Research Article Open Access
Antibiotic Resistance in Extended Spectrum Beta-Lactamases (Esbls) Salmonella Species Isolated from Patients with Diarrhoea in Calabar, Nigeria
Odafe James Oghenevo1, Bassey Enya Bassey1,2*, Nchawa Yangkam Yhiler1, Useh Monday Francis1 and Okocha-Ejeko Angela2
1Department of Medical Laboratory Science, Faculty of Allied Medical Science, University of Calabar, Calabar, Nigeria
2World Health Organization, UN HOUSE, Plot 617/618, Central Area District, FCT, Abuja, Nigeria
- *Corresponding Author:
- Bassey Enya Bassey
Department of Medical Laboratory Science
Faculty of Allied Medical Science
University of Calabar, Calabar, Nigeria
Email: yankgam@yahoo.com
Received Date: June 27, 2016; Accepted Date: July 16, 2016; Published Date: July 20, 2016
Citation: Oghenevo O, Bassey B, Yhiler N, Francis U, Angela O (2016) Antibiotic Resistance in Extended Spectrum Beta-Lactamases (Esbls) Salmonella Species Isolated from Patients with Diarrhoea in Calabar, Nigeria. J Clin Infect Dis Pract 1:107. doi: 10.4172/2476-213X.1000107
Copyright: © 2016 Bassey BE, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Clinical Infectious Diseases & Practice
Abstract
Objective: This study seeks to evaluate the prevalence of Salmonella producing ESBLs strains by phenotypic methods and their profile of drug resistance amongst patients with diarrhoea due to gastroenteritis and enteric Fever in Calabar, Nigeria.
Methods: Stool samples were collected from 256 patients in Calabar with diarrhoea due to enteric fever and gastroenteritis. They were examined for Salmonella infection. Isolation and detection of Salmonella species was done following the standard ISO 6579:2002/Amd2007 method. Modified double disc synergy and phenotypic confirmatory tests were used to determine ESBL-producing Salmonella species.
Results: Salmonella isolates were recovered from 44 (17.2%) stool samples; 24 (9.4%) were ESBLs producers and 20 (7.8%) were non-ESBLs producers. Eight (33.3%) and 3 (15.0%) strains of both ESBL producing and nonESBL producing respectively, demonstrated resistance against 7 of the 8 antibiotics used in this study. Resistance against 3rd generation cephalosporin was observed in 34 (77.3%) of the Salmonella strains against ceftazidime, 26 (59.1%) against cefotaxime while all 44 (100%) Salmonella strains were resistant against ceftriaxone.
Conclusion: The presence of ESBL Salmonella amongst isolated strains should not be overlooked. We recommend continuous surveillance of antimicrobial resistant strains and the rational use antimicrobials agents
Keywords
Salmonella; Antibiotic susceptibility; Multidrug resistance; ESBL; Double disk synergy; Calabar-Nigeria
Abbreviations:
ESBL: Extended Spectrum Beta Lactamase; UCTH: University of Calabar Teaching Hospital; GTH: General Hospital Calabar; NNH: Nigerian Navy Hospital Calabar
Introduction
The emergence and spread of antibiotic resistant strains particularly the detection of the Extended Spectrum Beta-lactamases (ESBLs) Salmonella species, is fast becoming an emerging world threat [1]. ESBLs frequently occur in K. pneumoniae but recent studies have increasingly reported its occurrence in the Salmonella specie and other gram-negative organisms [2,3]. Such Salmonella strains pose significant health problems worldwide by virtue of their acquisition of resistance against most beta-lactam antibiotics, which were previously used for treatment of Salmonella infections.
ESBLs are enzymes capable of hydrolyzing penicillin, cephalosporin and oxyiminino-β-lactam compounds (i.e. cefuroxime, third generation cephalosporin and aztreonam) with cephamycins and carbapenems as exceptions. Most ESBLs belong to the Ambler class “A” β-lactamases and are inhibited by β-lactamase inhibitors (clavulanate, sulbactam and tazobactam) [1]. ESBLs are plasmid-mediated and as a result, are easily transmitted among members of the Enterobacteriaceae family [2].
