Research Article Open Access
Association of Brain-Derived Neurotrophic Factor (BDNF) Gene Snps G196A and C270T with Alzheimer's Disease: A Meta-Analysis
Shovit R and Praveen KS*Central University of Jharkhand, Brambe, Ranchi, Jharkhand, India
- *Corresponding Author:
- Praveen KS
Centre for Life Sciences
Central University of Jharkhand, Brambe
Ranchi, Jharkhand, 835205, India
Tel: +91-8229813927
E-mail: pksharma.cuj@gmail.com
Received date: April 16, 2017; Accepted date: May 03, 2017; Published date: May 10, 2017
Citation: Shovit R, Praveen KS (2017) Association of Brain-Derived Neurotrophic Factor (BDNF) Gene Snps G196A and C270T with Alzheimer’s Disease: A Meta-Analysis. J Alzheimers Dis Parkinsonism 7:323. doi:10.4172/2161-0460.1000323
Copyright: © 2017 Shovit R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Alzheimers Disease & Parkinsonism
Abstract
Objective: To evaluate the association between diagnosis of Alzheimer’s Disease (AD) and brain-derived neurotrophic factor (BDNF) gene polymorphisms 196 G/A and 270 C/T. Methods: The authors conducted a meta-analysis along with Trial Sequential Analysis (TSA) of BDNF 196 G/A and 270 C/T polymorphisms and AD in all the available case-control studies on the topic published from 2002 to 2016. Results: Results showed no significant association between BDNF 196 G/A and 270 C/T polymorphisms and AD in overall and ethnicity specific studies. Conclusion: The present comprehensive meta-analysis suggested that further studies focusing on larger sample-size multi-ethnicity studies with homogeneous AD patients and well-matched controls are needed for future study to confirm the results of the study.
Keywords
BDNF; G196A; C270T; Alzheimer’s disease; Polymorphism; Meta-analysis
Introduction
Alzheimer's disease (AD) is a common age-associated neurodegenerative disorder, clinically characterized by progressive memory disorder and decline in cognitive function, which typically begins with dementia [1]. The number of people with AD worldwide in 2006 was estimated at 26.6 million and is predicted to nearly quadruple by 2050 [2].
The key pathological changes associated with AD brain tissue are the accumulation of intracellular neurofibrillary tangles (NFTs) and abnormally aggregated ‘reactive’ proteins like β amyloid (Aβ) plaques and tau [3]. Several elements, such as senile plaques, neurofibrillary tangles (NFTs), abnormally aggregated ‘reactive’ proteins like β amyloid (Aβ) and tau, brain inflammation and exposure to aluminum has already shown the development of AD [4]. Brain derived neurotrophic factor (BDNF) gene is supposed to be one of the important genes, playing a significant role in AD progression [5,6]. However, as a complex disorder, the neuropathological etiology of AD mentioned above are not due to the gene itself, but are also supposed to be associated with the combined interaction between genes and environmental factors.
BDNF, a member of the neurotrophic factor family, is encoded by a gene located on chromosome 11p13 [7]. The basal physiological role of BDNF is to play an important role in the growth, development, differentiation and regeneration of various types of neurons in the central nervous system [8]. Earlier studies have already shown reduced mRNA expression of BDNF in the hippocampus of AD patients [9]. Moreover, another study has also demonstrated that BDNF and its receptor level, tyrosine receptor kinase B level, were decreased in the frontal cortex and hippocampus of AD patients [10], suggesting the possible important role of BDNF in AD pathogenesis. The possible mechanism underlying this dysregulated BDNF expression may be due to reduced transport of BDNF from the Golgi region to appropriate secretory granules in neurons, resulting in the onset of AD.
Several single nucleotide polymorphisms (SNPs) in BDNF gene, such as rs11030104, rs16917204, rs7103411, rs6265 and rs2030324 are already been reported for their association with AD. However, only rs6265 and rs2030324 have been widely studied, with no linkage disequilibrium between them. The association between AD and the G196A and C270T polymorphism in the BDNF gene are already been investigated in several individual case-control studies, producing inconsistent results, which may be the result of limited power, different ethnic backgrounds or the different processes and status in AD patients. Since, individual studies have relatively small power to confirm this association and meta-analysis can strengthen the power by combining data from different individual studies and different ethnicities also, resulting in a more comprehensive conclusion [11- 13]. Moreover, we also conducted Trial Sequential Analysis (TSA) of all the published case–control studies for validating the result of meta-analysis.
Materials and Methods
Identification of eligible studies
Preferred Reporting Items for Systematic Reviews and Meta- Analyses (PRISMA) 2009 guidelines for systematic review and metaanalysis and the Cochrane Collaboration definition of both terms were followed for this work [14,15]. Literature search was carried out within PubMed (Medline), EMBASE and Science Direct database up to July, 2016, using the keywords-BDNF, gene, patient, polymorphism and Alzheimer’s disease. Then, potentially relevant publications and studies were retrieved by examining their titles and abstracts and matching the eligible criteria.
Inclusion and exclusion criteria
To facilitate the proper interpretation of results and to minimize heterogeneity, all eligible studies had to fulfill the following inclusion criteria like evaluation of BDNF gene 196 G>A and 270 C>T with AD risk; use of case control or cohort studies; recruitment of pathologically confirmed AD patients and healthy controls; and availability of genotypic frequency both in case and control. Moreover, when the case control study was included by more than one article using the same case series, then we selected the study that included the largest number of individuals.
The major reasons for exclusion of studies were overlapping data, case only studies; review articles, family-based studies and animal studies.
Data extraction and quality assessment
For each meta-analysis, the methodological quality assessment and data extraction were independently abstracted in duplicate using a standard protocol. Data accuracy was ensured using data collection form according to the inclusion and exclusion criteria listed above. In case of discrepancy on any item of the data collected from the retrieved studies, the problem was fully discussed to reach a consensus. Data extracted from each study included the name of first author, year of publication, ethnicity, number of cases and controls, types of study and genotyping methods and frequencies of the case and control.
Meta-analysis methods
The meta-analysis examined the overall association and ethnicity specific association of the A and T allele with the risk of AD relative to the G and C allele respectively, the contrast of homozygotes AA vs. GG; TT vs. CC, the contrast of heterozygotes AG vs. GG; TC vs. CC, the recessive model for the A allele: contrast AA vs. (AG+GG); TT vs. (TC+CC), and the dominant model for the A allele: contrast (AA+AG) vs. GG; (TT+TC vs. CC). All associations were indicated as odds ratios (ORs) with the corresponding 95% confidence interval (CI). A pooled OR was then estimated based on individual ORs.
Statistical Analysis
Hardy Weinberg equilibrium (HWE) was examined in the control subjects using a goodness of fit chi-square test for each study, Odds ratio (OR) with corresponding 95% confidence intervals (CI) was used to evaluate the association between the BDNF 196 G>A gene polymorphism and BDNF 270 C>T with AD risk separately. Heterogeneity was assessed by Chi-square based Q-Test [16]. If heterogeneity existed, then random effects model was used to calculate the overall pooled OR value [17]; otherwise, the fixed effect model was used [18]. Moreover, I2 statistics was used to quantify interstudy variability. It ranges between 0% and 100%, where a value of 0% indicates no observed heterogeneity, and larger values indicate an increasing degree of heterogeneity [19]. The HWE was examined in the control subjects using a goodness-of-fit chisquare test for each study. Begg’s funnel plots and Egger’s regression test were undertaken to evaluate the potential publication bias [20]. P-value less than 0.05 were judged significant. Publication bias was assessed by visual inspection of funnel plots in which the standard error of log (OR) of each study was plotted against its log (OR). An asymmetric plot suggests a possible publication bias. Funnel plot asymmetry was also assessed by the Egger’s linear regression test. The significance of the intercept was determined by the t-test (p<0.05 was considered representative of statistically significant publication bias) [21]. All the data analysis was performed using comprehensive meta-analysis (CMA) V2 software (Biostat, USA).
Trial sequential analysis (TSA)
According to Cochrane Handbook for systematic reviews of interventions, meta-analyses and systematic reviews are considered the best available evidence if all eligible trials are included. However, the best available evidence might not always be equal to strong sufficient evidence. It is well known that meta-analysis may result in increased risk of random errors when series of sparse data are analyzed and in reduplicative significance testing when new trials are updated in cumulative meta-analysis. Therefore, keeping mind on the issues raised above, we applied the TSA to increase the robustness of current conclusions by minimizing the random errors [22-24]. The methods of using TSA were based on the ‘User manual for Trial Sequential Analysis (TSA)’.
In the study, TSA was used to control the risk of random error by calculating the required information size and an adjusted threshold for statistical significance to make a robust conclusion [22,23,25]. The required information size was calculated with the assumption of a plausible relative risk of 20% with low risk bias, and the overall 5% risk for a type I error (α), 20% risk for a type II error (β) were adopted [26]. Based on required information size and risk for type I and type II errors, TSA monitoring boundaries were built. When the cumulative Z-curve crosses, the TSA monitoring boundary before the required information size is reached, a sufficient level of evidence might have been reached and further trials are not necessary. Otherwise, evidence to reach a conclusion is insufficient and further trials are necessary [27]. The software Trial Sequential Analysis Viewer (version 0.9.5.5 Beta) was used for the study and 95% CIs was adjusted for sparse data or repetitive testing, described as the TSA-adjusted 95% CIs.
