavirus, human calicivirus, astrovirus, and adenovirus type 40 and 41 infections among children with acute gastroenteritis in Dijon, France. J Clin Microbiol 37: 3055-3058.
- Gonzales-Gustavson E, Timoneda N, Fernandez-Cassi X, Caballero A , Abril J, et al. (2017) Identification of sapovirus GV. 2, astrovirus VA3 and novel anelloviruses in serum from patients with acute hepatitis of unknown aetiology. PLoS One 12: e0185911.
- Hargest V, Davis AE, Tan S, Cortez V, Schultz-Cherry S (2021) Human astroviruses: A tale of two strains. Viruses 13: 376.
- Janowski AB, Dudley H, Wang D (2020) Antiviral activity of ribavirin and favipiravir against human astroviruses. J Clin Virol 123: 104247.
- Zhang X, Ren D, Li T, Zhou H, X, et al. (2018) An emerging novel goose astrovirus associated with gosling gout disease, China. Emerg Microbes Infect 7: 1-8.
- Cortez V, Sharp B, Yao J, Livingston B, Vogel P, et al. (2019) Characterizing a murine model for astrovirus using viral isolates from persistently infected immunocompromised mice. J Virol 93(13) 00223-00319.
- Ali H, Lulla A, Nicholson AS, Hankinson J, Wignall-Fleming E B, et al. (2023) Attenuation hotspots in neurotropic human astroviruses. PLoS Biol 21: e3001815.
- Ren D, Li T, Zhang X, Yao X, Gao W, et al. (2020) OASL triggered by novel goose astrovirus via ORF2 restricts its replication. J Virol 94: e01767-e01820.
- Bub T, Hargest V, Tan S, Smith M, Vazquez-Pagan A, et al. (2023) Astrovirus replication is dependent on induction of double-membrane vesicles through a PI3K-dependent, LC3-independent pathway. J Virol 97: e0102523.
- Omosigho PO, Izevbuwa OE, Rotimi EO, Olalekan J, Othoigbe MO (2022) Prevalence and risk factors of Astrovirus gastroenteritis in children in Offa Kwara State. North central Nigeria. Microbes Infect 3: 596-605
- Kapusinszky B, Minor P, Delwart E (2012) Nearly constant shedding of diverse enteric viruses by two healthy infants. J Clin Microbiol 50: 3427-3434.
- Conner ME, Blutt SE (2013) The gastrointestinal frontier: IgA and viruses. Front immunol 28:4:402.
- Mitchell DK, Matson DO, Cubitt WD, Jackson LJ, Willcocks MM, et al. (1999) Prevalence of antibodies to astrovirus types 1 and 3 in children and adolescents in Norfolk, Virginia. Pediatr Infect Dis J, 18: 249-254.
- Selimovic-Hamza S, Boujon CL, Hilbe M, Oevermann A, Seuberlich T (2017) Frequency and pathological phenotype of bovine astrovirus CH13/NeuroS1 infection in neurologically-diseased cattle: Towards assessment of causality. Viruses 9: 12.
- Moser LA, Carter M, Schultz-Cherry S (2007) Astrovirus increases epithelial barrier permeability independently of viral replication. J Virol 81: 11937-11945.
- Koci MD, Moser LA, Kelley LA, Larsen D, Brown CC, et al. (2003). Astrovirus induces diarrhea in the absence of inflammation and cell death. J Virol 77:11798-11808.
- Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, et al. (2010) Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis 16: 918-925.
- Brown JR, Morfopoulou S, Hubb J, Emmett WA, Ip W, et al. (2015) Astrovirus VA1/HMO-C: An increasingly recognized neurotropic pathogen in immunocompromised patients. Clin Infect Dis 60:881-888.
- Sato M, Kuroda M, Kasai M, Matsui H, Fukuyama T, et al. (2016) Acute encephalopathy in an immunocompromised boy with astrovirus-MLB1 infection detected by next generation sequencing. J Clin Virol 78: 66-70.
- Cordey S, Vu DL, Schibler M, L Huillier AG, Brito F, et al. (2016) Astrovirus MLB2, a new gastroenteric virus associated with meningitis and disseminated infection. Emerg Infect Dis 22: 846-853.
- Naccache SN, Peggs KS, Mattes FM, Phadke R, Garson JA, et al. (2015) Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin Infect Dis 60: 919-923.
- Lum SH, Turner A, Guiver M, Bonney D, Martland T, et al. (2016) An emerging opportunistic infection: Fatal astrovirus (VA 1/HMO-C) encephalitis in a pediatric stem cell transplant recipient. Transplant Infectious Disease, 18: 960-964.