This potential exacerbates the spread of resistance against β lactams and other commonly used antibiotics including quinolones and aminoglycosides [4]. This has further limited the therapeutic options available and hence complicated the treatment and management of infections by such organism.
The determination of antibiotic susceptibility pattern and multidrug resistant pattern of infectious organisms is therefore necessary to provide a vivid guide for physicians to make informed drug choices during the management and treatment of patients. It is also important in ensuring the effective and quick treatment of Salmonella infections as well as other infections without aggravating the illness. This minimizes antibiotic resistance and cost of treatment [5].
However, there is a dearth of data on not only the Salmonella producing ESBL specie but also their multi drug resistance pattern amongst human subjects. The aim of this study was therefore to evaluate the prevalence of Salmonella producing ESBLs strains by phenotypic methods and their profile of drug resistance amongst patients with diarrhoea due to gastroenteritis and enteric fever in Calabar, Nigeria.
Methods
This study was carried out within the Calabar Municipal Local Government Area of Cross River State, Nigeria. Calabar has an area of 142 km² and a population of 179,392 by the 2006 census. The average annual rainfall is about 3070 mm with concentration between March and November while the dry season lasts from November to March. Calabar municipality is surrounded by large expanse of water bodies, yet faced with the problem of potable and good quality water. Most of the inhabitants of Calabar are traders [6].
Study design
This was a descriptive study, designed to investigate the presence of ESBLs among Salmonella strains isolated from patients attending three major hospitals within Calabar municipal. The sample size used in this study was calculated based on the formula by WHO using an expected prevalence of 16% with an absolute precision of 0.05 [7,8].
Sample collection
Stool samples were collected from 256 diarrhoeal patients between December 2014 and May 2015. Out of which, 42 attended the Calabar General Hospital while 80 and 134 patients were from the University of Calabar Teaching Hospital, and the Nigerian Navy Hospital, Calabar respectively. Faecal samples were collected from patients prior to any antibiotic therapy. Samples were collected in labeled sterile universal containers containing glycerol saline and were transported to the Laboratory for bacteriological analysis within 4 hours [9].
Ethical consideration
The protocol for this study was approved by the Ethical Committees of the selected hospitals in this study. Approval was also obtained from the Cross River State Ministry of Health, conveyed via CRS/MH/ CGSE-H/018/Vol/123 and the Health Research Ethical Committee of University of Calabar. Consent was sort from all study participants and guardians for those bellow 16 years of age prior to the collection of samples.
Isolation and identification of Salmonella species
The isolation and detection of Salmonella from the experimental samples was carried out based on the traditional ISO 6579: 2002/Amd: 2007 method (ISO, 2002). In the pre-enrichment stage, a 1 in 10 dilution of the stool sample was made in 10% modified buffered peptone water to promote the growth of all members of Enterobacteriaceae and to recover injured cells. This was carefully mixed and incubated at 35°C for 24-48 hours. Secondly, 0.1 ml of the pre-enrichment homogenate was transferred into 10 ml of Modified Semi-solid Rappaport Vassiliadis (a selective enrichment medium for Salmonella ). Following incubation at 40-41°C for 24 hours, streaking on Salmonella -Shigella (SS) agar, MacConkey’s agar (MAC) and Xylulose Lysine Deoxycholate Agar (XLDA) selectively plated a loopfull. This was incubated for 18-24 hours at 35-37°C. All the bacteriological media as well as reagents and some materials required were obtained from Hardy Diagnostics (HD) and BD Difco.
Confirmation of Salmonella isolates
Prior to the further confirmation of presumptive Salmonella isolates, typical Salmonella colonies on selective solid plates were subcultured on nutrient agar (NA) to obtain pure and distinct colonies and the isolated Salmonella strains were confirmed by various biochemical reactions [10]. The colonies were then picked by means of a sterile needle, stabbed and streaked on pre-prepared Triple Sugar Iron Agar (TSIA) and Christensen agar (CA) slants. Alkaline slope/ acid butt, with or without the production of H2S and gas, on TSIA slant and Urease negative on Christensen agar slant were suggestive of Salmonella . Further confirmation of Salmonella species was conducted based on the standard biochemical techniques in order to identify the isolates, which belonged to the genus Salmonella . This involved the use of Lysine Decarboxylation (LCD) test, β-galactosidase test, Acetone production test and Indole production test. Confirmation was further supplemented by using commercially available polyvalent Salmonella antisera kit specific for all group and type-factor Salmonella antigens. Here, a loop full from Salmonella isolates that satisfy all the confirmation procedures were then emulsified with a drop of normal saline (0.85% NaCl) on a microscopic glass slide [11]. The preparation was gently stirred and observed for auto-agglutination. If there was no auto-agglutination, a drop of the polyvalent antisera was added and gently agitated by rocking back and forth and observed for agglutination. Those that showed agglutination were considered to belong to the genus Salmonella . Those isolates that showed autoagglutination were not suitable for Salmonella confirmation.