Results
Eligible studies included in the meta-analysis
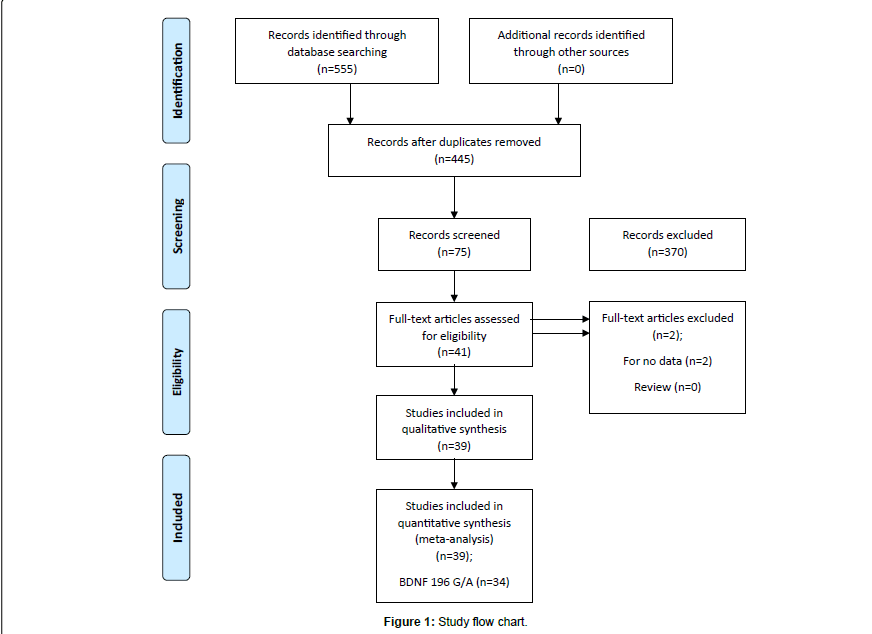
The literature review identified a total of 39 studies eligible for inclusion in our analysis as described in flow chart (Figure 1). Based on our preliminary search criteria, a total of 555 studies were identified in PubMed (Medline), EMBASE and Science Direct using the keywords- BDNF, gene, patient, polymorphism, Alzheimer disease and their combination. After careful review, finally, 39 potential studies were included. According to our inclusion criteria, four studies have not been included for estimating OR and 95% CI because they did not reported genotypic frequency of patients and healthy controls [28,29]. Finally, 39 eligible studies involving 9409 cases and 9522 controls were enrolled in the pooled analyses.
The populations came from 14 different countries, including Brazil, China, Colombia, Croatia, England, Finland, France, Germany, Hungary, India, Italy, Japan, Spain and USA. Detailed characteristics of all eligible studies included in meta-analysis are reported in Table 1. One overall study was conducted on BDNF 196GA polymorphism and 3 ethnicity specific studies were conducted, that includes 8 studies on American populations [30-36], 11 studies on Asian populations [37- 47] and 12 studies on European populations [35,48-58]. Moreover, one overall study was also conducted on BDNF 270CT polymorphism and 3 ethnicity specific studies were also possible, that includes 5 studies on American populations [30,31,33,34,59], 7 studies on Asian populations [37,39,41,42,44,46,60] and 7 studies on European populations [48,49,52,56,58,61,62]. Tables 2 and 3 reports genotypic distribution of G196A and C270T polymorphism of BDNF gene from each study. All studies observed HWE.
| Authors | Country | Ethnicity | Cases (AD) | Control (HC) | Genotyping | SNP | Association |
|---|---|---|---|---|---|---|---|
| Kunugi et al. [60] | Japan | Asian | 170 | 498 | PCR | C>T | Yes |
| Riemenschneider et al. [62] | Germany | European | 210 | 188 | PCR | C>T | Yes |
| Ventriglia et al. [57] | Italy | European | 130 | 111 | PCR | G>A | Yes |
| Bagnoli et al. [48] | Italy | European | 128 128 | 97 97 | PCR | G>A C>T | No |
| Combarros et al. [51] | Spain | European | 237 | 218 | PCR | G>A | No |
| Nacmias et al. [54] | Italy | European | 83 | 97 | PCR-RFLP | G>A | No |
| Nishimura et al. [59] | Brazil | American | 188 | 188 | PCR | C>T | No |
| Bian et al. [38] | China | Asian | 203 | 239 | PCR | G>A | No |
| Bodner et al. [30] | USA | American | 256 256 | 194 194 | ABI Prism 7900HT instrument | G>A C>T | No |
| Desai et al. [31] | USA | American | 995 719 | 671 523 | Pyrosequencing | G>A C>T | No |
| Desai et al. [31] | USA | African | 64 58 | 45 42 | Pyrosequencing | G>A C>T | No |
| Lee et al. [34] | USA | American | 95 106 | 70 73 | PCR-RFLP | G>A C>T | No |
| Li et al. [35] | England | European | 359 | 396 | Allele-specific Real Time PCR | G>A | No |
| Li et al. [35] | USA | American | 188 | 361 | Allele-specific Real Time PCR | G>A | No |
| Li et al. [35] | USA | American | 388 | 349 | Allele-specific Real Time PCR | G>A | No |
| Matsushita et al. [41] | Japan | Asian | 487 487 | 471 471 | PCR | G>A C>T | Yes |
| Nishimura et al. [42] | Japan | Asian | 172 172 | 275 275 | PCR-RFLP | G>A C>T | Yes |
| Olin et al. [69] | USA | Mixed (Non-Hispanic American) | 212 | 202 | PCR | C>T | Yes |
| Vepsalainen et al. [58] | Finland | European | 375 375 | 460 460 | PCR | G>A C>T | No |
| Akatsu et al. [37] | Japan | Asian | 95 95 | 108 108 | PCR-RFLP | G>A C>T | No |
| Forero et al. [67] | Colombia | Mixed | 101 | 168 | PCR | G>A | Yes |
| Saarela et al. [56] | Finland | European | 97 97 | 101 101 | PCR | G>A C>T | No |
| Tsai et al. [46] | China | Asian | 175 175 | 189 189 | PCR | G>A C>T | Yes |
| Zhang et al. [68] | USA | Mixed (European-American) | 295 295 | 250 250 | PCR-RFLP | G>A C>T | No |
| He et al. [40] | China | Asian | 513 | 575 | Allele-specific PCR | G>A | No |
| Huang et al. [33] | USA | American | 220 220 | 128 128 | PCR | G>A C>T | No |
| Cozza et al. [52] | Italy | European | 251 251 | 97 97 | PCR | G>A C>T | No |
| Qian et al. [44] | China | Asian | 105 105 | 105 105 | PCR | G>A C>T | No |
| Yu et al. [47] | China | Asian | 99 | 99 | PCR | G>A | No |
| Feher et al. [53] | Hungary | European | 160 | 164 | PCR | G>A | Yes |
| Qi et al. [43] | China | Asian | 80 | 86 | PCR-RFLP | G>A | No |
| Fukumoto et al. [39] | Japan | Asian | 657 657 | 525 525 | Taqman PCR Assay | G>A C>T | Yes |
| Cousin et al. [61] | France | European | 425 | 470 | PCR | C>T | No |
| Pivac et al. [55] | Croatia | European | 211 | 402 | Taqman based Allele-specific PCR Assay | G>A | No |
| Borroni et al. [50] | Italy | European | 234 | 162 | PCR | G>A | Yes |
| Boiocchi et al. [49] | Italy | European | 191 192 | 408 384 | PCR-RFLP | G>A C>T | Yes |
| Sonali et al. [45] | India | Asian | 57 | 63 | PCR-RFLP | G>A | No |
| Vieira et al. [36] | Brazil | American | 269 | 114 | Taqman Real Time PCR Assay | G>A | No |
| Gomar et al. [32] | USA | American | 222 | 175 | Illumina Human610-Quad BeadChip | G>A | Yes |
Table 1: Main characteristics of all studies included in meta-analysis.
| First author | Cases (AD) | Controls (HC) | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Val66Met/G196A Genotype rs6265 |
Minor Allele Frequency (MAF) |
Val66Met/G196A Genotype rs6265 |
Minor Allele Frequency (MAF) |
||||||
| GG | GA | AA | GG | GA | AA | P-value | |||
| Ventriglia et al. [57] | 85 | 33 | 12 | 0.219 | 54 | 48 | 9 | 0.297 | 0.712 |
| Bagnoli et al. [48] | 62 | 60 | 6 | 0.281 | 55 | 38 | 4 | 0.237 | 0.414 |
| Combarros et al. [51] | 149 | 78 | 10 | 0.206 | 143 | 67 | 8 | 0.190 | 0.965 |
| Nacmias et al. [54] | 48 | 29 | 6 | 0.246 | 55 | 38 | 4 | 0.237 | 0.414 |
| Bian et al. [38] | 49 | 113 | 41 | 0.480 | 73 | 115 | 51 | 0.453 | 0.649 |
| Bodner et al. [30] | 163 | 85 | 8 | 0.197 | 126 | 62 | 6 | 0.190 | 0.623 |
| Desai et al. [31] | 662 | 299 | 34 | 0.184 | 456 | 197 | 18 | 0.173 | 0.549 |
| Desai et al. [31] | 59 | 5 | 0 | 0.039 | 42 | 3 | 0 | 0.033 | 0.817 |
| Lee et al. [34] | 45 | 47 | 3 | 0.278 | 32 | 30 | 8 | 0.328 | 0.810 |
| Li et al. [35] | 239 | 105 | 15 | 0.188 | 269 | 114 | 13 | 0.176 | 0.828 |
| Li et al. [35] | 109 | 73 | 6 | 0.226 | 235 | 110 | 16 | 0.196 | 0.497 |
| Li et al. [35] | 251 | 126 | 11 | 0.190 | 237 | 105 | 7 | 0.170 | 0.234 |
| Matsushita et al. [41] | 171 | 247 | 69 | 0.395 | 150 | 223 | 98 | 0.444 | 0.368 |
| Nishimura et al. [42] | 61 | 85 | 26 | 0.398 | 88 | 140 | 47 | 0.425 | 0.493 |
| Vepsalainen et al. [58] | 280 | 87 | 8 | 0.137 | 342 | 109 | 9 | 0.138 | 0.926 |
| Akatsu et al. [37] | 25 | 58 | 12 | 0.431 | 35 | 53 | 20 | 0.430 | 0.993 |
| Forero et al. [67] | 72 | 27 | 2 | 0.153 | 131 | 34 | 3 | 0.119 | 0.648 |
| Saarela et al. [56] | 62 | 32 | 3 | 0.195 | 81 | 17 | 3 | 0.113 | 0.095 |
| Tsai et al. [46] | 43 | 92 | 40 | 0.491 | 64 | 95 | 30 | 0.410 | 0.592 |
| Zhang et al. [68] | 178 | 108 | 9 | 0.213 | 166 | 74 | 10 | 0.188 | 0.629 |
| He et al.[40] | 155 | 245 | 113 | 0.459 | 165 | 285 | 125 | 0.465 | 0.925 |
| Huang et al. [33] | 150 | 66 | 4 | 0.168 | 98 | 25 | 5 | 0.136 | 0.050 |
| Cozza et al. [52] | 152 | 84 | 15 | 0.227 | 60 | 33 | 4 | 0.211 | 0.839 |
| Qian et al. [44] | 28 | 52 | 25 | 0.485 | 25 | 56 | 24 | 0.495 | 0.493 |
| Yu et al. [47] | 31 | 41 | 27 | 0.479 | 28 | 51 | 20 | 0.459 | 0.712 |
| Feher et al. [53] | 94 | 56 | 10 | 0.237 | 52 | 79 | 33 | 0.442 | 0.763 |
| Qi et al. [43] | 27 | 32 | 21 | 0.462 | 27 | 48 | 11 | 0.406 | 0.147 |
| Fukumoto et al. [39] | 218 | 319 | 120 | 0.425 | 197 | 249 | 79 | 0.387 | 0.982 |
| Pivac et al. [55] | 135 | 59 | 17 | 0.220 | 268 | 118 | 16 | 0.186 | 0.509 |
| Borroni et al. [50] | 128 | 87 | 19 | 0.267 | 89 | 63 | 10 | 0.256 | 0.794 |
| Boiocchi et al. [49] | 113 | 63 | 15 | 0.243 | 231 | 150 | 27 | 0.25 | 0.692 |
| Sonali et al. [45] | 3 | 32 | 22 | 0.666 | 12 | 23 | 28 | 0.626 | 0.081 |
| Vieira et al. [36] | 205 | 59 | 5 | 0.128 | 84 | 25 | 5 | 0.153 | 0.095 |
| Gomar et al. [32] | 153 | 69 | 0 | 0.155 | 120 | 55 | 0 | 0.157 | 0.013 |
Table 2: Genotypic distribution of BDNF gene rs6265 polymorphism included in meta-analysis.
