Association Mapping of Seedling and Adult Plant Resistance for Stripe Rust Resistance in Spring Bread Wheat (Triticum aestivum L.)
Received: 20-Jun-2018 / Accepted Date: 05-Jul-2018 / Published Date: 12-Jul-2018 DOI: 10.4172/2329-8863.1000383
Keywords: Association mapping analysis; Bread wheat; SNPS; Stripe rust; Genotypes
Introduction
Wheat is the most widely grown cereal crop globally and feeds 4.5 billion people in 95 developing countries [1]. The most common species grown are Triticum aestivum L. (common wheat) and Triticum turgidum var. durum L. (durum wheat). Common wheat accounts for 95% of the total wheat consumed worldwide [2]. It is also one of the major cereal crop to ensure food security in Ethiopia. In 2013/14, Ethiopia’s wheat production covered 1.61 million hectares of land and produced 3.93 million tons of wheat, almost twice the quantity produced in 2010. However, this level of productivity is still below the global average yield (3 t/ha) principally due to yellow/stripe rust caused by Puccinia striiformis f.sp.tritici [3,4]. Most popular commercial bread wheat cultivars such as Kubsa and Dashen were susceptible to stripe rust resulting 70-100% yield loss in Ethiopia [5].
Continuous search for new sources of resistance ahead of changing pathogen and pyramiding of more resistance genes in single cultivars is important to control stripe rust and to avoid the ‘boom and bust cycle’ of cultivar performance. Field evaluation of the level of resistance of various genotypes and multi-locational disease testing of germplasm is used to obtain data to support breeding strategies aimed at broadening the genetic base of resistance in International Maize and Wheat Improvement Centre (CIMMYT) and ICARDA. On the other hand, the development of molecular markers that are closely associated with the respective resistance genes would facilitate effective and successful gene pyramiding [6].
Single-Nucleotide Polymorphism (SNP) markers seem the best to meet needs of marker-assisted management of genetic resources, and of diversity studies and marker-assisted selection in breeding programs [6]. Association mapping analysis has the potential to identify a single polymorphism within a gene that is responsible for the difference in phenotype. It has the promise of higher mapping resolution through exploitation of historical recombination events at the population level that may enable gene level mapping on non-model organisms where linkage-based approaches would not be feasible and also utilizes ancestral recombination and natural genetic diversity within a population to dissect quantitative traits and is built on the basis of linkage disequilibrium [7].
This study was carried out in order to identify resistant spring wheat genotypes against stripe rust both at seedling and adult plant stages and to identify SNP markers associated with yellow rust resistance using genome wide association mapping.
Materials and Methods
A total of 198 elite spring bread wheat genotypes and 2 checks (Pastor-2 and Attila-7) were received from ICARDA. These genotypes were planted using alpha lattice design in two replications in a plot size of 2.5 m length, placed in four rows with 0.2 m spacing between rows at Kulumsa and Meraro research station during 2015 cropping season for identifying resistance genotypes for stripe rust. Trials were managed as per the recommended agronomic management practices. Kulumsa is located in 39°09'11'' E longitudes and 08°01'10'' North latitudes with an altitude of 2200 m.a.s.l and mean annual rainfall of 820 mm. Meraro represents extreme highland and cold area. It is located at an altitude of 2990 m.a.s.l and 39°14'56'' E longitudes and 07°24'27'' North latitude.
The stripe rust assessment
Mixed races of stripe rust spore was harvested from the field during the previous growing season (September-October 2014) and maintained at Karc greenhouse and multiplied in the greenhouse using universal susceptible wheat cultivars (Morocco and Kubsa) during (July-September, 2015) and used for inoculation of the 198 elite spring bread wheat genotypes and two checks.
The 198 genotypes and two checks were evaluated against different mixed stripe rust races at adult-plant stage. Spreader rows were planted as mixtures of the most susceptible bread wheat cultivars and the dominant varieties (Morocco and Kubsa) in adjacent to the 198 elite genotypes and 2 checks on both sides of each block, bordering the trials to ensure production of sufficient inoculum to provide uniform stripe rust infection.
The inoculation of spreader row was carried out during tillering stage by spraying method and during stem elongation stage by injection methods at 50 cm interval. Spraying of stripe rust on spreader row during tillering stage was done by mixing stripe rust spore with mineral oil and then sprayed to spreader row using pumping (Knapsak spray) machine. Stripe rust injection to spreader row was conducted by mixing stripe rust spore with distil water and applied to the spreader row by injecting stem at stem elongation stage using injection syringe.
Adult-plant responses for the major infection types were recorded according to Roelfs et al. Disease severity as a percentage of covered areas was assessed following a modified Cobb’s scale. Field responses were recorded 2 times and the final scoring at soft-dough stage was considered for the AM analysis. The data on disease severity and host reaction was combined to calculate the coefficient of infection (CI) following Pathan and Park (2006), by multiplying the severity value by a value of 0, 0.2, 0.4, 0.6, 0.8 or 1.0 for host response ratings of immune(I), resistant (R), moderately resistant (MR), intermediate (M), moderately susceptible (MS) or susceptible (S), respectively.
Seedling stage: Four to five seeds of each genotype were planted in a 7 cm × 7 cm × 7 cm plastic pots. Each pot was filled with a potting mix which consists of soil, sand and compost at a ratio of 2:1:1 (v/v/v). One week after planting, when the first leaves were fully expanded, the seedlings were inoculated by spraying the most virulent and dominant varieties Kubsa/Attila and mixed-race isolates of urediospores suspended in mineral oil using an atomizer.