Antimicrobial susceptibility testing
Antimicrobial susceptibility was determined by Kirby-Bauer disc diffusion method based on the Clinical and Laboratory Standards Institute (CLSI) in Mueller-Hinton agar plates with commercially available antibiotic discs (Hardy Disc-HDx) [12]. The confirmed Salmonella isolates were cultured on pre-prepared Brain Heart Infusion (BHI) agar (HARDY Diagnostics) plates and incubated at 37oC for 24 hours in to obtain confluent growth for sensitivity test. A loop-full of isolates from BHI plates was dispensed in sterile normal saline to match the 0.5 McFarland Turbidity Standard (1.0 × 108 CFU/μl) for sensitivity tests as described elsewhere [13]. 100 μl of the bacterial suspension was then inoculated on the iso-sensitivity test agar plates, and the excess was siphoned by means of sterile Pasteur pipettes. The plates were allowed to dry at room temperature in a laminar flow. The pre-determined antibiotic disks were then dispensed into the bacterial lawn by means of a sterile pair of forceps and gently pressed to ensure complete contact with the agar. The discs were positioned 15 mm way from the edge of the plate and 25 mm away from each other. The plates were incubated at 35-37°C for 18 to 24 hrs. The diameters of the zones of inhibition were read and interpreted in accordance with standards approved by the Clinical and laboratory standard institute [14]. The antibiotic discs used included: chloramphenicol (30 μg), sparfloxacin (5 μg), amoxicillin (30 μg), gentamicin (10 μg), ceftriaxone (30 μg), cefotaxime (30 μg), ceftazidime (30 μg) and amoxicillin/clavulanic acid (25/5 μg).
Screening for ESBLs producing Salmonella strains
Salmonella isolates that showed resistance against third generation cephalosporin by disc diffusion method were selected for further detection of ESBL production by double disc synergy test (DDST) [15]. A disc of amoxillin-clavulanic acid (30 μg) was placed on the centre of the Muller-Hinton agar (HardyDiagnostics) plate, which was previously inoculated with resistant strain. Each cephalosporin disc of ceftriaxone-30 μg, cefotaxime-30 μg and ceftazidime-30 μg was placed around the amoxillin-clavulanic acid disc 20 mm apart and incubated for 18-24 hours at 37°C. A clear extension of the edge of the inhibition zone of any of the antibiotics towards the central disc (amoxillinclavulanic acid) disc was interpreted as positive for ESBL production [16]. K. pneumoniae 700603 was used as a control strain for a positive ESBLs production while E. coli 25922 was used as a negative control for the ESBLs production.
Phenotypic disc diffusion method for ESBLs confirmation
Confirmation of ESBL phenotype was done by phenotypic confirmatory disc diffusion method, using both ceftazidime alone and in combination with amoxillin-clavulanic acid. The ESBL positive Salmonella strains as determined by double disk synergy test were used for confirmation. The Salmonella strains were confirmed as ESBLs producers if they showed a ≥ 5 mm increase in the zone diameter of inhibition when ceftazidime was used in combination with clavulanic acid compared to the zone diameter of inhibition when ceftazidime was used alone. Screening for ESBL was considered positive if the zone diameter of inhibition for ceftazidime, cefotaxime and ceftriaxone were respectively ≤ 27 mm, ≤ 22 mm and ≤ 25 mm [17].
Salmonella strains that showed resistance against more than three different groups of antimicrobials such as fluoroquinolones, aminoglycosides, cephalosporin, were considered multi drug resistant strains [18].