| First author | Cases (AD) | Control (HC) | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|
| C270T Genotype rs2030324 | Minor Allele Frequency (MAF) | C270T Genotype rs2030324 | Minor Allele Frequency (MAF) | ||||||
| CC | CT | TT | CC | CT | TT | P-value | |||
| Kunugi et al. [60] | 150 | 19 | 1 | 0.061 | 477 | 21 | 0 | 0.021 | 0.630 |
| Riemenschneider et al. [62] | 185 | 24 | 1 | 0.061 | 175 | 13 | 0 | 0.034 | 0.623 |
| Bagnoli et al. [48] | 113 | 14 | 1 | 0.062 | 83 | 14 | 0 | 0.072 | 0.443 |
| Nishimura et al. [59] | 175 | 13 | 0 | 0.034 | 170 | 17 | 1 | 0.050 | 0.429 |
| Bodner et al. [30] | 230 | 26 | 0 | 0.050 | 175 | 19 | 0 | 0.048 | 0.473 |
| Desai et al. [31] | 629 | 86 | 4 | 0.065 | 454 | 69 | 0 | 0.065 | 0.106 |
| Desai et al. [31] | 54 | 4 | 0 | 0.034 | 38 | 4 | 0 | 0.047 | 0.745 |
| Lee et al. [34] | 102 | 4 | 0 | 0.018 | 66 | 7 | 0 | 0.047 | 0.666 |
| Matsushita et al. [41] | 457 | 30 | 0 | 0.030 | 438 | 33 | 0 | 0.035 | 0.430 |
| Nishimura et al. [42] | 154 | 18 | 0 | 0.052 | 264 | 11 | 0 | 0.02 | 0.735 |
| Olin et al. [69] | 173 | 36 | 3 | 0.099 | 189 | 13 | 0 | 0.032 | 0.636 |
| Vepsalainen et al. [58] | 90 | 199 | 86 | 0.494 | 124 | 239 | 97 | 0.470 | 0.359 |
| Akatsu et al. [37] | 89 | 6 | 0 | 0.031 | 101 | 7 | 0 | 0.032 | 0.727 |
| Saarela et al. [56] | 88 | 9 | 0 | 0.046 | 81 | 19 | 1 | 0.103 | 0.922 |
| Tsai et al. [46] | 151 | 24 | 0 | 0.068 | 167 | 20 | 2 | 0.063 | 0.129 |
| Zhang et al. [68] | 271 | 22 | 2 | 0.044 | 220 | 30 | 0 | 0.06 | 0.312 |
| Huang et al. [33] | 202 | 16 | 2 | 0.045 | 113 | 15 | 0 | 0.058 | 0.481 |
| Cozza et al. [52] | 212 | 35 | 4 | 0.085 | 80 | 15 | 2 | 0.097 | 0.218 |
| Qian et al. [44] | 104 | 1 | 0 | 0.004 | 96 | 8 | 1 | 0.047 | 0.101 |
| Fukumoto et al. [39] | 611 | 45 | 1 | 0.035 | 490 | 34 | 1 | 0.034 | 0.613 |
| Cousin et al. [61] | 370 | 54 | 1 | 0.065 | 419 | 50 | 1 | 0.055 | 0.698 |
| Boiocchi et al. [49] | 55 | 93 | 44 | 0.471 | 103 | 192 | 89 | 0.481 | 0.979 |
Table 3: Genotypic distribution of BDNF gene rs2030324 polymorphism included in meta-analysis.
Association of BDNF SNP rs6265 polymorphisms with AD
Overall, the meta-analysis results based on different genetic models (Allelic, Homozygote, Heterozygote, Dominant and Recessive) revealed no association between BDNF 196 G/A allele in overall studies. Moreover, no association were identified between BDNF 196 G/A polymorphism and AD in ethnicity specific studies (i.e., American, Asian and European) also.
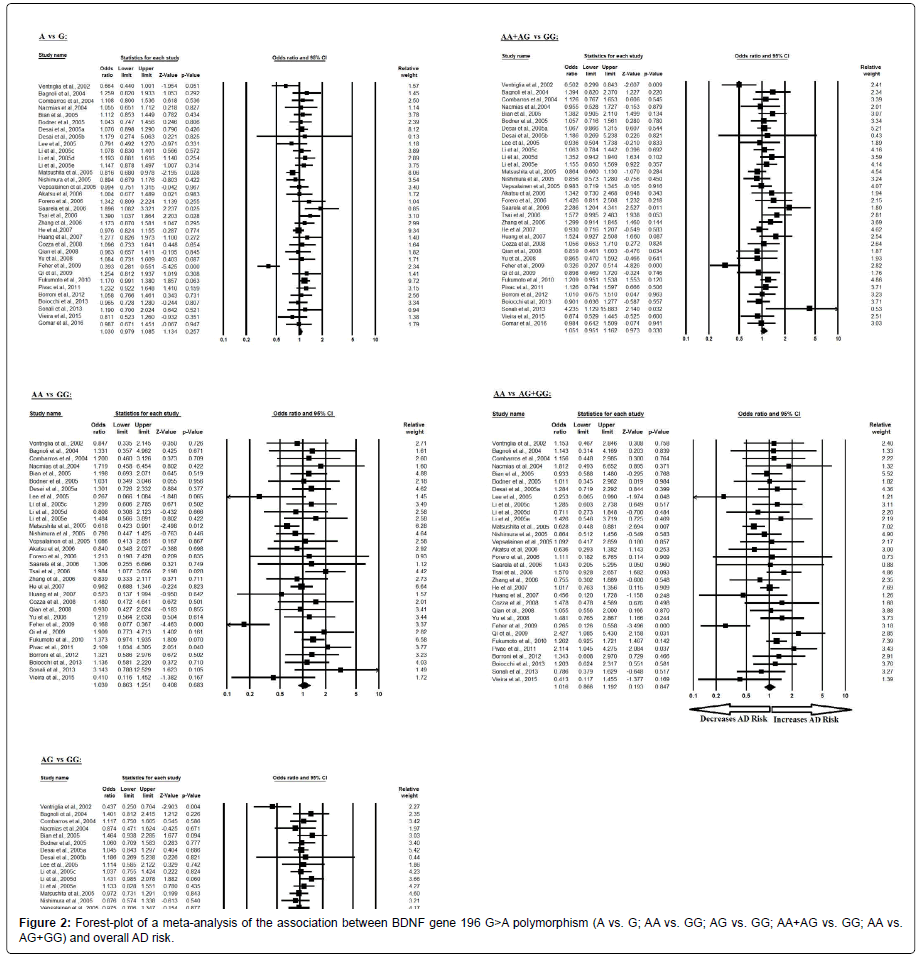
The pooled ORs of overall study analysis revealed that BDNF G>A gene polymorphism is not associated with AD risk in allelic (A vs. G: p=0.257; OR=1.030, 95% CI=0.979 to 1.085) genetic models; homozygous (AA vs. GG: p=0.683; OR=1.039, 95% CI=0.863 to 1.251) genetic models; heterozygous (AG vs. GG: p=0.329; OR=1.052, 95% CI=0.950 to 1.165) genetic models; dominant (AA+AG vs. GG: p=0.330; OR=1.051, 95% CI=0.951 to 1.162) genetic models; and recessive (AA vs. AG+GG: p=0.847; OR=1.016, 95% CI=0.866 to 1.192) genetic models (Figure 2). All ORs were pooled through a random effect models (Table 4).
| Comparisons | Egger’s regression analysis | Heterogeneity analysis | Model used for meta-analysis | ||||
|---|---|---|---|---|---|---|---|
| Intercept | 95% Confidence Interval | P value | Q value | Pheterogeneity | I2 (%) | ||
| A vs. G | 0.282 | -1.120 to 1.685 | 0.684 | 65.777 | 0.001 | 49.831 | Random |
| AA vs. GG | 0.035 | -1.107 to 1.177 | 0.950 | 55.024 | 0.005 | 43.661 | Random |
| AG vs. GG | 0.385 | -0.985 to 1.755 | 0.570 | 61.076 | 0.002 | 45.969 | Random |
| AA+AG vs. GG | 0.360 | -1.052 to 1.773 | 0.606 | 64.671 | 0.001 | 48.973 | Random |
| AA vs. AG+GG | -0.080 | -1.098 to 0.936 | 0.872 | 48.938 | 0.021 | 36.655 | Random |
Table 4: Statistics to test publication bias and heterogeneity in meta-analysis (rs6265-Overall).