Inoculated plants were allowed to dry for 5 minutes and were finemisted with water and placed in a wet plastic cage with a small amount of water at the bottom. The inoculated seedlings were incubated at 10°C for 24 hours in a dew chamber with relative humidity close to 100%. Seedlings were transferred to a greenhouse with mean temperature of about 18°C at the Ethiopian Institute of Agricultural Research, Kulumsa Agricultural Research Center (KARC), greenhouse lab. Disease assessment was carried out on the 15th days after inoculation using 0-4 scale based on the infection types. Low infection types (LITs=0-2) were considered resistant and infection type=2+ as intermediate while high infection types (HITs=3-4) were rated susceptible.
Area Under Disease Progress Curve (AUDPC) was calculated in order to compare the genotypes susceptibility and resistance. The AUDPC was calculated using the midpoint rule method.
Genotyping
DNA was extracted from four to five leaves of each genotype from two weeks old seedlings at ICARDA-AGERI laboratory. The leaves were placed in 2 ml microfuge tubes and tightly tied with Parafilm. Each eppendorf containing the fresh leaves was then dried in freeze dryer at about -32°C for 72 hours. When all samples have been dried, two 4 mm diameter stainless steel grinding balls were placed in each tube and crushed using electrical mortar. DNA extraction was performed according to Ogbonnaya et al. [8] and genotyping was carried out using 1 k SNP at Trait Genetics, Germany.
Population structure
The genetic structure of the 197 genotypes including the checks was investigated using 64 unlinked SNPs markers distributed across the whole bread wheat genome with at least two loci on each wheat chromosome [9]. The remaining three elite spring bread wheat genotypes failed to amplify due to degradation of DNA.
Genetic distance between pairs of chosen markers on the same chromosome was more than 50 cm to minimize LD caused by tightly linked markers. A Bayesian clustering method was applied to identify clusters of genetically similar individuals using the software structure version 2.3.4 [9]. To infer the number of subpopulations (K), five runs for each k value from 2 to 15 were made. Both the length of burn-in period and the number of iterations were set at 105 (to minimize the effect of starting configuration) and 106 respectively.
To reach the appropriate k value, the estimated normal logarithm of the probability of fit [lnP(D)] provided in the structure output was plotted against k. This value reaches a plateau when the minimal number of groups that best describe the population substructure has been reached [9]. The output from structure was analyzed in structure harvester. The Δk statistics based on the rate of change in the logarithm of the probability of likelihood [LnP(D)] value between successive k values was used to predict the optimum number of subpopulations.
Linkage disequilibrium
Genome-wide LD analysis was performed across A, B and D genomes for the complete association mapping set. TASSEL 5.0.8 software was used to estimate LD between pairs of SNP markers as squared allele frequency correlation estimates (R2) and to measure the significance of R2 at P values ≤ 0.005 for each pair of loci on different chromosomes. Locally weighed polynomial regression (LOESS) curves were then fitted into the scatter plot using function ‘smooth spline’ of R (R Development Core Team, 2015). Only SNP markers with known chromosomal position were used in the estimation of LD.
Association mapping (AM)
The CI data from Kulumsa amd Meraro locations plus the seedling data in the greenhouse were used for the AM analysis. TASSEL version 5.0.8 was used to perform association mapping analysis. Both the General Linear Model (GLM) and Mixed Linear Model (MLM) methods were used to assess the associations. The two different GLM models: the model with no control for population structure and kinship (naïve model) and the model with population structure (the Q model) were used.
For MLM the model that considers the familial relatedness between accessions (the K model) and the model that takes into account both the population structure and the familial relatedness i.e., the Q+K model were used. The compressed mixed linear models (MLM) which takes into consideration kinship matrix (K) and population structure (Q) as a covariate and P3D algorithms to reduce computing time was the best model and therefore selected.
Results
Response of genotypes for stripe rust in field condition
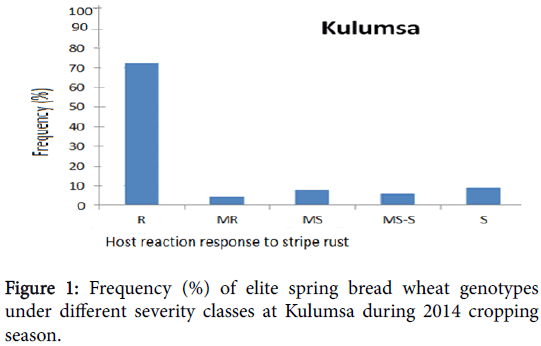
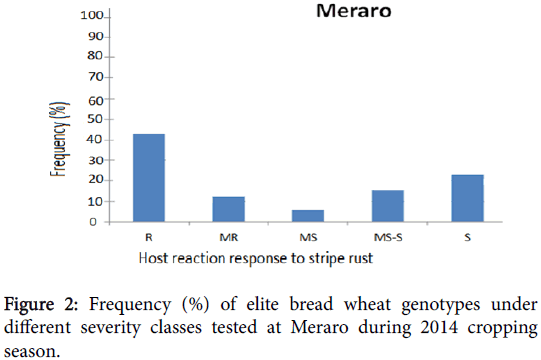
Phenotypic variation was observed at both environments for infection types and level of stripe rust severity for the 198 ICARDA elite spring bread wheat genotypes and two checks. Terminal score ranged from 0 (immune) to 100 S (highly susceptible). Reaction response to stripe rust for these genotypes at Kulumsa and Meraro locations are summarized in Figures 1 and 2 and Table 1. Continuous variation was observed for reaction response to stripe rust across both locations. More disease severity/pressure was observed at Meraro than at Kulumsa. The checks showed variable reaction responses from moderately resistant to susceptible and a severity level ranging from 10 to 90% (Table 1).