- Wunderli W, Meerbach A, Guengoer T, Berger C, Greiner O, et al. (2011) Astrovirus infection in hospitalized infants with severe combined immunodeficiency after allogeneic hematopoietic stem cell transplantation. PloS One 6: e27483.
- Janowski A B, Bauer IK, Holtz LR, Wang D (2017) Propagation of astrovirus VA1, a neurotropic human astrovirus, in cell culture. J Virol 91: e00740-17.
- Baez-Navarro C, Quevedo IR, López S, Arias CF, Isa P (2022) The association of human astrovirus with extracellular vesicles facilitates cell infection and protects the virus from neutralizing antibodies. J Virol 96: e0084822.
- Burbelo PD, Ching KH, Esper F, Iadarola MJ, Delwart E, et al. (2011) Serological studies confirm the novel astrovirus HMOAstV-C as a highly prevalent human infectious agent. PloS One 6: e22576.
- Janowski AB, Klein RS, Wang D (2019) Differential in vitro infection of neural cells by astroviruses. mBio 10: e01455-19.
- Li ML, Chen BS, Shih SR (2020) Editorial: Viral encephalitis. Front Microbiol11, 599257.
- Koukou G, Niendorf S, Hornei B, Schlump JU and Jenke AC, et al. (2019) Human astrovirus infection associated with encephalitis in an immunocompetent child: A case report. J Med Case Rep, 13(1), 341.
- Wu W, Qiu S, Huang H, Xu R, Bao E, et al. (2021) Immune-related gene expression in the kidneys and spleens of goslings infected with goose nephritic astrovirus. Poult Sci 100:100990
- Wu W, Xu R, Lv Y, Bao E (2020) Goose astrovirus infection affects uric acid production and excretion in goslings. Poult Sci 99: 1967-1974.
- Zaccaria G, Lorusso A, Hierweger MM, Malatesta D, Defourny SV, et al. (2020) Detection of Astrovirus in a cow with neurological signs by nanopore technology, Italy. Viruses 12: 530.
- Wu Q, Li J, Wang W, Zhou J, Wang D, et al. (2021) Next-generation sequencing reveals four novel Viruses associated with calf diarrhea. Viruses 13: 1907.
- Kauer RV, Koch MC, Hierweger MM, Werder S, Boujon CL, et al. (2019) Discovery of novel astrovirus genotype species in small ruminants. Peer J 7: e7338.
- Liu C, Sun M, Liao M (2022) A review of emerging goose astrovirus causing gout. Biomed Res Int 1635373.
- Lee SY, Kim JH, Kim YJ, Kim YS, Roh, SG, et al. (2021) Astrovirus infection in cattle with nonsuppurative meningoencephalitis in South Korea. Viruses 13: 1941.
- Adebiyi AI, Mcilwaine K, Oluwayelu DO, Smyth VJ (2021) Detection and characterization of chicken astrovirus associated with hatchery disease in commercial day-old turkeys in southwestern Nigeria. Arch Virol 166:1607-1614.
- Wang AP, Zhang S, Xie J, Gu LL, Wu S, et al. (2021) Isolation and characterization of a goose astrovirus 1 strain causing fatal gout in goslings, China. Poult Sci 100: 101432.
- Pakbin B, Rossen JWA, Brück WM, Montazeri N, Allahyari S, et al. (2022) Prevalence of foodborne and zoonotic viral pathogens in raw cow milk samples. FEMS Microbiol Lett 369: fnac108.
[Crossref] [Google Scholar] [PubMed]
- Ingle H, Lee S, Ai T, Orvedahl A, Rodgers R, et al. (2019) Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon. Nature Microbiol 4(7), 1120–1128.
[Crossref] [Google Scholar] [PubMed]
- Willcocks M, Carter M, Laidler F, Madeley C (1990) Growth and characterisation of human faecal astrovirus in a continuous cell line. Arch Virol 113: 73-81.
[Crossref] [Google Scholar] [PubMed]
- Darling NJ, Mobbs CL, Hau GAL, Freer MA, Przyborski S (2020) Bioengineering novel in vitro co-culture models that represent the human intestinal mucosa with improved Caco-2 structure and barrier function. Front bioeng 8: 992.
[Crossref] [Google Scholar] [PubMed]
- Janowski AB, Wang D (2019) Infection and propagation of astrovirus VA1 in cell culture. Current Prot Microbiol 52: e73.
[Crossref] [Google Scholar] [PubMed]
- Tassoni L, Zamperin G, Schiavon E, Vendramin V, Cavicchio L, et al. (2019) First whole genome characterization of porcine astrovirus detected in swine faeces in Italy. Vet Ital 55: 221-229.
[Crossref] [Google Scholar] [PubMed]
- Martella V, Moschidou P, Lorusso E, Mari V, Camero M, et al. (2011) Detection and characterization of canine astroviruses. J General Virol 92: 1880-1887.