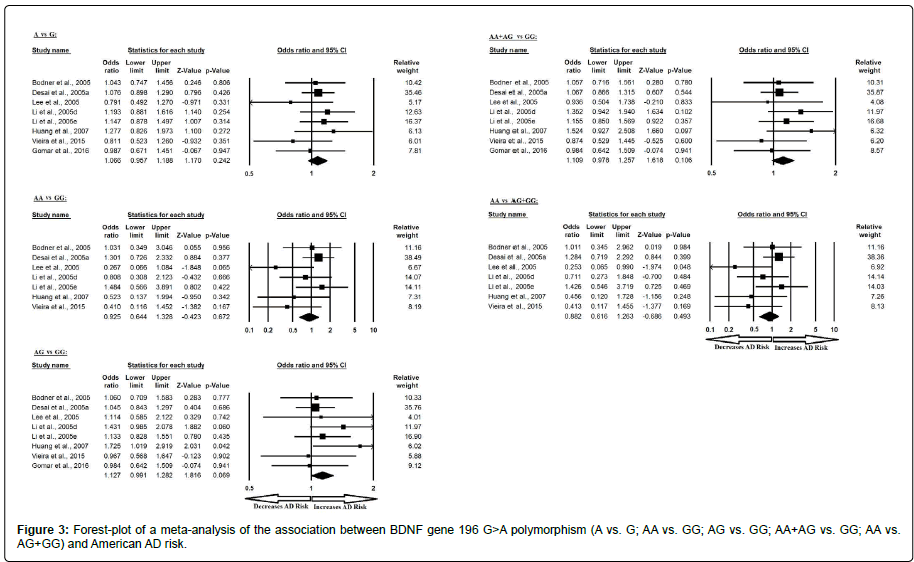
Similarly, the pooled ORs of American study analysis revealed that BDNF G>A gene polymorphism is not associated with AD risk in allelic (A vs. G: p=0.242; OR=1.066, 95% CI=0.957 to 1.188) genetic models; homozygous (AA vs. GG: p=0.672; OR=1.280, 95% CI=0.644 to 1.328) genetic models; heterozygous (AG vs. GG: p=0.069; OR=1.127, 95% CI=0.991 to 1.282) genetic models; dominant (AA+AG vs. GG: p=0.106; OR=1.109, 95% CI=0.978 to 1.257) genetic models; and recessive (AA vs. AG+GG: p=0.493; OR=0.882, 95% CI=0.616 to 1.263) genetic models (Figure 3). All ORs were pooled through a fixed effect models (Table 5).
| Comparisons | Egger’s regression analysis | Heterogeneity analysis | Model used for meta-analysis | ||||
|---|---|---|---|---|---|---|---|
| Intercept | 95% Confidence Interval | P value | Q value | Pheterogeneity | I2 (%) | ||
| A vs. G | -0.983 | -3.139 to 1.172 | 0.307 | 4.664 | 0.701 | 0.000 | Fixed |
| AA vs. GG | -2.776 | -5.099 to -0.452 | 0.027 | 7.660 | 0.264 | 21.671 | Fixed |
| AG vs. GG | 0.751 | -1.581 to 3.084 | 0.460 | 3.218 | 0.617 | 0.000 | Fixed |
| AA+AG vs. GG | -0.028 | -2.278 to 2.220 | 0.975 | 4.784 | 0.729 | 0.000 | Fixed |
| AA vs. AG+GG | -2.945 | -5.356 to -0.533 | 0.025 | 8.386 | 0.211 | 28.451 | Fixed |
Table 5: Statistics to test publication bias and heterogeneity in meta-analysis (rs6265- American).
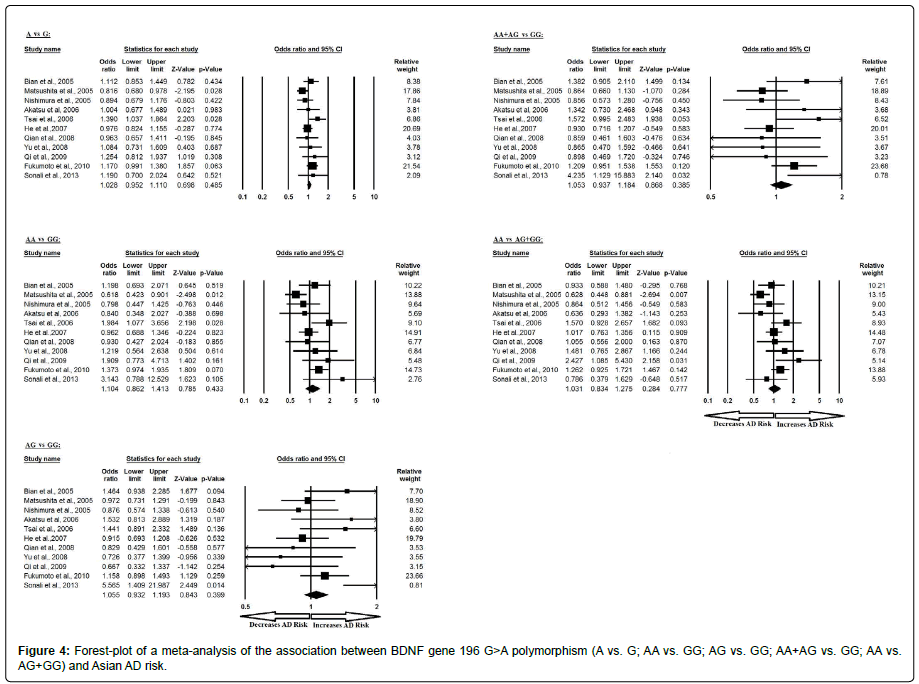
Additionally, the pooled ORs of Asian study analysis revealed that BDNF G>A gene polymorphism is not associated with AD risk in allelic (A vs. G: p=0.485; OR=1.028, 95% CI=0.952 to 1.110) genetic models; homozygous (AA vs. GG: p=0.433; OR=1.104, 95% CI=0.862 to 1.413) genetic models; heterozygous (AG vs. GG: p=0.399; OR=1.055, 95% CI=0.932 to 1.193) genetic models; dominant (AA+AG vs. GG: p=0.385; OR=1.053, 95% CI=0.937 to 1.184) genetic models; and recessive (AA vs. AG+GG: p=0.777; OR=1.031, 95% CI=0.834 to 1.275) genetic models (Figure 4). All ORs were pooled through a fixed effect models except for homozygous and recessive genetic models (Table 6).
| Comparisons | Egger’s regression analysis | Heterogeneity analysis | Model used for meta-analysis | ||||
|---|---|---|---|---|---|---|---|
| Intercept | 95% Confidence Interval | P value | Q value | Pheterogeneity | I2 (%) | ||
| A vs. G | 0.869 | -1.485 to 3.225 | 0.425 | 15.621 | 0.111 | 35.986 | Fixed |
| AA vs. GG | 1.278 | -1.164 to 3.721 | 0.266 | 19.996 | 0.029 | 49.991 | Random |
| AG vs. GG | 0.779 | -1.429 to 2.988 | 0.445 | 16.666 | 0.082 | 39.996 | Fixed |
| AA+AG vs. GG | 0.989 | -1.092 to 3.070 | 0.310 | 15.706 | 0.108 | 36.330 | Fixed |
| AA vs. AG+GG | 0.737 | -2.142 to 3.616 | 0.576 | 20.385 | 0.026 | 50.945 | Random |
Table 6: Statistics to test publication bias and heterogeneity in meta-analysis (rs6265-Asian).
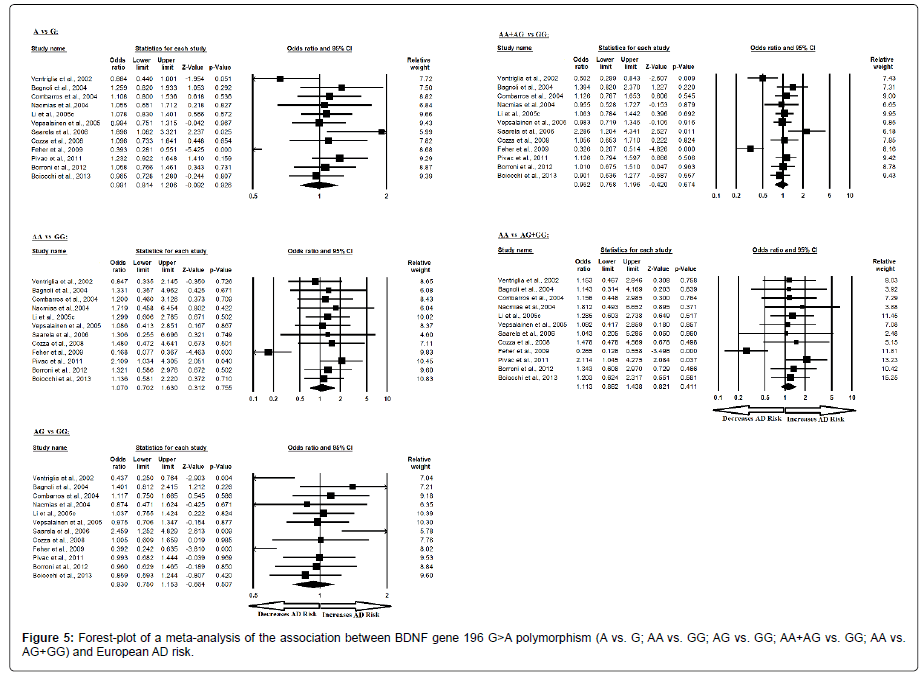
Moreover, the pooled ORs of European study analysis revealed that BDNF G>A gene polymorphism is not associated with AD risk in allelic (A vs. G: p=0.926; OR=0.991, 95% CI=0.814 to 1.206) genetic models; homozygous (AA vs. GG: p=0.755; OR=1.070, 95% CI=0.702 to 1.630) genetic models; heterozygous (AG vs. GG: p=0.507; OR=0.930, 95% CI=0.750 to 1.153) genetic models; dominant (AA+AG vs. GG: p=0.674; OR=0.952, 95% CI=0.758 to 1.196) genetic models; and recessive (AA vs. AG+GG: p=0.411; OR=1.113, 95% CI=0.862 to 1.438) genetic models (Figure 5). All ORs were pooled through a random effect models except for recessive genetic model (Table 7).
| Comparisons | Egger’s regression analysis | Heterogeneity analysis | Model used for meta-analysis | ||||
|---|---|---|---|---|---|---|---|
| Intercept | 95% Confidence Interval | P value | Q value | Pheterogeneity | I2 (%) | ||
| A vs. G | 0.935 | -5.248 to 7.119 | 0.743 | 42.336 | 0.000 | 74.017 | Random |
| AA vs. GG | 0.754 | -3.555 to 5.064 | 0.704 | 26.706 | 0.005 | 58.810 | Random |
| AG vs. GG | -0.080 | -5.035 to 4.873 | 0.971 | 31.298 | 0.001 | 64.854 | Random |
| AA+AG vs. GG | -0.142 | -5.701 to 5.416 | 0.955 | 38.830 | 0.000 | 71.671 | Random |
| AA vs. AG+GG | 0.517 | -3.044 to 4.079 | 0.752 | 18.667 | 0.067 | 41.074 | Fixed |
Table 7: Statistics to test publication bias and heterogeneity in meta-analysis (rs6265-European).