| Chromosome | Number of loci | Number of loci with position | Average Distance (cm) | LD decay |
|---|---|---|---|---|
| 1A | 416 | 191 | 4.9 | 31.4 |
| 1B | 725 | 328 | 5.3 | 25.8 |
| 1D | 296 | 79 | 2.4 | 74.1 |
| 2A | 496 | 243 | 3. 2 | 34.9 |
| 2B | 888 | 545 | 7.2 | 24.7 |
| 2D | 366 | 106 | 9.3 | 52.7 |
| 3A | 415 | 233 | 5.8 | 47.5 |
| 3B | 621 | 421 | 14.5 | 27.4 |
| 3D | 61 | 29 | - | - |
| 4A | 314 | 182 | 6.2 | 38.3 |
| 4B | 313 | 161 | 6.5 | 16.8 |
| 4D | 43 | 10 | - | - |
| 5A | 606 | 299 | 11 | 36 |
| 5B | 764 | 418 | 17.1 | 45.8 |
| 5D | 134 | 49 | 8.1 | 78.5 |
| 6A | 571 | 263 | 6.7 | 33.7 |
| 6B | 667 | 237 | 1.7 | 35.6 |
| 6D | 146 | 48 | 4 | 61.9 |
| 7A | 490 | 300 | 9.1 | 40.2 |
| 7B | 546 | 329 | 6.3 | 41.6 |
| 7D | 88 | 51 | 8.7 | 54.6 |
Table 1: Genetic marker statistics: number of markers, number of markers with position on the consensus SNPs map and the average distance between each two adjacent markers for each chromosome and LD decay based on specific chromosomes.
The frequency of these elite spring bread wheat genotypes and the checks under different severity classes at Kulumsa and Meraro is presented on Figures 1 and 2, respectively, according to the coefficient of infection (CI) score. At Kulumsa of the total 198 elite spring bread wheat genotypes and two checks evaluated, 145 (72.5%) including the check (Pastor-2 exhibited resistance reaction response (CI=0 to 20); nine genotypes (4.5%) were moderately resistant (CI=20 to 30); sixteen (8%) were moderately susceptible (CIs=30 to 40), twelve (6%) genotypes with Attila-7) were moderately susceptible to susceptible (CI=40 to 60) and 18 (9%) the remaining were susceptible (CI=60 to 100).
At Meraro, eighty-six (43%) elite genotypes exhibited resistance reaction response (CI=0 to 20); twenty-four (12%) moderately resistance (CI=20 to 30); 12 (6%) moderately susceptible (CI=30 to 40); 30 (15%) including the check pastor-2, were moderately susceptible to susceptible (CI=40 to 60) and 48 (24%) were found to be susceptible (CI=60 to 100).
After the final score 74 genotypes (37%) out of the 198 spring bread wheat genotypes showed similar reaction response at both environments, they were resistant to stripe rust (CI from 0 to 20); 27 of these genotypes had CI less than 2 at both locations and were almost immune to the disease. Disease severity development was increased gradually through time from 0 to 100% depending upon differences in stripe rust reaction response of the genotypes. AUDPC computed for each genotype varied from 0 to 2490 and from 0 to 1956 for Kulumsa and Meraro, respectively. The stripe rust disease development intensity through time and AUDPC at both locations are given in Table 1. Thirteen genotypes (6.5%) were susceptible to stripe rust (CI>60) at both locations.
Generally, the AUDPC showed that the disease severity development at Meraro was higher than at Kulumsa, which indicated the availability of more virulent races, high disease pressure and/or suitable environment at Meraro than at Kulumsa.
Seedling stage screening in greenhouse
For Kubsa and one mixed stripe rust isolates races the 200 elite spring bread wheat genotypes exhibited different reaction type the 2 checks at seedling stage. Among them, mixed stripe rust isolates were the more virulent than Kubsa isolates. Out of the elite spring bread wheat genotypes tested in the greenhouse, 53% of the genotypes showed susceptible reaction (IT=3-4) for the mixed stripe rust isolates and 43% of the genotypes showed susceptible reaction (IT=3-4) for Kubsa isolates.
Reaction of elite spring bread wheat genotypes and checks against Kubsa and mixed isolated at seedling stage is shown in Table 1. Nearly 47% of the genotypes exhibited resistance reaction response (IT=0-2), only one genotypes showed intermediate reaction (2+) for mixed stripe rust isolates and 57% were resistance for kubsa isolate. Out of 198 bread wheat genotypes tested in the greenhouse sixty-two (31%) exhibited common resistance reaction response for both (Kubsa and mixed) stripe rust isolate.
Marker coverage, population structure and linkage disequilibrium
All of the 198 elite spring bread wheat genotypes and the two checks were genotyped using 13006 SNP markers. However, 3381 (26%) SNP markers were monomorphic, 645 (5%) SNPs were of poor quality and thus were excluded from analysis. The remaining 8980 (69%) SNPs were further reduced to 8966 (68.9%) by eliminating markers with minor allele effect. Thus, 8966 (68.9%) high quality SNPs were used in association analysis for stripe rust disease resistant using the Q+K MLM and Q+GLM methods (Table 2).