[Crossref] [Google Scholar] [PubMed]
- Wildi N, Seuberlich T (2021) Neurotropic astroviruses in animals. Virus 13: 1201.
[Crossref] [Google Scholar] [PubMed]
- Frémond M-L, Perot P, Muth E, Cros G, Dumarest M, et al. (2015) Next-generation sequencing for diagnosis and tailored therapy: A case report of astrovirus-associated progressive encephalitis. J Pediatric Infect Dis Soc 4: e53-e57.
[Crossref] [Google Scholar] [PubMed]
- Vu D-L, Bosch A, Pintó RM, Ribes E, Guix S (2019) Human astrovirus MLB replication in vitro: Persistence in extraintestinal cell lines. J Virol 93: e00557-005519.
[Crossref] [Google Scholar] [PubMed]
- Kolawole AO, Mirabelli C, Hill DR, Svoboda SA, Janowski AB, et al. (2019) Astrovirus replication in human intestinal enteroids reveals multi-cellular tropism and an intricate host innate immune landscape. PLoS Pathog 15: e1008057.
[Crossref] [Google Scholar] [PubMed]
- Saxena K, Blutt SE, Ettayebi K, Zeng X-L, Broughman JR, et al. (2016) Human intestinal enteroids: A new model to study human rotavirus infection, host restriction, and pathophysiology. J Virol 90: 43-56.
[Crossref] [Google Scholar] [PubMed]
- Owusu IA, Mirabelli C, Kolawole AO (2020) Intestinal enteroid culture for human astroviruses. Bio-Protocol 10: e3687-e3687.
[Crossref] [Google Scholar] [PubMed]
- Espinosa AC, Mazari-Hiriart M, Espinosa R, Maruri-Avidal L, Méndez E, (2008) Infectivity and genome persistence of rotavirus and astrovirus in groundwater and surface water. Water Res 42: 2618-2628.
[Crossref] [Google Scholar] [PubMed]
- He X, Cheng L, Zhan, D, Xie X, Wang D, et al. (2011) One-year monthly survey of rotavirus, astrovirus and norovirus in three sewage treatment plants in Beijing, China and associated health risk assessment. Water Sci Technol 63: 191-198.
[Crossref] [Google Scholar] [PubMed]
- Zhou N, Lin X, Wang S, Wang H, Li W, et al. (2014) Environmental surveillance for human astrovirus in Shandong Province, China in 2013. Sci Rep 4: 1-5.
[Crossref] [Google Scholar] [PubMed]
- Meleg E, Jakab F, Kocsis B, Banyai K, Melegh B, et al. (2006) Human astroviruses in raw sewage samples in Hungary. J App Microbiol 101: 1123-1129.
[Crossref] [Google Scholar] [PubMed]
- Shaheen MNF, El-Daim SEA, Ahmed NI, Elmahdy ME (2018) Molecular detection of three gastroenteritis viruses in an urban sewage treatment plant and river water in Egypt. Egy J Aqua Biol Fish 22: 615-627.
- Masachessi G, Pisano MB, Prez VE, Martínez L, Michelena J, et al. (2018) Enteric viruses in surface waters from Argentina: Molecular and viable-virus detection. App Environ Microbiol 84: e02327-e02317.
[Crossref] [Google Scholar] [PubMed]
- Guimarães FR, Ferreira FFM, Vieira CB, Fumian TM, Shubo T, et al. (2008) Molecular detection of human astrovirus in an urban sewage treatment plant in Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz 103: 819-823.
[Crossref] [Google Scholar] [PubMed]
- Lizasoain A, Tort L, Garcia M, Gomez M, Leite J, et al. (2015) Environmental assessment reveals the presence of MLB-1 human astrovirus in Uruguay. J Appl Microbiol 119: 859-867.
[Crossref] [Google Scholar] [PubMed]
- Prevost B, Lucas FS, Ambert-Balay K, Pothier P, Moulin L, et al. (2015) Deciphering the diversities of astroviruses and noroviruses in wastewater treatment plant effluents by a high-throughput sequencing method. Appl Environ Microbiol 81: 7215-7222.
[Crossref] [Google Scholar] [PubMed]
- Prevost B, Lucas F, Goncalves A, Richard F, Moulin L, et al. (2015) Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ Inter 79: 42-50.
[Crossref] [Google Scholar] [PubMed]
- Hata A, Kitajima M, Haramoto E, Lee S, Ihara M, et al. (2018) Next-generation amplicon sequencing identifies genetically diverse human astroviruses, including recombinant strains, in environmental waters. Sci Rep 8: 1-9.