Association of BDNF SNP rs2030324 polymorphisms with AD
The meta-analysis results based on different genetic models revealed no association between BDNF 270 C/T allele in overall studies for all genetic models i.e. T vs. C allelic contrast, TT vs. CC homozygous genotype, TC vs. CC heterozygous genotype, dominant TT+TC vs. CC genotype and recessive TT vs. TC+CC genotype. Ethnicity specific studies also not found to be associated with BDNF 270 C/T polymorphism and AD.
The pooled ORs of overall study analysis revealed that BDNF C>T gene polymorphism is not associated with AD risk in allelic (T vs. C: p=0.538; OR=1.059, 95% CI=0.881 to 1.274) genetic models; homozygous (TT vs. CC: p=0.419; OR=1.125, 95% CI=0.845 to 1.496) genetic models; heterozygous (TC vs. CC: p=0.748; OR=1.034, 95% CI=0.842 to 1.270) genetic models; dominant (TT+TC vs. CC: p=0.662; OR=1.047, 95% CI=0.852 to 1.287) genetic models; and recessive (TT vs. TC+CC: p=0.499; OR=1.088, 95% CI=0.853 to 1.387) genetic models (Figure 6). All ORs were pooled through a random effect models except for homozygous and recessive genetic models (Table 8).
| Comparisons | Egger’s regression analysis | Heterogeneity analysis | Model used for meta-analysis | ||||
|---|---|---|---|---|---|---|---|
| Intercept | 95% Confidence Interval | P value | Q value | Pheterogeneity | I2 (%) | ||
| T vs. C | -0.362 | -1.780 to 1.055 | 0.599 | 52.111 | 0.000 | 59.701 | Random |
| TT vs. CC | 0.191 | -0.415 to 0.797 | 0.509 | 10.175 | 0.809 | 0.000 | Fixed |
| TC vs. CC | -0.707 | -2.507 to 1.093 | 0.422 | 49.386 | 0.000 | 57.478 | Random |
| TT+TC vs. CC | -0.723 | -2.528 to 1.081 | 0.412 | 51.664 | 0.000 | 59.353 | Random |
| TT vs. TC+CC | -0.215 | -0.333 to 0.764 | 0.413 | 9.348 | 0.859 | 0.000 | Fixed |
Table 8: Statistics to test publication bias and heterogeneity in meta-analysis (rs2030324-Overall).
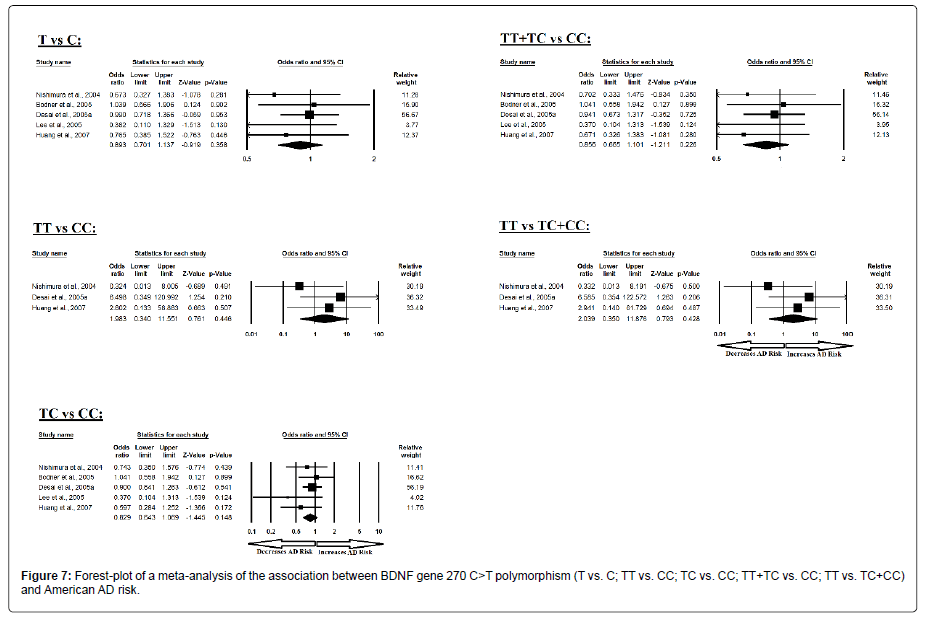
Similarly, the pooled ORs of American study analysis revealed that BDNF C>T gene polymorphism is not associated with AD risk in allelic (A vs. G: p=0.358; OR=0.893, 95% CI=0.701 to 1.137) genetic models; homozygous (AA vs. GG: p=0.446; OR=1.983, 95% CI=0.340 to 11.551) genetic models; heterozygous (AG vs. GG: p=0.148; OR=0.829, 95% CI=0.643 to 1.069) genetic models; dominant (AA+AG vs. GG: p=0.226; OR=0.856, 95% CI=0.665 to 1.101) genetic models; and recessive (AA vs. AG+GG: p=0.428; OR=2.039, 95% CI=0.350 to 11.876) genetic models (Figure 7). All ORs were pooled through a fixed effect models (Table 9).
| Comparisons | Egger’s regression analysis | Heterogeneity analysis | Model used for meta-analysis | ||||
|---|---|---|---|---|---|---|---|
| Intercept | 95% Confidence Interval | P value | Q value | Pheterogeneity | I2 (%) | ||
| T vs. C | -1.640 | -3.489 to 0.208 | 0.066 | 3.206 | 0.524 | 0.000 | Fixed |
| TT vs. CC | -20.859 | -65.562 to 23.843 | 0.106 | 1.909 | 0.385 | 0.000 | Fixed |
| TC vs. CC | -1.441 | -3.790 to 0.906 | 0.145 | 3.135 | 0.535 | 0.000 | Fixed |
| TT+TC vs. CC | -1.553 | -3.601 to 0.494 | 0.094 | 3.083 | 0.544 | 0.000 | Fixed |
| TT vs. TC+CC | -20.819 | -68.169 to 26.529 | 0.112 | 1.905 | 0.386 | 0.000 | Fixed |
Table 9: Statistics to test publication bias and heterogeneity in meta-analysis (rs2030324-American).
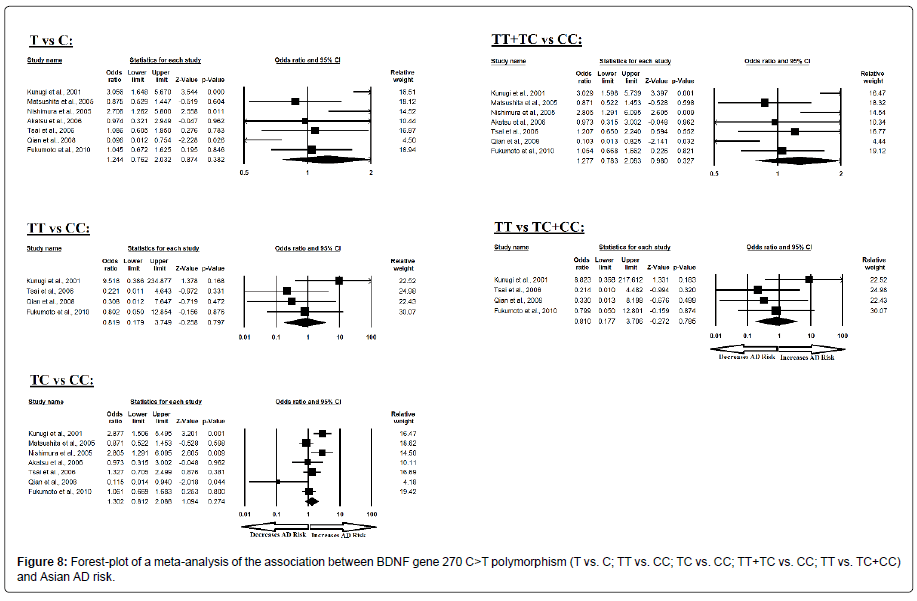
Additionally, the pooled ORs of Asian study analysis revealed that BDNF C>T gene polymorphism is not associated with AD risk in allelic (A vs. G: p=0.382; OR=1.244, 95% CI=0.762 to 2.032) genetic models; homozygous (AA vs. GG: p=0.797; OR=0.819, 95% CI=0.179 to 3.749) genetic models; heterozygous (AG vs. GG: p=0.274; OR=1.302, 95% CI=0.812 to 2.088) genetic models; dominant (AA+AG vs. GG: p=0.327; OR=1.277, 95% CI=0.783 to 2.083) genetic models; and recessive (AA vs. AG+GG: p=0.785; OR=0.810, 95% CI=0.177 to 3.706) genetic models (Figure 8). All ORs were pooled through a random effect models except for homozygous and recessive genetic models (Table 10).
| Comparisons | Egger’s regression analysis | Heterogeneity analysis | Model used for meta-analysis | ||||
|---|---|---|---|---|---|---|---|
| Intercept | 95% Confidence Interval | P value | Q value | Pheterogeneity | I2 (%) | ||
| T vs. C | -0.930 | -6.291 to 4.429 | 0.673 | 20.906 | 0.002 | 71.300 | Random |
| TT vs. CC | 3.105 | -41.558 to 47.770 | 0.793 | 3.316 | 0.345 | 9.534 | Fixed |
| TC vs. CC | -0.780 | -5.834 to 4.273 | 0.707 | 18.038 | 0.006 | 66.737 | Random |
| TT+TC vs. CC | -0.864 | -6.112 to 4.382 | 0.689 | 19.666 | 0.003 | 69.490 | Random |
| TT vs. TC+CC | 3.128 | -40.505 to 46.761 | 0.786 | 3.169 | 0.366 | 5.326 | Fixed |
Table 10: Statistics to test publication bias and heterogeneity in meta-analysis (rs2030324-Asian).