| Markers | Chromosome | Position | P value | R2 | SNP | Effect |
|---|---|---|---|---|---|---|
| BS00022733_51 | 1A | 69.8 | 0.00167 | 0.039 | [A/G] | 8.41 |
| RAC875_rep_c112044_340 | 1A | 72.1 | 0.00203 | 0.037 | [A/G] | 7.79 |
| TA003955-1138 | 1A | 72.1 | 0.002 | 0.037 | [A/G] | 7.66 |
| Kukri_c30322_414 | 1A | 72.1 | 0.0021 | 0.037 | [A/G] | 7.66 |
| CAP12_c1906_217 | 1A | 72.1 | 0.00210 | 0.037 | [A/G] | 7.66 |
| wsnp_Ex_rep_c108951_9195419 | 1A | 72.1 | 0.0049 | 0.031 | [A/G] | 8.31 |
| BS00022717_51 | 2B | 82.7 | 0.00299 | 0.052 | [A/G] | 9.06 |
| BS00079621_51 | 2B | 80.4 | 0.00276 | 0.05 | [T/G] | 9.23 |
| Kukri_c19751_873 | 2B | 78.8 | 0.00279 | 0.053 | [A/G] | 9.19 |
| Kukri_c36747_195 | 5A | 13.6 | 0.00182 | 0.057 | [T/C] | 10.02 |
| Kukri_c5501_1515 | 5A | 13.6 | 0.00241 | 0.054 | [A/G] | 9.47 |
| BS00022500_51 | 5A | 13.6 | 0.00273 | 0.035 | [T/C] | 11.03 |
| BS00005311_51 | 5A | 13.6 | 0.00279 | 0.052 | [A/G] | 9.19 |
| Tdurum_contig102312_245 | 5A | 13.6 | 0.0033 | 0.033 | [T/G] | 8.98 |
| Excalibur_c7180_862 | 5A | 13.6 | 0.0031 | 0.052 | [T/C] | 8.98 |
| GENE-4859_218 | 7A | 81.9 | 0.00026 | 0.073 | [A/G] | 13.86 |
| GENE-4953_139 | 7A | 81.9 | 0.00043 | 0.067 | [A/G] | 12.86 |
| CAP7_c10133_40 | 7A | 81.9 | 0.00096 | 0.064 | [T/C] | 11.28 |
| wsnp_JD_c38071_27729378 | 7A | 81.9 | 0.00099 | 0.065 | [A/G] | 11.21 |
| BobWhite_c24063_231 | 7A | 81.9 | 0.00114 | 0.063 | [T/C] | 10.93 |
| BobWhite_c911_127 | 7A | 81.9 | 0.00126 | 0.061 | [A/G] | 10.72 |
| RAC875_c32895_304 | 7A | 81.9 | 0.00126 | 0.062 | [T/C] | 10.74 |
| Kukri_c39894_178 | 7A | 81.9 | 0.00131 | 0.061 | [A/G] | 10.67 |
| Kukri_rep_c70389_57 | 7A | 81.9 | 0.00138 | 0.061 | [A/G] | 10.56 |
| BS00030911_51 | 7A | 81.9 | 0.00146 | 0.06 | [A/G] | 10.45 |
| RAC875_rep_c117475_289 | 7A | 81.9 | 0.00148 | 0.06 | [T/C] | 10.43 |
| BS00066651_51 | 7A | 81.9 | 0.00154 | 0.059 | [T/C] | 10.34 |
| Kukri_c27692_822 | 7A | 81.9 | 0.00154 | 0.059 | [T/C] | 10.34 |
| wsnp_Ra_c31751_40835513 | 7A | 81.9 | 0.00154 | 0.059 | [A/G] | 10.34 |
| wsnp_JD_c18167_16742264 | 7A | 81.9 | 0.00289 | 0.051 | [A/G] | 9.12 |
| BS00031923_51 | 7B | 129.6 | 0.00062 | 0.064 | [A/G] | 12.13 |
| Tdurum_contig82534_311 | 7B | 129.6 | 0.0008 | 0.06 | [A/C] | 11.61 |
| Tdurum_contig93425_441 | 7B | 129.6 | 0.00095 | 0.06 | [A/G] | 11.3 |
| Tdurum_contig82534_237 | 7B | 129.6 | 0.00099 | 0.06 | [A/G] | 11.19 |
| Tdurum_contig28884_460 | 7B | 129.6 | 0.00145 | 0.056 | [A/C] | 10.45 |
| Tdurum_contig67161_99 | 7B | 129.6 | 0.00155 | 0.054 | [T/C] | 0.32 |
| Tdurum_contig28884_379 | 7B | 129.6 | 0.00168 | 0.053 | [T/G] | 10.16 |
| Tdurum_contig52079_1174 | 7B | 129.6 | 0.00171 | 0.053 | [T/C] | 10.12 |
Table 2: Chromosome location, P values, and R2, values of significantly associated (SNP) markers with stripe rust (YR) resistance at Kulumsa, indicate the year.
The genetic framework map of the whole 21 wheat chromosomes was constructed using the 8,966 polymorphic SNP markers based on the consensus SNPs map previously produced and resulting an average of 427 markers per chromosome. However, the marker density for the D genome was relatively poor that were 53.14 markers per chromosome. In total, the markers spanned a genetic distance of 31,112 cm with an average density of 3.47 cm per marker.
Population structure
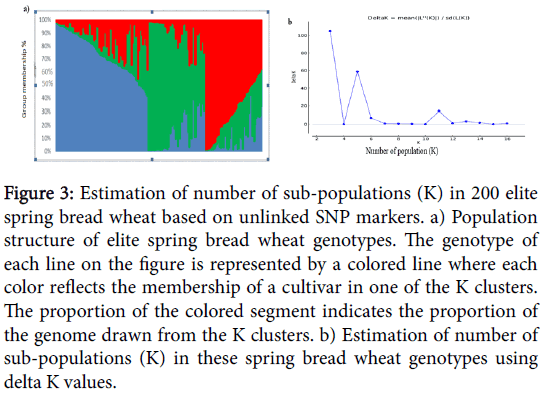
Analysis of population structure showed that the logarithm of the data likelihood (Ln P(D)) on average continued to increase with increasing numbers of assumed subpopulations (K) from 2 to 15 with exception of the depression at K5, K6 and K7. However, these significant changes at higher K values do not truly reflect the actual number of sub populations. The ad-hoc quantity based on the second order rate of change in the log probability (DK) showed a clear peak at K=3, which confirmed that a K value of three was the most probable prediction for the number of subpopulations.