- Hata A, Katayama H, Kitajima M, Furumai H (2015) Wastewater analysis indicates that genetically diverse astroviruses, including strains belonging to novel clades MLB and VA, are circulating within Japanese populations. Appl Environ Microbiol 81: 4932-4939.
[Crossref] [Google Scholar] [PubMed]
- Kaas L, Ogorzaly L, Lecellier G, Berteaux-Lecellier V, Cauchie H-M, et al. (2019) Detection of human enteric viruses in French Polynesian wastewaters, environmental waters and giant clams. Food and Environ Virol 11: 52-64.
[Crossref] [Google Scholar] [PubMed]
- Ghalyoun F, Alcay AÜ (2018) Investigation of rotavirus, adenovirus and astrovirus in mussels and shrimps using multiplex real-time PCR. Kafkas Univ Vet Fak Derg 24.
- Lanave G, Loconsole D, Centrone F, Catella C, Capozza P, et al. (2021) Astrovirus VA1 in patients with acute gastroenteritis. Transbound Emerg Dis 69: 864-869.
[Crossref] [Google Scholar] [PubMed]
- Fusco G, Di Bartolo I, Cioffi B, Ianiro G, Palermo P, et al. (2017) Prevalence of foodborne viruses in mussels in Southern Italy. Food Environ Virol 9: 187-194.
[Crossref] [Google Scholar] [PubMed]
- Fusco G, Anastasio A, Kingsley DH, Amoroso MG, Pepe T, et al. (2019) Detection of hepatitis A virus and other enteric viruses in shellfish collected in the Gulf of Naples, Italy. Inter J Environ Res Public Health 16: 2588.
[Crossref] [Google Scholar] [PubMed]
- Elamri D, Aouni M, Parnaudeau S, Le Guyader F (2006) Detection of human enteric viruses in shellfish collected in Tunisia. Letters App Microbiol 43: 399-404.
[Crossref] [Google Scholar] [PubMed]
- Iritani N, Kaida A, Abe N, Kubo H, Sekiguchi J-I, et al. (2014) Detection and genetic characterization of human enteric viruses in oyster-associated gastroenteritis outbreaks between 2001 and 2012 in Osaka City, Japan. J Med Virol 86: 2019-2025.
[Crossref] [Google Scholar] [PubMed]
- Nakagawa-Okamoto R, Arita-Nishida T, Toda S, Kato H, Iwata H, et al. (2009) Detection of multiple sapovirus genotypes and genogroups in oyster-associated outbreaks. Japan J Infect Dis 62: 63-66.
[Crossref] [Google Scholar] [PubMed]
- Suffredini E, Le Q, Di Pasquale S, Pham T, Vicenza T, et al. (2020) Occurrence and molecular characterization of enteric viruses in bivalve shellfish marketed in Vietnam. Food Control 108:8 106828.
- Abad FX, Villena C, Guix S, Caballero S, Pintó RM, et al. (2001) Potential role of fomites in the vehicular transmission of human astroviruses. Appl Environ Microbiol 67: 3904-3907.
[Crossref] [Google Scholar] [PubMed]
- Racki N, Kramberger P, Steyer A, Gaspersic J and Strancar A, et al. (2015) Methacrylate monolith chromatography as a tool for waterborne virus removal. J Chromatogr A 1381:118-124.
- El-Senousy WM, El-Gamal MS, Mousa A, El-Hawary S, Fathi M N (2014) Prevalence of Noroviruses among detected enteric viruses in Egyptian aquatic environment. World Appl Sci J 32: 2186-2205.
- Morsy El-Senousy W, Guix S, Abid I, Pintó RM, Bosch A (2007) Removal of astrovirus from water and sewage treatment plants, evaluated by a competitive reverse transcription-PCR. Appl Environ Microbiol 73:164-167.
- Shaheen MN, Elmahdy EM (2019) Environmental monitoring of astrovirus and norovirus in the Rosetta branch of the River Nile and the El-Rahawy drain, Egypt. Water Supply 19: 1381-1387. .
- Gabrieli R, Macaluso A, Lanni L, Saccares S, Di Giamberardino F, et al. (2007) Enteric viruses in molluscan shellfish. Microbiologica-Quarterly J Microbiol 30: 471-476.
- Govaris A, Pexara A (2021) Inactivation of foodborne viruses by High-Pressure Processing (HPP). Foods 10: 215.
Citation: Beikpour F, Fathabadi F, Shahrivar RY, Sadeghi M (2023) Astrovirus: A Challenge for Humans, Animals and the Environment. J Infect Dis Ther 11: 575 DOI: 10.4172/2332-0877.575
Copyright: © 2023 Farzad B, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 221
- [From(publication date): 0-0 - Nov 23, 2024]
- Breakdown by view type
- HTML page views: 188
- PDF downloads: 33