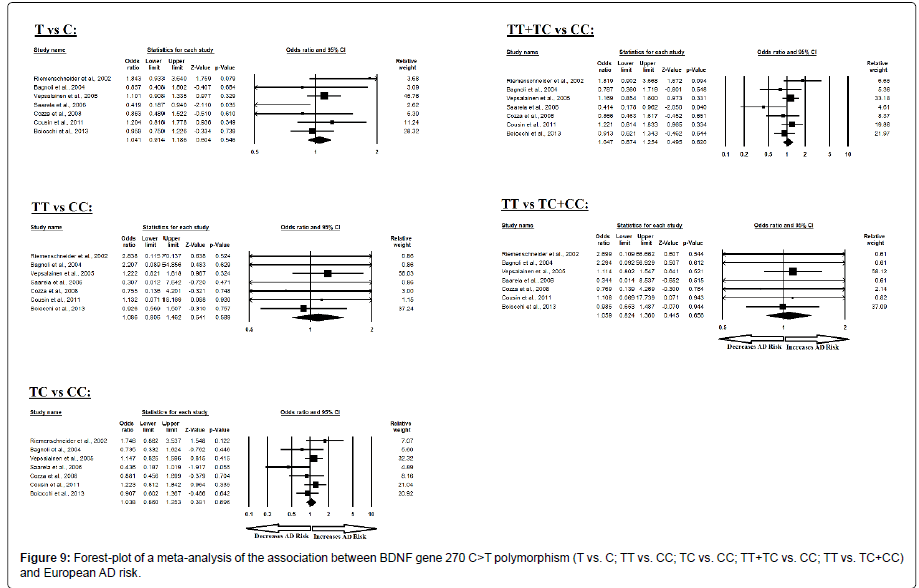
Moreover, the pooled ORs of European study analysis revealed that BDNF C>T gene polymorphism is not associated with AD risk in allelic (A vs. G: p=0.546; OR=1.041, 95% CI=0.914 to 1.186) genetic models; homozygous (AA vs. GG: p=0.589; OR=1.086, 95% CI=0.806 to 1.462) genetic models; heterozygous (AG vs. GG: p=0.696; OR=1.038, 95% CI=0.860 to 1.253) genetic models; dominant (AA+AG vs. GG: p=0.620; OR=1.047, 95% CI=0.874 to 1.254) genetic models; and recessive (AA vs. AG+GG: p=0.656; OR=1.059, 95% CI=0.824 to 1.360) genetic models (Figure 9). All ORs were pooled through a fixed effect models (Table 11).
| Comparisons | Egger’s regression analysis | Heterogeneity analysis | Model used for meta-analysis | ||||
|---|---|---|---|---|---|---|---|
| Intercept | 95% Confidence Interval | P value | Q value | Pheterogeneity | I2 (%) | ||
| T vs. C | -0.637 | -3.265 to 1.991 | 0.560 | 9.550 | 0.145 | 37.172 | Fixed |
| TT vs. CC | -0.057 | -0.957 to 0.841 | 0.875 | 2.047 | 0.915 | 0.000 | Fixed |
| TC vs. CC | -1.504 | -4.871 to 1.862 | 0.302 | 8.455 | 0.207 | 29.038 | Fixed |
| TT+TC vs. CC | -1.425 | -4.930 to 2.078 | 0.343 | 9.412 | 0.152 | 36.249 | Fixed |
| TT vs. TC+CC | -0.004 | -0.711 to 0.702 | 0.988 | 1.363 | 0.968 | 0.000 | Fixed |
Table 11: Statistics to test publication bias and heterogeneity in meta-analysis (rs2030324-European).
Evaluation of publication bias
No between study heterogeneity was found in analyses of the BDNF 196 G/A and 270 C/T polymorphisms in the overall, American, Asian or European study populations. Begg’s Funnel Plot and Egger’s Test were performed to evaluate the publication bias among the included studies for this meta-analysis. The shape of funnel plots did not reveal any evidence of obvious symmetry in all comparisons and the Egger’s regression test was used to provide statistical evidence of funnel plot. The results of Egger’s regression analysis did not show any evidence of publication bias in all genetic models except for homozygous and recessive genetic models of BDNF 196 G/A polymorphism in the American study populations (Egger’s regression test p values>0.05; (Tables 4-11).
Quantitative sensitivity analysis
Sensitivity analysis was conducted to verify the robustness of our results. It is also used to ascertain whether modification of the inclusion criteria of the meta-analysis affected the final results. The effect of each study included in this meta-analysis assessed by sensitivity analysis of each individual study on the pooled OR by eliminating each single casecontrol study was done for each BDNF polymorphism [rs6265(G>A), rs2030324(C>T)] to evaluate the influence. Outcomes of sensitivity analysis revealed that no individual genetic model influenced the pooled ORs significantly in all the BDNF variants, which suggest the credibility and stability of this meta-analysis.
Trial sequential analysis (TSA)
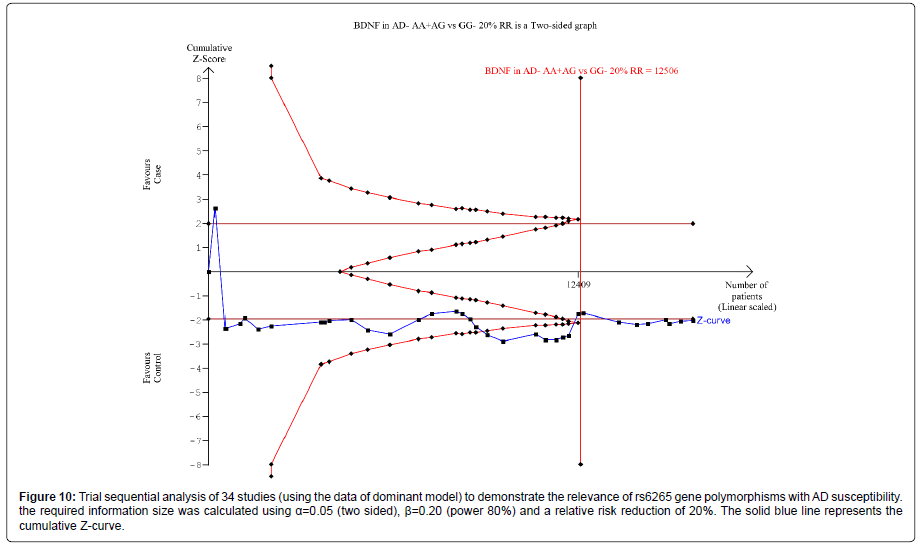
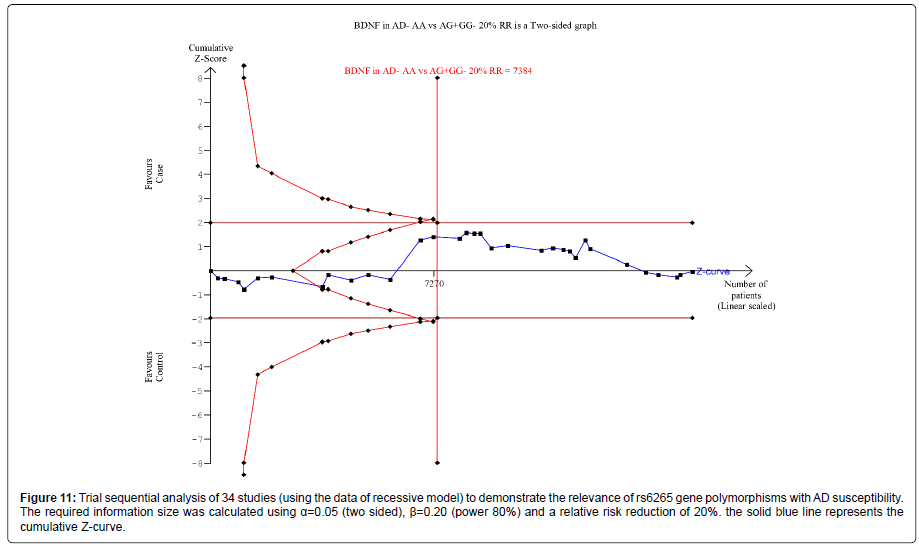
Thirty nine trials (27329 subjects) were used to investigate the association of rs6265 and rs2030324 gene polymorphisms with AD risk. Using the data of dominant model for rs6265 (including 34 trials with 16165 subjects) as an example, the TSA was performed and found that the required information size (RIS) is 12506 subjects to demonstrate the issue. The cumulative Z-curve crosses the TSA monitoring boundary before reaching RIS, indicating that the cumulative evidence is sufficient and further trials are not necessary (Figure 10). However, the cumulative Z-curve does not crossed with TSA monitoring boundary when we performed the analysis using the data of recessive model, confirming that cumulative evidence is insufficient and further relevant trials are necessary (Figure 11).
Figure 10: Trial sequential analysis of 34 studies (using the data of dominant model) to demonstrate the relevance of rs6265 gene polymorphisms with AD susceptibility.the required information size was calculated using a=0.05 (two sided), ì?=0.20 (power 80%) and a relative risk reduction of 20%. The solid blue line represents the cumulative Z-curve.
Figure 11: Trial sequential analysis of 34 studies (using the data of recessive model) to demonstrate the relevance of rs6265 gene polymorphisms with AD susceptibility. The required information size was calculated using a=0.05 (two sided), ì?=0.20 (power 80%) and a relative risk reduction of 20%. the solid blue line represents the cumulative Z-curve.
Similarly, for rs2030324, we chose the data of five models to perform TSA. The cumulative Z-curve have not crossed with TSA monitoring boundaries before the required information size is reached, indicating that cumulative evidence is insufficient and further trials are necessary (figures were not shown).
Moreover, when we performed the sub-analysis based on the ethnicity (American, Asian and European) for all models for both BDNF gene polymorphisms, the cumulative Z-curve does not crossed with TSA monitoring boundary, confirming that cumulative evidence is insufficient and further relevant trials are necessary in this regard (figures were not shown).