Analysis of population structure showed that the logarithm of the data likelihood (Ln P(D)) on average continued to increase with increasing numbers of assumed subpopulations (K) from 2 to 15 but showed a plateau after K=3 and thereafter tended to fluctuate (Figure 3). Thus, the best probability for k value was determined to be 3, which appeared to be the most stable prediction of LnP(D) over the fifteen repetitions.
Figure 3: Estimation of number of sub-populations (K) in 200 elite spring bread wheat based on unlinked SNP markers. a) Population structure of elite spring bread wheat genotypes. The genotype of each line on the figure is represented by a colored line where each color reflects the membership of a cultivar in one of the K clusters. The proportion of the colored segment indicates the proportion of the genome drawn from the K clusters. b) Estimation of number of sub-populations (K) in these spring bread wheat genotypes using delta K values.
Marker-Trait associations and linkage disequilibrium
In this study, markers associated with resistance to stripe rust both at adult plant stage under field condition and seedling stage in greenhouse were identified in elite spring bread wheat genotypes. Of those 8966 (69%) polymorphic SNP markers, 4522 (50.4%) were of known position on the consensus map in which 1711, 2439 and 372 were specific to the A, B, and D genomes, respectively (Table 2).
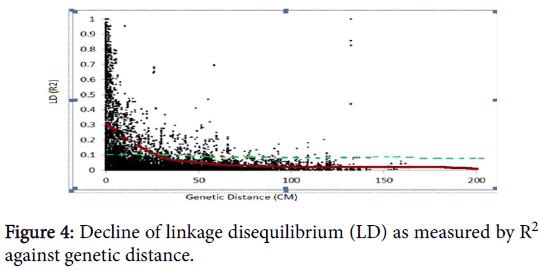
Another 4444 (49.6%) markers have no position on the consensus map. Chromosomes with the largest number of markers were 2B (545 markers) followed by 3B (421 markers) and 5B (418 markers). Chromosomes 3D and 4D showed the least number of markers, 29 and 10, respectively (Table 2). A scatter plot of LD (R2) values versus genetic distances between all markers across the genome abruptly declined to 0.2 within less than 50 cm when all mapped SNPs with chromosome position were analyzed (Figure 4). This result is expected for self-pollinated crop species such as wheat. The estimated genomewide LD decay in this study ranged from 0-50 cm.
Fifty-three SNP markers in ten genomic regions were significantly (P<0.005) associated with resistant to stripe rust disease and detected on chromosomes 1A, 2A, 2B, 2D, 3B, 4B, 4D, 5A, 7A and 7B (Tables 3 and 4).
| Markers | Chromosome | Position | P-value | R2 | SNP | effect |
|---|---|---|---|---|---|---|
| wsnp_Ex_rep_c69738_68695568 | 1A | 65.9 | 0.0031 | 0.05 | [A/G] | 8.97 |
| wsnp_Ku_c33374_4287754 | 2A | 4 | 0.00078 | 0.06 | [A/G] | 11.67 |
| BS00093111_51 | 2D | 13.9 | 0.00026 | 0.07 | [T/C] | 13.9 |
| wsnp_Ra_c41135_48426638 | 3B | 5.4 | 0.00214 | 0.05 | [T/C] | 9.69 |
| Kukri_c35140_75 | 4B | 58 | 0.00129 | 0.06 | [A/G] | 10.68 |
| wsnp_Ex_c25373_34639805 | 4B | 61 | 0.00146 | 0.05 | [T/C] | 10.43 |
| Excalibur_c17607_542 | 4B | 56.4 | 0.0016 | 0.06 | [A/G] | 10.26 |
| wsnp_JD_c1549_21853 | 4B | 56.4 | 0.00162 | 0.06 | [T/C] | 10.24 |
| RFL_Contig3563_1130 | 4B | 61 | 0.00223 | 0.05 | [A/G] | 9.61 |
| Excalibur_c56787_95 | 4B | 56.4 | 0.0024 | 0.05 | [A/C] | 9.48 |
| wsnp_Ex_c4358_7854194 | 4B | 61 | 0.00274 | 0.05 | [T/C] | 9.22 |
| wsnp_Ex_c12450_19850925 | 4D | 56.5 | 0.00214 | 0.05 | [T/C] | 9.69 |
| RAC875_rep_c105718_304 | 4D | 47 | 0.0005 | 0.07 | [A/C] | 12.55 |
| RAC875_rep_c105718_585 | 4D | 47 | 0.0009 | 0.06 | [A/C] | 11.37 |
Table 3: Chromosome location, P values, and R2, values of significantly associated (SNP) markers with stripe rust resistance at Meraro, indicate the year.
| Markers | Chromosome | Position | P value | R2 | SNP | Effect |
|---|---|---|---|---|---|---|
| BS00018764_51 | 7B | 117.1 | 0.0031 | 0.062 | [A/G] | 8.80 |
| Tdurum_contig31235_99 | 3A | 162.9 | 0.0024 | 0.068 | [T/C] | 9.13 |
| IAAV3570 | 2A | 72.1 | 0.0032 | 0.066 | [A/G] | 9.05 |
| RAC875_c102123_187 | 2A | 63.2 | 0.0031 | 0.066 | [A/G] | 9.00 |
| tplb0043c20_1046 | 3B | 14.7 | 0.0026 | 0.054 | [A/G] | 8.70 |
| BS00064002_51 | 4B | 82.7 | 0.0031 | 0.064 | [A/G] | 7.00 |
| BS00106571_51 | 4D | 80.4 | 0.0018 | 0.069 | [A/G] | 7.50 |
| Kukri_rep_c101341_425 | 2B | 78.8 | 0.0020 | 0.065 | [A/G] | 7.35 |
| wsnp_CAP12_rep_c4379_1995966 | 5A | 13.6 | 0.0032 | 0.065 | [T/C] | 8.25 |
| BS00093841_51 | 1B | 41.3 | 0.0034 | 0.037 | [A/G] | 7.35 |
| Excalibur_rep_c104532_80 | 5A | 13.6 | 0.0020 | 0.076 | [T/C] | 9.15 |
| Excalibur_rep_c67448_528 | 3B | 13.6 | 0.0020 | 0.065 | [T/G] | 8.80 |
| Excalibur_c7610_143 | 3A | 162.9 | 0.0020 | 0.068 | [T/G] | 8.50 |
Table 4: Chromosome location, P values, R2, SNPs, and effect of significantly associated (SNP) markers with stripe rust resistance at seedling stage for mixed isolates.