Availability of data
The data supporting our findings cannot be share now because it is a product of an on-going study.
Statistical analysis
The data obtained in this study were analysed by means of the IBM SPSS Statistics 20.0. The difference in drug resistance between ESBL and non-ESBL producing Salmonella strains was evaluated by means of the Chi-square test. P-value less than 0.05 was considered statistically significant.
Results
Stool samples were obtained from 256 patients presenting with diarrhoea due to enteric Fever and gastroenteritis. These patients attended 3 major hospitals in Calabar. In total, Salmonella was recovered from 44 patients (17.2%) of the 256 sampled. A breakdown of the isolates from the patients by hospital attended is shown in Table 1. The percentage number of Salmonella isolated from all 3 hospitals showed that the University of Calabar Teaching Hospital (UCTH) isolated the most strains from the samples. The Nigerian Navy Hospital Calabar (NNHC) follows this while General Hospital Calabar (GHC) isolated the least. There was significant difference in the isolation rate of Salmonella species in the three hospitals in Calabar (X2;=9.890, df=2, p=0.007) (Table 1).
| Hospitals | No. Of samples Collected | No. (%) Of Confirmed Salmonella species |
|---|---|---|
| UCTH | 42 | 14(33.3) |
| GHC | 80 | 9(11.3) |
| NNHC | 134 | 21(15.7) |
| Total | 256 | 44(17.2) |
Table 1: Distribution of Salmonella species by the three hospitals in Calabar.
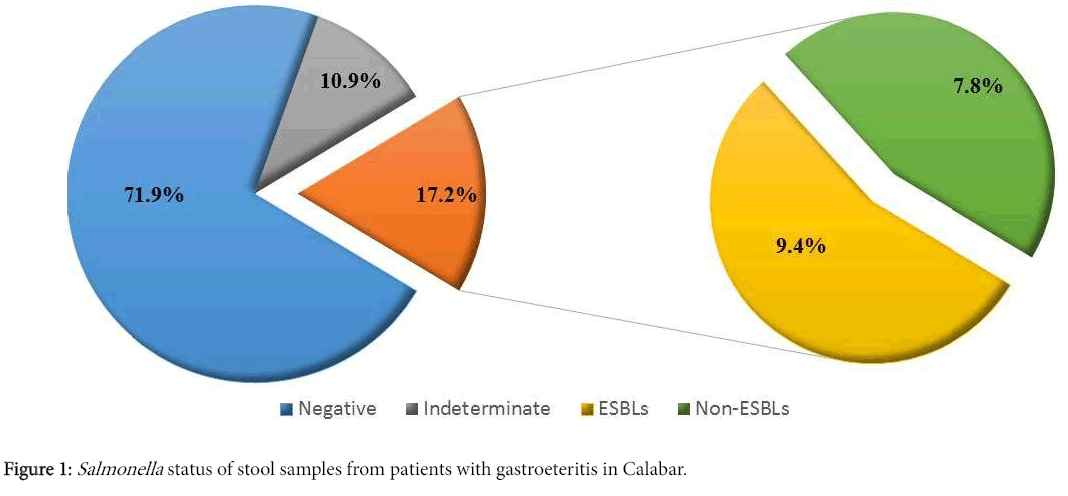
Of the 256 stool samples obtained, 184 (71.9%) did not reveal Salmonella species and 28 (10.9%) were not suitable for the Salmonella confirmation, therefore, 212 (82.8%) were considered negative for Salmonella . However, 44 (17.2%) were confirmed positive for Salmonella , out of which 24 (9.4%) demonstrated ESBLs production based on the double disc synergy test while 20 (7.8%) did not (Figure 1). All 24 presumptive ESBLs producing Salmonella isolates were confirmed to be ESBL producers as revealed by the phenotypic confirmatory test.