Discussion
In the present study, 39 studies covering 9,409 cases and 9,522 controls were carried out to investigate the association of the BDNF G196A and BDNF C270T variants with the risk of AD. 34 articles related to rs6265 and 22 articles related to rs2030324 were included in our meta-analysis. However, the results showed no significant associations between BDNF G196A and BDNF C270T with AD in overall studies along with American, Asian and European ethnic studies.
There are few evidences supporting altered mRNA and protein expression as a consequence of the gene polymorphisms 196 G/A and 270 C/T. For instance, earlier studies have shown that the BDNF Val66Met polymorphism has been shown to impair the secretion of BDNF [63], that changes brain morphology and cognitive function [64], and these polymorphisms has been shown to affect BDNF or neuronal function by reducing transport of BDNF from the Golgi region to appropriate secretory granules in neurons [65]. Moreover, the A allele of rs6265 has also found to be associated with poorer episodic memory, abnormal hippocampal activation, and lower hippocampal n-acetyl aspartate (NAA) in human subjects [66]. Individual studies have reported 10 positive results [32,39,41,42,46,49,50,53,57,67] and 22 negative results [30,31,33-38,40,45,47,48,51,54-56,58,59,68] among the previous association studies between the BDNF G196A polymorphism and AD. In the present meta-analysis, no significant association was found between BDNF G196A and AD (P>0.05, Figures 2-5). The present meta-analysis of BDNF G196A included 34 articles. In addition, meta analyses were performed under various genetic models, including allelic, homozygous, heterozygous, dominant and recessive models. Subgroup meta-analysis by ethnicity were also conducted, although no statistically significant association results were obtained.
Similarly, literature has already reported that the BDNF C270T polymorphism is located in a non-coding region and may affect the BDNF expression in the neural soma, dendrites or axonal regions [29]. Moreover, the heterozygous carriers of the T allele of rs2030324 tend to have a greater risk of developing AD in comparison to noncarriers [49]. Individual studies have reported a total of 8 significant results [39,41,42,46,49,60,62,69] and 12 non-significant results [3 0,31,33,34,37,48,56,58,59,61,68] among the previous association studies between the BDNF C270T polymorphism and AD. In the present meta-analysis, no significant association was found between the BDNF C270T polymorphism and risk of AD (P>0.05, Figures 6-9). The present meta-analysis of BDNF G196A included 22 articles.
In addition, meta analyses were performed under various genetic models, including allelic, homozygous, heterozygous, dominant and recessive models. Subgroup meta-analysis by ethnicity were also conducted, although no statistically significant association results were obtained.
As we know the multifactorial nature of AD, so combined effects between gene variants and environmental factors, as well as their possible interaction, may be potential contributors to this disease. We further performed a sensitivity analysis, the results of which were consistent and strongly identified the stability of our results. Moreover, no publication bias was observed in any of the above-mentioned genetic models for both two polymorphisms except for homozygous and recessive genetic models of BDNF 196 G/A polymorphism in the American study populations. Besides, we also performed the TSA and the results of TSA showed that the conclusions in this meta-analysis are robust. All the studies included in the present meta-analysis met the HWE and were done under various genetic models with subgroup meta-analysis stratified by ethnicity. With stringent inclusion and exclusion criteria, a larger sample size and more comprehensive analysis, the present meta-analysis of BDNF G196A and C270T showed a more reliable conclusion.
Advantages
The major advantage of our meta-analysis is that the results were based on the larger number of studies, resulting into a greater chance of getting definitive conclusions. Moreover, we also performed subgroup analyses for the potential sources of heterogeneity, and sensitivity analysis for ensuring the stability of our results. However, our metaanalysis also had certain limitations. Firstly, publication bias may occur, due to the less likely published or even missed negative result studies. Secondly, limited number of ethnic studies in other populations like Africans and the most of the studies were carried out in the European, Asian and American ethnic populations. So, future studies in other ethnic populations are required in this regard. Thirdly, since AD is a complex disease, so different statuses in AD may affect the results of the study; but no detailed information of the AD diagnostic criteria was available in the previous individual studies. Therefore, there is a need for future case control studies with more comprehensive information, as different diagnostic criteria may have possible influence on the diagnosis of AD. Fourthly, there is numerous numbers of polymorphisms in BDNF and our study only focused on two polymorphisms of BDNF, which may not fully illustrate the function of this gene. Hence, studies investigating a wider range of polymorphisms are required in this regard.
Conclusion
In conclusion, the present comprehensive meta-analysis revises the previous incomplete data and suggests no significant association between the BDNF G196A and BDNF C270T polymorphisms with the risk of AD under any genetic models and in any ethnic populations. Since potential biases and confounders could not be ruled out completely in this study, further studies focusing on a wider range of ethnic populations are required to confirm the results of the study.
Acknowledgement
This work was partially supported by Department of Biotechnology; Government of India BUILDER project (BT/PR-9028/INF/22/193/2013) granted to Centre for Life Sciences, Central University of Jharkhand and UGC Central University Fellowship to S.R. Authors declare no conflict of Interest.
References
- Mucke L (2009) Neuroscience: Alzheimer's disease. Nature 461: 895-897.
- Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM (2007) Forecasting the global burden of Alzheimer's disease. Alzheimers Dement 3: 186-191.
- Reitz C, Brayne C, Mayeux R (2011) Epidemiology of Alzheimer disease. Nat Rev Neurol 7: 137-152.
- Armstrong RA (2013) What causes Alzheimerâ¬?¬?s disease? Folia Neuropathol 51: 169-188.
- Borroni B, Costanzi C, Padovani A (2010) Genetic susceptibility to behavioural and psychological symptoms in Alzheimer disease. Curr Alzheimer Res 7: 158-164.
- Serretti A, Olgiati P, De Ronchi D (2007) Genetics of Alzheimer's disease. A rapidly evolving field. J Alzheimers Dis 12: 73-92.
- Maisonpierre PC, Le Beau MM, Espinosa R, Ip NY, Belluscio L, et al. (1991) Human and rat brain-derived neurotrophic factor and neurotrophin-3: Gene structures, distributions and chromosomal localizations. Genomics 10: 558-568.
- Fahnestock M, Garzon D, Holsinger RM, Michalski B (2002) Neurotrophic factors and Alzheimer's disease: Are we focusing on the wrong molecule? J Neural Transm Suppl 62: 241-252.
- Connor B, Young D, Yan Q, Faull RL, Synek B, et al. (1997) Brain-derived neurotrophic factor is reduced in Alzheimer's disease. Brain Res Mol Brain Res 49: 71-81.
- Ferrer I, Marin C, Rey MJ, Ribalta T, Goutan E, et al. (1999) BDNF and full-length and truncated TrkB expression in Alzheimer disease. Implications in therapeutic strategies. J Neuropathol Exp Neurol 58: 729-739.
- Dai D, Wang Y, Wang L, Li J, Ma Q, et al. (2014) Polymorphisms of DRD2 and DRD3 genes and Parkinson's disease: A meta-analysis. Biomed Rep 2: 275-281.
- Xu X, Wang Y, Wang L, Liao Q, Chang L, et al. (2013) Meta-analyses of 8 polymorphisms associated with the risk of the Alzheimer's disease. PLoS ONE 8: e73129.
- Zhou J, Huang Y, Huang RS, Wang F, Xu L, et al. (2012) A case-control study provides evidence of association for a common SNP rs974819 in PDGFD to coronary heart disease and suggests a sex-dependent effect. Thromb Res 130: 602-606.
- Green S, McDonald S (2005) Cochrane collaboration: More than systematic reviews? Intern Med J 35: 3-4.
- Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group (2009) Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med 6: e1000097.
- Wu R, Li B (1999) A multiplicative-epistatic model for analyzing interspecific differences in outcrossing species. Biometrics 55: 355-365.
- DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177-188.
- MANTEL N, HAENSZEL W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719-748.
- Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327: 557-560.
- Harbord RM, Egger M, Sterne JA (2006) A modified test for small-study effects in meta-analyses of controlled trials with binary endpoints. Stat Med 25: 3443-3457.
- Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629-634.
- Brok J, Thorlund K, Wetterslev Jr, Gluud C (2008) Apparently conclusive meta-analyses may be inconclusiveââ¬?¬â¬?¬Ětrial sequential analysis adjustment of random error risk due to repetitive testing of accumulating data in apparently conclusive neonatal meta-analyses. Int J Epidemiol 38: 287-298.
- Wetterslev Jr, Thorlund K, Brok J, Gluud C (2008) Trial sequential analysis may establish when firm evidence is reached in cumulative meta-analysis. J Clin Epidemiol 61: 64-75.
- Xie S, Shan XF, Shang K, Xu H, He J, et al. (2014) Relevance of LIG4 gene polymorphisms with cancer susceptibility: Evidence from a meta-analysis. Sci Rep 4: 6630.
- Turner RM, Bird SM, Higgins JP (2013) The impact of study size on meta-analyses: Examination of underpowered studies in Cochrane reviews. PLoS ONE 8: e59202.
- Wetterslev Jr, Thorlund K, Brok J, Gluud C (2009) Estimating required information size by quantifying diversity in random-effects model meta-analyses. BMC Med Res Methodol 9: 86.
- Holst LB, Petersen MW, Haase N, Perner A, Wetterslev Jr (2015) Restrictive versus liberal transfusion strategy for red blood cell transfusion: Systematic review of randomised trials with meta-analysis and trial sequential analysis. BMJ 350: h1354.
- Lim YY, Villemagne VL, Laws SM, Pietrzak RH, Snyder PJ, et al. (2015) APOE and BDNF polymorphisms moderate amyloid beta-related cognitive decline in preclinical Alzheimer's disease. Mol Psychiatry 20: 1322-1328.
- Nagata T, Shinagawa S, Nukariya K, Ochiai Y, Kawamura S, et al. (2011) Association between brain-derived neurotrophic factor (BDNF) gene polymorphisms and executive function in Japanese patients with Alzheimer's disease. Psychogeriatrics 11: 141-149.
- Bodner SM, Berrettini W, van Deerlin V, Bennett DA, Wilson RS, et al. (2005) Genetic variation in the brain derived neurotrophic factor gene in Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet 134B: 1-5.