Of the 53 SNP markers, 39 SNPs in five genomic regions located on wheat chromosome 1A, 2B, 5A, 7A and 7B were associated with resistant to stripe rust evaluated at Kulumsa and fourteen SNPs in six genomic regions: 1A, 2A, 2D, 3B, 4B and 4D were associated with resistant reaction to stripe rust evaluated at Meraro (Figure 5).
For seedling stage test 21 SNP markers in ten genomic regions located on wheat chromosomes 1B, 2A, 2B, 3A, 3B, 4B, 4D, 5A, 6B and 7B were associated with resistant to stripe rust for seedling stage condition tested in greenhouse. Almost all listed SNPs markers were significantly (p>0.005) linked to stripe rust resistance in elite spring bread wheat at seedling stage (Tables 4 and 5).
| Markers | Chromosome | Position | P value | R2 | SNP | Effect |
|---|---|---|---|---|---|---|
| Tdurum_contig31201_380 | 1B | 41.3 | 0.0033 | 0.039 | [A/G] | 9.44 |
| RAC875_c62325_320 | 6B | 72.1 | 0.0025 | 0.037 | [A/C] | 8.23 |
| Kukri_c62848_106 | 1B | 41.3 | 0.0030 | 0.037 | [A/G] | 9.45 |
| Excalibur_rep_c67448_528 | 3B | 13.6 | 0.0020 | 0.065 | [T/G] | 8.80 |
| Ku_c31251_565 | 1B | 41.3 | 0.0017 | 0.037 | [A/G] | 8.72 |
| BS00093841_51 | 1B | 41.3 | 0.0034 | 0.037 | [A/G] | 7.35 |
| BS00106571_51 | 4D | 80.4 | 0.0018 | 0.069 | [A/G] | 7.50 |
| BS00060029_51 | 3A | 101.7 | 0.0030 | 0.031 | [T/C] | 8.44 |
Table 5: Chromosome location, P values, R2, SNPs and effect of significantly associated (SNP) markers with stripe rust resistance at seedling stage for Kubsa isolates.
The percentage of phenotypic variation explained (R2) by the SNPs markers for resistance to stripe rust at seedling stage ranged from 3.1 to 7.6%. These were located on chromosomes 1B, 2A, 2B, 3A, 3B, 4B, 4D, 5A, 6B and 7B. Of these, the locus on 5A was strongly associated with stripe rust resistance at seedling stage explaining the phenotypic variation of 7.6% preceded by 6.9% on 5A and 4D. Some markers are common on both (Kubsa and mixed) isolates; BS00093841_51, BS00106571_51 and Excalibur_rep_c67448_528.
Discussion
Phenotypic and genotypic variability for resistance to stripe rust
Knowledge of the genetic basis of stripe rust resistance is very essential because it will facilitate the incorporation of resistance genes into high yielding and locally adapted bread wheat cultivars and release new stripe rust resistant varieties for large scale production by end users/ farmers. According to Chen et al. [10] considerable numbers of virulent races of the stripe rust have appeared through somatic recombination or mutation. Somatic recombination plays a major role in variation of stripe rust populations and formation of new races with combinations of previously existing virulence’s.
Ayele et al. [3] also reported that stripe rust isolates with virulence factors on Yr8 and Yr9 were detected in Ethiopia. In Ethiopia, stripe rust often causes substantial yield loss in higher elevation (>2400 m.a.s.l), however, in 2010, the disease was wide spread reaching even to the lower elevations as a result of virulence to Yr27 present in the most widely grown cultivar, ‘Kubsa’. The country previously experienced yellow rust epidemics resulting in significant yield losses to farmers [3].
This study was undertaken with the objectives of screening 198 elite spring bread wheat genotypes from ICARDA along with two checks under field (adult plant stage) and greenhouse (seedling stage) conditions for resistance against Ethiopian phatho types of stripe rust; identifying a set of markers associated with resistant genes against the prevailing virulent stripe rust races in Ethiopia.
Results of these testing in 2015 revealed that many of the elite spring bread wheat genotypes (72.5% at Kulumsa, 43% at Meraro, 47% for mixed isolates and 31% for kubsa isolates) showed resistance reaction responses to stripe rust disease based on coefficient of infection (CI). Seventy-two genotypes (36%) showed resistant reaction at both locations in field condition for adult plant stage (CI<20). Thirteen genotypes (6.5%) were immune to the disease at both locations (CI=0). In general, higher disease severity level was observed at Meraro as compared to that at Kulumsa (mean CI of 36.1 vs 18.9).