The result of multidrug resistance was recorded for both groups of ESBLs and non-ESBLs producers. The drug resistance of Salmonella isolates to antimicrobial agents commonly used in the treatment of diarrhoea is shown in Table 2. All the isolates demonstrated resistance against ceftriaxone.
| Antibiotics | ESBL producing | Non-ESBL producing | Total No. (%) | P-value |
|---|---|---|---|---|
| Salmonella spp. (n=24) | Salmonella spp. (n=20) | (N = 44) | ||
| Chloramphenicol (30μg) | 18 (75.0) | 5 (25.0) | 23(52.3) | 0.001* |
| Sparfolxacin (5μg) | 16 (66.7) | 3 (15.0) | 19(43.2) | 0.001* |
| Amoxicillin (30μg) | 20 (83.3) | 7 (35.0) | 27(61.4) | 0.001* |
| Gentamicin (10μg) | 8 (33.3) | 11 (55.0) | 19(43.2) | 0.127 |
| Ceftriaxone (30μg) | 24 (100.0) | 20 (100.0) | 44(100) | NA |
| Cefotaxime (30μg) | 19 (79.1) | 7 (35.0) | 26 (59.1) | 0.004* |
| Ceftazidime (30μg) | 24 (100.0) | 10 (50) | 34 (77.3) | 0.000* |
| Amoxicillin/Clavulanate (25/5µg) | 0 (0) | 0 (0) | 0 (0) | NA |
Table 2: Pattern of antimicrobial resistance between ESBL and non- ESBL producing salmonella species.
The Salmonella isolates (both ESBLs and non-ESBLs producers) were resistant against ceftriaxone while all Salmonella strains were susceptible to amoxicillin/clavulanic acid. Resistance against 3rd generation cephalosporin was observed in 34 (77.3%) patients for ceftazidime in 34 (77.3%), 26 (59.1%) for cefotaxime and 44 (100%) for ceftriaxone in all Salmonella strains.
Amongst the 24 ESBL producers, 6 (25.0%) strains were sensitive to chloramphenicol, while 16 (66.7%) were resistant to sparfloxacin. Amongst non-ESBL producing Salmonella isolates, resistance was observed against cefotaxime and ceftazidime in 7 (35.0%) and 10 (50%) strains, respectively. Also, Amoxicillin resistance was seen in 20 (83.3%) and 7 (35.0%) strains of ESBL and non-ESBL producers respectively, while gentamicin resistance was observed in 8 (33.3%) and 11 (55.0%) strains of ESBL and non-ESBL producers respectively.
Among the Salmonella strains, ESBL producers demonstrated significantly more antimicrobial resistance against all the antibiotics used in this study (with the exception of gentamicin) compared to non-ESBL producers.
Among the 24 ESBL producers, 8 (33.3%) Salmonella strains exhibited resistance against 7 of the 8 antibiotics used in this study whereas only 3 (15.0%) Salmonella strains demonstrated resistance to 7 out of the 8 antibiotics used among the 20 non-ESBL producers.
Discussion
Non-typhoidal salmonellosis poses a potential threat to human health and still persists as an important enteric infection in humans, particularly in the young, the elderly and the immunocompromised [19]. Although the disease is most of the time self-limiting and confined within the bounds of the intestinal tract, in some cases the disease has been known to exhibit dangerous consequences when it spreads beyond the intestines or when the immune system of the host is compromised in one way or the other [5].
The injudicious and indiscriminate use of antibiotics particularly in the developing countries has led to an increasing incidence of antibiotic resistance among Salmonella strains, worldwide [16]. Recently, ESBLs have emerged as an important driver of drug resistance.
In spite of the widespread distribution of ESBLs, the prevalence and phenotypic characteristic among clinical isolates may differ with different geographical locations [2]. Prevalence of ESBLs between E. coli and Klebsiella has been frequently demonstrated in several countries, but it is worth recognising the emergence of ESBLs in Salmonella , which now confers serious clinical problem [4,20].
This study revealed a prevalence of 17.2% Salmonella species isolated from stool samples from patients presenting with gastroenteritis in Calabar (Figure 1). This is relatively low when compared with the results of other studies such as the 24.56% recorded in west Bengal, India, the 32.1% recorded in North Karnataka, India, the 53% in Mumbai and the 62.3% recorded in Pondicherry [21-23]. However, a recent study in Bangladesh reported that the distribution of Salmonella species shows distinct seasonality, with higher prevalence occurring during the raining season (May to October) [3]. Therefore, the relatively low prevalence of Salmonella as observed in this study could account for the fact that the period of collection of samples fell within the dry season (December 2014 to May 2015).