- Desai P, Nebes R, DeKosky ST, Kamboh MI (2005) Investigation of the effect of brain-derived neurotrophic factor (BDNF) polymorphisms on the risk of late-onset Alzheimer's disease (AD) and quantitative measures of AD progression. Neurosci Lett 379: 229-234.
- Gomar JJ, Conejero-Goldberg C, Huey ED, Davies P, Goldberg TE (2016) Lack of neural compensatory mechanisms of BDNF val66met met carriers and APOE E4 carriers in healthy aging, mild cognitive impairment and Alzheimer's disease. Neurobiol Aging 39: 165-173.
- Huang R, Huang J, Cathcart H, Smith S, Poduslo SE (2007) Genetic variants in brain-derived neurotrophic factor associated with Alzheimer's disease. J Med Genet 44: e66.
- Lee J, Fukumoto H, Orne J, Klucken J, Raju S, et al. (2005) Decreased levels of BDNF protein in Alzheimer temporal cortex are independent of BDNF polymorphisms. Exp Neurol 194: 91-96.
- Li Y, Rowland C, Tacey K, Catanese J, Sninsky J, et al. (2005) The BDNF Val66Met polymorphism is not associated with late onset Alzheimer's disease in three case-control samples. Mol Psychiatry 10: 809-810.
- Vieira RN, Magalhaes JD, Sant'Anna J, Moriguti MM, de Paula JJ, et al. (2015) The GAB2 and BDNF polymorphisms and the risk for late-onset Alzheimer's disease in an elderly Brazilian sample. Int Psychogeriatr 27: 1687-1692.
- Akatsu H, Yamagata HD, Kawamata J, Kamino K, Takeda M, et al. (2006) Variations in the BDNF gene in autopsy-confirmed Alzheimer's disease and dementia with Lewy bodies in Japan. Dement Geriatr Cogn Disord 22: 216-222.
- Bian JT, Zhang JW, Zhang ZX, Zhao HL (2005) Association analysis of brain-derived neurotrophic factor (BDNF) gene 196 A/G polymorphism with Alzheimer's disease (AD) in mainland Chinese. Neurosci Lett 387: 11-16.
- Fukumoto N, Fujii T, Combarros O, Kamboh MI, Tsai SJ, et al. (2010) Sexually dimorphic effect of the Val66Met polymorphism of BDNF on susceptibility to Alzheimer's disease: New data and meta-analysis. Am J Med Genet B Neuropsychiatr Genet 153: 235-242.
- He XM, Zhang ZX, Zhang JW, Zhou YT, Tang MN, et al. (2007) Lack of association between the BDNF gene Val66Met polymorphism and Alzheimer disease in a Chinese Han population. Neuropsychobiology 55: 151-155.
- Matsushita S, Arai H, Matsui T, Yuzuriha T, Urakami K, et al. (2005) Brain-derived neurotrophic factor gene polymorphisms and Alzheimer's disease. J Neural Transm (Vienna) 112: 703-711.
- Nishimura M, Kuno S, Kaji R, Kawakami H (2005) Brain-derived neurotrophic factor gene polymorphisms in Japanese patients with sporadic Alzheimer's disease, Parkinson's disease and multiple system atrophy. Mov Disord 20: 1031-1033.
- Qi HM, Sheng YL, Qian Y (2009) Association of brain-derived neurotrophic factor and apolipoprotein gene polymorphisms with sporadic Alzheimerâ¬?¬?s disease. Heilongjiang Medical Journal 33: 724-726.
- Qian ZZ, Zhang XB, Yong Y, Gui Y, Yu H, et al. (2008) Haplotype analysis of Alzheimerâ¬?¬?s disease BDNF gene G196A and C270T polymorphisms. Journal of Southeast University 27: 161-165.
- Sonali N, Tripathi M, Sagar R, Vivekanandhan S (2013) Val66Met polymorphism and BDNF levels in Alzheimer's disease patients in North Indian population. Int J Neurosci 123: 409-416.
- Tsai SJ, Hong CJ, Liu HC, Liu TY, Liou YJ (2006) The brain-derived neurotrophic factor gene as a possible susceptibility candidate for Alzheimer's disease in a Chinese population. Dement Geriatr Cogn Disord 21: 139-143.
- Yu H, Zhang Z, Shi Y, Bai F, Xie C, et al. (2008) Association study of the decreased serum BDNF concentrations in amnestic mild cognitive impairment and the Val66Met polymorphism in Chinese Han. J Clin Psychiatry 69: 1104-1111.
- Bagnoli S, Nacmias B, Tedde A, Guarnieri BM, Cellini E, et al. (2004) Brain-derived neurotrophic factor genetic variants are not susceptibility factors to Alzheimer's disease in Italy. Ann Neurol 55: 447-448.
- Boiocchi C, Maggioli E, Zorzetto M, Sinforiani E, Cereda C, et al. (2013) Brain-derived neurotrophic factor gene variants and Alzheimer disease: An association study in an Alzheimer disease Italian population. Rejuvenation Res 16: 57-66.
- Borroni B, Bianchi M, Premi E, Alberici A, Archetti S, et al. (2012) The brain-derived neurotrophic factor Val66Met polymorphism is associated with reduced hippocampus perfusion in frontotemporal lobar degeneration. J Alzheimers Dis 31: 243-251.
- Combarros O, Infante J, Llorca J, Berciano J (2004) Polymorphism at codon 66 of the brain-derived neurotrophic factor gene is not associated with sporadic Alzheimer's disease. Dement Geriatr Cogn Disord 18: 55-58.
- Cozza A, Melissari E, Iacopetti P, Mariotti V, Tedde A, et al. (2008) SNPs in neurotrophin system genes and Alzheimer's disease in an Italian population. J Alzheimers Dis 15: 61-70.
- Fehér A, Juhász A, Rimanóczy A, Kálmán J, Janka Z (2009) Association between BDNF Val66Met polymorphism and Alzheimer disease, dementia with Lewy bodies and Pick disease. Alzheimer Dis Assoc Disord 23: 224-228.
- Nacmias B, Piccini C, Bagnoli S, Tedde A, Cellini E, et al. (2004) Brain-derived neurotrophic factor, apolipoprotein E genetic variants and cognitive performance in Alzheimer's disease. Neurosci Lett 367: 379-383.
- Pivac N, Nikolac M, Nedic G, Mustapic M, Borovecki F, et al. (2011) Brain derived neurotrophic factor Val66Met polymorphism and psychotic symptoms in Alzheimer's disease. Prog Neuropsychopharmacol Biol Psychiatry 35: 356-362.
- Saarela MS, Lehtimaki T, Rinne JO, Huhtala H, Rontu R, et al. (2006) No association between the brain-derived neurotrophic factor 196 G>A or 270 C>T polymorphisms and Alzheimer's or Parkinson's disease. Folia Neuropathol 44: 12-16.
- Ventriglia M, Bocchio Chiavetto L, Benussi L, Binetti G, Zanetti O, et al. (2002) Association between the BDNF 196 A/G polymorphism and sporadic Alzheimer's disease. Mol Psychiatry 7: 136-137.
- Vepsäläinen S, Castren E, Helisalmi S, Iivonen S, Mannermaa A, et al. (2005) Genetic analysis of BDNF and TrkB gene polymorphisms in Alzheimer's disease. J Neurol 252: 423-428.
- Nishimura AL, Oliveira JR, Mitne-Neto M, Guindalini C, Nitrini R, et al. (2004) Lack of association between the brain-derived neurotrophin factor (C-270T) polymorphism and late-onset Alzheimer's disease (LOAD) in Brazilian patients. J Mol Neurosci 22: 257-260.
- Kunugi H, Ueki A, Otsuka M, Isse K, Hirasawa H, et al. (2001) A novel polymorphism of the brain-derived neurotrophic factor (BDNF) gene associated with late-onset Alzheimer's disease. Mol Psychiatry 6: 83-86.
- Cousin E, Mace S, Rocher C, Dib C, Muzard G, et al. (2011) No replication of genetic association between candidate polymorphisms and Alzheimer's disease. Neurobiol Aging 32: 1443-1451.
- Riemenschneider M, Schwarz S, Wagenpfeil S, Diehl J, Muller U, et al. (2002) A polymorphism of the brain-derived neurotrophic factor (BDNF) is associated with Alzheimer's disease in patients lacking the Apolipoprotein E epsilon4 allele. Mol Psychiatry 7: 782-785.
- McHughen SA, Rodriguez PF, Kleim JA, Kleim ED, Marchal Crespo L, et al. (2010) BDNF val66met polymorphism influences motor system function in the human brain. Cereb Cortex 20: 1254-1262.
- Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, et al. (2004) The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci 24: 10099-10102.
- del Toro D, Canals JM, Gines S, Kojima M, Egea G, et al. (2006) Mutant huntingtin impairs the post-Golgi trafficking of brain-derived neurotrophic factor but not its Val66Met polymorphism. J Neurosci 26: 12748-12757.
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, et al. (2003) The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 112: 257-269.
- Forero DA, Benitez B, Arboleda G, Yunis JJ, Pardo R, et al. (2006) Analysis of functional polymorphisms in three synaptic plasticity-related genes (BDNF, COMT AND UCHL1) in Alzheimer's disease in Colombia. Neurosci Res 55: 334-341.
- Zhang H, Ozbay F, Lappalainen J, Kranzler HR, van Dyck CH, et al. (2006) Brain derived neurotrophic factor (BDNF) gene variants and Alzheimer's disease, affective disorders, post-traumatic stress disorder, schizophrenia and substance dependence. Am J Med Genet B Neuropsychiatr Genet 141B: 387-393.
- Olin D, MacMurray J, Comings DE (2005) Risk of late-onset Alzheimer's disease associated with BDNF C270T polymorphism. Neurosci Lett 381: 275-278.
Relevant Topics
- Advanced Parkinson Treatment
- Advances in Alzheimers Therapy
- Alzheimers Medicine
- Alzheimers Products & Market Analysis
- Alzheimers Symptoms
- Degenerative Disorders
- Diagnostic Alzheimer
- Parkinson
- Parkinsonism Diagnosis
- Parkinsonism Gene Therapy
- Parkinsonism Stages and Treatment
- Stem cell Treatment Parkinson
Recommended Journals
Article Tools
Article Usage
- Total views: 3861
- [From(publication date):
June-2017 - Apr 02, 2025] - Breakdown by view type
- HTML page views : 3043
- PDF downloads : 818