The AUDPC result also confirmed the availability of more disease severity/pressure and suitable environment for stripe rust development at Meraro than at Kulumsa (mean AUDPC of 567.9 vs 371.4). This may be attributed to variation of environmental conditions that favor the incidence, level of disease expressions and presence of more stripe rust races and greater rust pressure at Meraro. In fact, Meraro’s environment is very cool with high humidity that is suitable for stripe rust spore germination and multiplication. Chen [11] reported that high humidity with cool environment and low temperature promotes stripe rust disease by favoring spore germination. Several sources of durable stripe rust resistance have been reported in wheat lines from Europe, Northwest USA, and China and in cultivars released from CIMMYT.
Wang et al. [12] indicated that field resistance in the CIMMYT wheat Pavon-76 which has been grown in Ethiopia for the last many decades remained effective under high stripe rust pressure. Pavon-76 contains three to four genes for APR that are different from Yr18. Two QTLs in Pavon-76 have been designated as Yr29 (chromosome 1BL) and Yr30 (chromosome 3BS). Host plant resistance is the most economically effective option to manage stripe rust in developing countries.
According to Tadesse et al. [5] most of the spring bread wheat genotypes introduced to Ethiopia from CIMMYT and ICARDA possess adult plant resistance to stripe and leaf rust based on several genes with minor effects, there is significant diversity for genes that have minor to intermediate additive effects on stripe rust resistance; in the case of seedling stage test sixty-two (31%) of the tested genotypes were resistance for both isolates (Kubsa and Mixed) (Table 1). There were more susceptible genotypes in the mixed isolate than Kubsa isolate, these mostly true the mixed races would attack more genotypes than one single race; due to more genes would be attack by more race than single race.
Marker-trait associations (MAT)
One of the major factors in the success of AM analysis is good marker coverage of the genome because sparse coverage reduces the power for marker identification [13] and results from AM are strongly influenced by the choice of germplasms, size of the population under study [14]. There are several examples of association mapping studies for disease resistance QTL discovery. Crossa et al. [15] used 813 DArT and 530 SSR and sequence tagged site (STS) markers on 170 CIMMYT wheat germplasms for AM studies.
Neumann et al. [16] used 574 DArT markers for AM studies on 96 winter wheat germplasms while Emebiri et al. [17] employed 395 DArT markers for AM studies using 91 synthetic hexaploid wheat germplasms. Mulki et al. used 667 DArT markers to identify known and potentially new genomic regions associated with resistance to soil borne pathogens in synthetic hexaploid wheat and Joukhadar et al. [13] used 2518 DArT markers for AM studies of resistance to the five most destructive pests on 134 wheat genotypes.
Tadesse et al. [5] employed 3051 DArT markers for AM studies using 167 winter wheat cultivars and elite genotypes and Habtemariam et al. [18] used 2590 SNP markers for GWAS on 181 synthetic hexaploid wheat genotypes.
In the current study, 8,966 SNP markers were polymorphic, of which 4522 (50.4%) were of known map position and covered about 311048 cm with an average distance of 3.47 cm, a comparatively greater coverage than previously reported in other studies. Generally, the power of association mapping depends on accurate estimation of the population structure using the admixture model to avoid type I errors [9].
In this study, the result obtained using structure 2.3.4 software indicated that subpopulations exist in the association panel and three subpopulations were adequately separated into appropriate clusters. Depending on the diversity of the genotypes different numbers of subpopulations have been reported. Accordingly, Sukumaran et al. reported two subpopulations for Genome-wide association study; Gurung et al. [19] identified six subpopulations for GWAS Study of Spring Wheat; Habtemariam et al. [18] reported eight subpopulations while Joukhadar et al. [13] identified six subpopulations.
The extent of LD in these elite spring bread wheat genotypes was examined, considering all pairs of SNP markers. The general trend was an extremely high LD with a slight decline even at intervals extending over 50 cm. A medium range up to 30-40 cm was reported by Crossa et al. [15] and Dreisigacker et al. [20]. Smaller ranges up to 20 cm have also been reported [17,21]. Ranges covering wider distances such as 10-100 cm were also reported by Joukhadar et al. [13].
Successful application of MAS in traditional wheat breeding programs requires the identification of molecular markers tightly linked to the gene of interest (Figure 6). Furthermore, since selection for resistance to stripe rust is difficult, closely linked markers provide an alternative means for the selection of resistant gene(s)/QTLs in breeding programs in the absence of pathogens [22-27].
Conclusion
Using MLM corrected for population structure and familial relatedness adjusted for false discovery rate at P-values of ≤ 0.005, a total of 53 SNP markers for adult plant stage, a total of 21 SNP markers for seedling stage conditions were identified respectively in this study. These were significantly associated with 10 different QTLs conferring resistance to stripe rust and were located on different wheat chromosomes 1B, 2A, 2B, 3A, 3B, 4B, 4D, 5A, 6B and 7B. The SNP markers linked to QTLs for resistance on 7AL (GENE-4859_218 and GENE-4953_139) and 2DL (BS00093111_51) were highly significant and explained up to 6.7 and 7.3% of the variation for resistance, respectively. The percent variation explained by these SNP markers is quite high, suggesting that both can be major QTLs and/or major genes responsible for coding stripe rust resistance. Earlier studies have reported different QTLs and genes associated with stripe rust resistance. Maccaferri et al. reported markers on several chromosome regions (1B, 2A, 2B, 2D, 3B, 3D, 4A, 4B, 4D, 5A, 5B, 5D, 6B, 7A, 7B, and 7D) that were significantly associated with stripe rust resistant genes/quantitative trait loci (QTLs). In this study, several significant SNP markers were found in regions where stripe rust resistance genes are located (on wheat chromosomes 1A, 2A, 2B, 2D, 3B, 4B, 4D, 5A, 7A and 7B). The locus on chromosome 1AL between 65.9-72.1 cm has not been reported so far and may be a new gene for yellow rust resistance. The significantly associated markers in the current study could be used for marker assisted selection after validation analysis using a set of different elite spring wheat genotypes.