This study also demonstrated a 9.4% prevalence rate of ESBLs producing Salmonella strains by using the phenotypic confirmatory test method (Figure 1). A limited number of studies have been carried out to determine the prevalence of ESBLs producing Salmonella strains with zero to very low prevalence such as in the study carried out in Abuja, Nigeria [24]. However, the prevalence of ESBLs producing Salmonella species was lower in this study compared to other species of the Enterobacteriaceae family [4,25,26].
Furthermore, antimicrobial resistance was significantly higher among ESBL producers than non-ESBL producers (Table 2). Ceftazidime and ceftriaxone resistance in this study were found to be particularly related to ESBLs production in Salmonella strains. This corroborates the recent finding that indicates that most detected ESBLs have special affinity to degrade ceftazidime [2]. Another report also shows ceftazidime to be efficient in screening isolates as potential ESBLs producers [20].
Also, resistance due to ESBLs was demonstrated by using the phenotypic confirmatory disc diffusion method, which is relatively cheaper and easy to carry out. However, the sensitivity of this method is inferior when considering Epsilometer test strip (an exponential gradient method of determination of antimicrobial resistance) and molecular detection techniques, which are more sensitive and reproducible for confirmation of false positive results. Nonetheless, they are very expensive and were not used in this study.
The increasing antibiotic resistance among ESBL producing Salmonella species is a major public health concern worldwide therefore, surveillance of antimicrobial resistant strains and rational use of antimicrobial agents is necessary for effective treatment and prediction of occurrence of resistant populations.
Funding source
None
Competing Interests
The authors declare that they have no competing interests
Author’s Contributions
OJO, BEB initiated planned and completed the study and wrote the initial manuscript. NYY, AOE and UMF analysed data and also reviewed the manuscript. All authors read and approved the final manuscript.
Acknowledgement
We would like to thank the staff of the University of Calabar Teaching Hospital, Calabar; General Hospital Calabar; and Nigerian Navy Hospital Calabar for their support and help in this study.
References
- Ranjbar R, Glammanco GM, Aleo A, Plaro MR, Naghomi A, et al. (2010) Characterization of the first extended spectrum beta-lactamase–producing nontyphoidal Salmonella strains isolated in Tehran, Iran. Foodborne Pathog Dis 7: 91-95.
- Bradford PA (2001) Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology and detection of this important resistance threat. Clin Microbiol Rev 14: 933-951.
- Dilruba A, Abu I, Salah U, Syeda l, Razib M, et al. (2014) Emergence of blaTEM type extended spectrum β- lactamase producing Salmonella species in the urban area of Bangladesh. ISRN Microbiology 1: 1-3.
- Kocagoz S, Budak F, Gur D (2006) Evaluation of a chlorogenic medium for rapid detection of extended spectrum beta-lactamases producing Salmonella spp. Indian J Med Res 124: 443-446.
- Youri A (2008) ESBL-An Emerging Worldwide Threat. J Clin Microbiol 1: 42-52.
- Ottong JG, Ering SO, Akpan FU (2010) The Population Situation in Cross-River State of Nigeria and Its Implication for Socio-Economic Development: Observations from the 1991 and 2006 Censuses. J Emerging Trends in Educational Research and Policy Studies 1: 36-42.
- Niang NN (2003) Determination of sample size for researches. Malays J Med Sci 10: 84-86.
- Bharat P, Jonak k, Rajan D, Shyam M, Prem K et al. (2006) Multidrug resistant and extended spectrum beta–lactamase (ESBLs)–producing Salmonella enteric (serotypes, typhi and paratyphi) from blood isolates in Nepal: Surveillance of resistance and a search for newer alternatives. Int J Infect Dis 10: 434-438.
- Rall V, Rall R, Aragon L, De- Silva M (2005) Evaluation of three enrichment broths and five plating media for Salmonella detection in poultry. Braz J Microbiol 36: 147-150.
- Collee JG, Miles RS, Watt B (1996) Mackie and McCartney: Practical Medical Microbiology. (13edn), Churchill Livingstone.
- OIE Terrestrial Manual: World organisation for animal health manual of diagnostic tests and vaccines for terrestrial animals (mammals, birds and bees). (6edn), Paris, France.