Acknowledgements
This study was carried out through the financial support from Ethiopian Institute of Agricultural Research (EIAR) and International Centre for Agricultural Research in Dry Areas (ICARDA).
References
- Braun HJ, Atlin G, Payne T (2010) Multi-location testing as a tool to identify plant response to global climate change. Climate Change and Crop Production 1: 115-138.
- Randhawa HS, Asif M, Pozniak C, Clarke JM, Graf RJ, et al. (2013) Application of molecular markers to wheat breeding in Canada. Plant Breeding 132: 458-471.
- Ayele B, Stubbs RW, Van Ginkel M, Getinet G (1990) Identification of resistance Genes to Puccinia striiformis in seedlings of Ethiopian and CIMMYT bread wheat varieties and lines. Neth J Plant Pathol 96: 199-210.
- Singh RP, Nelson JC, Sorrells ME (2000) Mapping Yr28 and other genes for resistance to stripe rust in wheat. Crop Science 40: 1148-1155.
- Tadesse W, Ogbonnaya FC, Jighly A, Nazari K, Rajaram S, et al. (2014) Association mapping of resistance to yellow rust in winter wheat cultivars and elite genotypes. Crop Science 54: 607-616.
- Khan MH, Bukhari A, Dar ZA, Rizvi SM (2013) Status and strategies in breeding for rust resistance in wheat. Agricultural Sciences 4: 292.
- Ersoz ES, Yu J, Buckler ES (2007) Applications of linkage disequilibrium and association mapping in crop plants. In Genomics-Assisted Crop Improvement, pp: 97-119.
- Ogbonnaya FC, Imtiaz M, Bariana HS, McLean M, Shankar MM, et al. (2008) Mining synthetic hexaploids for multiple disease resistance to improve bread wheat. Australian Journal of Agricultural Research 59: 421-431.
- Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945-959.
- Chen X, Moore M, Milus EA, Long DL, Line RF, et al. (2002) Wheat stripe rust epidemics and races of Puccinia striiformis f. sp. tritici in the United States in 2000. Plant Disease 86: 39-46.
- Chen XM (2005) Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Canadian Journal of Plant Pathology 27: 314-337.
- Wang L, Ma J, Zhou R, Wang X, Jia J (2002) Molecular tagging of the yellow rust resistance gene Yr10 in common wheat, PI 178383 (Triticum aestivum L.). Euphytica 124: 71-73.
- Joukhadar R, El-Bouhssini M, Jighly A, Ogbonnaya FC (2013) Genome-wide association mapping for five major pest resistances in wheat. Molecular Breeding 32: 943-960.
- Kulwal P, Ishikawa G, Benscher D, Feng Z, Yu LX, et al. (2012) Association mapping for pre-harvest sprouting resistance in white winter wheat. Theoretical and Applied Genetics 125: 793-805.
- Crossa J, Burgueno J, Dreisigacker S, Vargas M, Herrera-Foessel SA, et al. (2007) Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics.
- Neumann K, Kobiljski B, DenÄić S, Varshney RK, Börner A (2011) Genome-wide association mapping: a case study in bread wheat (Triticum aestivum L.). Molecular Breeding 27: 37-58.
- Emebiri LC, Oliver JR, Mrva K, Mares D (2010) Association mapping of late maturity α-amylase (LMA) activity in a collection of synthetic hexaploid wheat. Molecular Breeding 26: 39-49.
- Zegeye H, Rasheed A, Makdis F, Badebo A, Ogbonnaya FC (2014) Genome-wide association mapping for seedling and adult plant resistance to stripe rust in synthetic hexaploid wheat. PLoS One 9: e105593.
- Gurung S, Mamidi S, Bonman JM, Xiong M, Brown-Guedira G, et al. (2014) Genome-wide association study reveals novel quantitative trait loci associated with resistance to multiple leaf spot diseases of spring wheat. PLoS One 9: e108179.
- Dreisigacker S, Kishii M, Lage J, Warburton M (2008) Use of synthetic hexaploid wheat to increase diversity for CIMMYT bread wheat improvement. Australian Journal of Agricultural Research 59: 413-420.
- Chao S, Zhang W, Dubcovsky J, Sorrells M (2007) Evaluation of genetic diversity and genome-wide linkage disequilibrium among US wheat (Triticum aestivum L.) germplasm representing different market classes. Crop Science 47: 1018-1030.
- Philippou OA (2007) Targeting quantitative trait loci for adult plant stripe rust resistance in wheat.
- Badebo A (2002) Breeding bread wheat with multiple disease resistance and high yield for the Ethiopian highlands: broadening the genetic basis of yellow rust and tan spot resistance. Cuvillier Verlag, Goettingen, Germany.
- Chen XM (2007) Challenges and solutions for stripe rust control in the United States. Australian Journal of Agricultural Research 58: 648-655.
- CSA (Central Statistical Agency) (2014) The 2013/14 agricultural sample survey, report on area and production of crops. Addis Ababa, Ethiopia.
- ICARDA (International Center for Agricultural Research in the Dry Areas). (2013) Tackling the threat of stripe rust in Ethiopia. ICARDA Project brief.
- Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Bhavani S, et al. (2011) The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annual Review of Phytopathology 49: 465-481.
Citation: Shewaye Y, Taddesse W (2018) Association Mapping of Seedling and Adult Plant Resistance for Stripe Rust Resistance in Spring Bread Wheat (Triticum aestivum L.). Adv Crop Sci Tech 6:383. DOI: 10.4172/2329-8863.1000383
Copyright: © 2018 Shewaye Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 3628
- [From(publication date): 0-2018 - Apr 03, 2025]
- Breakdown by view type
- HTML page views: 2683
- PDF downloads: 945