- Bauer W, Kirby W, Sherries J (1996) Antibiotics susceptibility testing by standard disc method. Am J Clin Pathol 106: 45-46.
- Cheesbrough M (2006) District Laboratory Practice in Tropical Countries Part 2.(3edn). Cambridge University Press
- Clinical and Laboratory Standards institute (CLSI): Performance standards for antimicrobial susceptibility testing (2006), Wayne, Pennsylvania, USA.
- Jarlier V, Nicolas MH, Fournier G, Philippon A (1998) Extended broad spectrum beta lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriacae: hospital prevalence and susceptibility patterns. Rev Infect Dis 10: 867-878.
- Mathew AW, Franklin RC, William AC, Michael ND, George ME, et al (2007) Performance Standard for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standard Institute 27: 1-82.
- Rodriues C, Joshi P, Jani H, Alphonse M, Radhakrishnan R, et al (2004) Detection of β-Lactamases in nosocomial Gram-negative clinical isolates. Indian J Med Microbiol 22: 247-250.
- Taneja N, Mohan B, Khurana S, Sharma M (2004) Antimicrobial resistance in selected bacterial enteropathogens in north India. Indian J Med Res 120: 39-43.
- Winokur PL, Canton R, Casellas JM, Legakis N (2001) Variations in the prevalence of strains expressing an extended-spectrum β- lactamase phenotype and characterization of isolates from Europe, the Americas, and the Western Pacific region. Clin Infect Dis 1: 94-103.
- Vahaboglu H, Fuzi S, Cetin S, Gundes S, Ujhelyi E et al. (2001) Characterization of extended beta-lactamase producing strains of Salmonella enterica serovar typhimurium with diverse resistance phenotypes. J Clin Microbiol 39: 791-793.
- Das N, Borthakur AK (2012) Antibiotic co-resistance among extended-spectrum beta lactamase- producing urinary isolates in a tertiary medical center: A prospective study. Chronological Young Science 1: 53-56.
- Metri BC, Jyothi P, Peerapur BV (2011) The prevalence of ESBL among Enterobacteriaceae in a tertiary care hospital of North Karnataka, India. J Clin Diag Res 5: 470-475.
- Rodrigues C, Joshi P, Jani SH, Alphonse M, Radhakrishnan R, Mehta A (2004) Detection of β-Lactamases in nosocomial Gram negative clinical isolates. Indian J Med Microbiol 22: 247-250.
- Rudresh SM, Nagarathnamma T (2011) Extended spectrum β-lactamase producing Enterobacteriaceae and antibiotic co-resistance. Indian J Med Res 133: 116-118.
- Ahmed A, Abu I, Salah U, Syeda l, Razib M, et al (2014) Emergence of blaTEM type extended spectrum β- lactamase producing Salmonella species in the urban area of Bangladesh. ISRN Microbiol 1: 1-3.
- Casmir CI, Bassey EB, Nkiruka I, Nazek A (2014) Molecular characterization and antibiotic resistance of Salmonella in children with acute gastroenteris in Abuja, Nigeria. J Infect Developing Countries 8: 712-719.
- Katsanis GP, Spargo J, Ferraro MJ, Sutton L, Jacoby GA (1994) Detection of Klebsiella pneumoniae and Escherichia coli strains producing extended-spectrum beta-lactamases. J Clin Microbiol 32: 691-696.
- Mathur P, Kapil A, Das B, Dhawan B (2002) Prevalence of extended spectrum beta lactamase producing gram negative bacteria in a tertiary care hospital. Indian J Med Res 115: 153-157.
Relevant Topics
- Antibiotics and Resistance
- Antifungal
- Antiviral therapy
- Bacteremia
- Bacterial diseases
- Broad Spectrum of Antibiotics
- Clinical Infectious Diseases
- Diagnosis of Pathogenic microorganisms
- Emerging infections
- Natural Antibiotics
- Opportunistic Pathogens
- Parasitic Diseases
- Pertussis Vaccines
- Prevention of infection
- Septicemia
- Viral Infections
- Viremia
Recommended Journals
Article Tools
Article Usage
- Total views: 12761
- [From(publication date):
August-2016 - Apr 03, 2025] - Breakdown by view type
- HTML page views : 11717
- PDF downloads : 1044