Research Article Open Access
Antagonistic Interactions among Bacteria Isolated from either the Same or from Different Sponges Native to the Brazilian Coast
Marinella S Laport1,3*, Juliana F Santos-Gandelman1, Guilherme Muricy2, Marcia Giambiagi-deMarval1 and Isabelle George31Laboratory Molecular Bacteriology and Marine Institute of Microbiology Paulo de Goes, Federal University of Rio de Janeiro (UFRJ), Av. Carlos Chagas Filho, 373, University City, 21941-590, Rio de Janeiro, Brazil
2Laboratory of Porifera, National Museum, Federal University of Rio de Janeiro (UFRJ), Av. Bartolomeu Gusmão s/ n, Quinta da Boa Vista, 20940-040, Rio de Janeiro, Brazil
3Laboratoire of Marine Biology, Department of Biology of Organisms, Université Libre de Bruxelles (ULB), Solbosch Campus, Av. F. Roosevelt 50, 1050 Brussels, Belgium
- *Corresponding Author:
- Marinella S Laport
Laboratory Molecular Bacteriology and
Marine Institute of Microbiology Paulo de Goes
Federal University of Rio de Janeiro (UFRJ)
Av. Carlos Chagas Filho, 373
University City, 21941-590, Rio de Janeiro, Brazil
Tel: +552125608028
E-mail: marinella@micro.ufrj.br
Received Date: February 09, 2016; Accepted Date: February 22, 2016; Published Date: February 29, 2016
Citation: Laport MS, Santos-Gandelman JF, Muricy G, deMarval MG, George I (2016) Antagonistic Interactions among Bacteria Isolated from either the Same or from Different Sponges Native to the Brazilian Coast. J Marine Sci Res Dev 6:185. doi: 10.4172/2155-9910.1000185
Copyright: © 2016 Laport MS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Marine Science: Research & Development
Abstract
Marine sponges are sessile and filter-feeding organisms that harbor dense and diverse microbial communities of considerable ecological and biotechnological importance. They represent an important target for the study of bacterial interactions in marine ecosystems. The purpose of this study was to examine the frequency of antagonistic interactions among the culturable microbial communities associated with sponges from the Brazilian coast. The specimens were collected over six years at Cagarras Archipelago, Praia Vermelha Beach and Urca square, Rio de Janeiro State, SE Brazil. Fifty-six bacterial isolates representing four classes of cultivable sponge-associated bacteria were studied for their ability to produce inhibitory substances. Antagonistic interactions occurred among isolates from both, the same and different sponge species. Most isolates (98.2%) were able to inhibit growth of at least one indicator bacterium. In contrast, there were few antagonistic interactions among bacteria obtained from the same sponge specimen. Our results suggest that chemical antagonism could play a significant role in shaping the bacterial communities within sponge tissues.
Keywords
Antagonism; Cultivable bacteria; Inhibitory substances; Sponge microbial communities
Introduction
Associations between sponges and bacteria have existed for over 600 million years and are one of the most ancient of all symbioses between microbes and metazoa [1]. Most sponges host diverse and abundant communities of microorganisms, which contribute to host health, ecology and evolution [2,3]. The relationship between sponges and their associated microbial communities is so important that microorganisms can contribute to more than 35% of the sponge biomass [4] and may undertake diverse functional roles including nutrition, cycling of metabolites and host defense [5].
The phylum Porifera includes approximately 8,700 valid species known worldwide [6]. Of these, 515 species have so far been identified in Brazil [7]. There is a lack of reliable baseline data on the composition and stability of symbiotic microbial communities for most sponge species. This knowledge gap makes it difficult to determine the role of microorganisms in health, diseases and mortality of sponges. This role has been studied for coral-associated bacteria [8], where inhibitory activities measured towards known coral pathogens have led to the hypothesis that bacteria associated with healthy corals play a protective role for the coral holobionts [9]. An ecological role of antagonism has also been suggested for bacteria associated with Antarctic sponges [10], brittle stars [11], and marine aggregates [12].
In this study, we tested the hypothesis that chemical antagonism is common among cultivable bacteria associated with sponges. For this purpose, a total of 27 sponge samples representing 13 species were collected (between 2005 and 2011) at the coast of Rio de Janeiro city, Brazil. We measured the frequency of antagonistic interactions among the culturable microbial communities associated with either the same or different sponge species from the same area. More specifically, we aimed to address the following questions: How frequent is antagonism between sponge-associated bacteria? Is this frequency modulated by taxonomic identity, sponge specimen, sampling site and/or sampling date of the sponge?
Material and Methods
Sponge collection and bacterial isolation and cultivation
A total of 27 sponge samples representing 13 sponge species were collected between 2005 and 2011 by scuba diving at depths of 4-20 m, at 18–25°C in the Cagarras Archipelago (CA) (23°01’S, 43°11’W), Praia Vermelha beach (PV) (22°57’S, 43°09’W) and Urca square (Us) (22°95’S, 43°16’W), located at the coast of Rio de Janeiro, Brazil (Table 1) [13].
These three different sites (CA, PV and Us) are located within a perimeter of about 10 km. QU and PV are dynamic ecosystems located at the interface between the Guanabara Bay eutrophic (polluted) estuarine waters and the adjacent coastal Atlantic Ocean seawaters. CA is located further away from the bay and is under the influence of the Brazil Current and the South Atlantic Central waters [14].
Specimens were macerated at room temperature (RT, 25 ± 2°C) in brain-heart infusion medium (BHI) (Difco). Subsequently, macerates were serially 10-fold diluted, inoculated in replicates on BHI agar and incubated for up to seven days at RT. Bacteria were purified from the primary culture and kept in slant cultures at -20°C [15].
16S rRNA sequence analysis of bacterial isolates
Bacterial DNA was recovered by a thermal lysis protocol consisting in re-suspending cellular material from each colony in 25 μl sterile PCR grade water and boiling the suspension at 100°C for 15 min. PCR amplification was performed by adding 3 μl DNA solution to 47 μl containing 1× buffer GO TAQ green master mix (Promega), BSA 0.4 mg/ml, Igepal 0.05%, and 20 pmol of each universal primer, 27F (5′-GAGTTTGATCMTGGCTCAG- 3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) [16], Cycle conditions consisted in an initial denaturation step at 94°C for 6 min, followed by 30 cycles at 94°C for 30 s, 55°C for 1 min 30 s and 72°C for 2 min 30 s, and a final elongation step at 72°C for 5 min.
PCR products were confirmed by electrophoresis on a 0.8% agarose gel, purified using the QIAquick PCR Purification Kit (Qiagen), and sequenced using the universal primer 338F (5′-ACTCCTACGGGAGGCAGC-3′) at Beckman Coulter Genomics (Takeley, UK). 16S rRNA gene sequences obtained for the isolates were aligned and classified using the online portal of the SILVA SINA alignment service of the ARB-Silva database (http://www.arb-silva.de/ aligner/) [17].
Antagonistic interactions among bacterial isolates
Bacterial strains were screened for antagonistic interactions by a previously described method to evaluate production of antimicrobial substances [18]. Hereafter, bacterial strains tested for antimicrobial substance production will be termed ‘‘producer’’ strains, whereas those used as targets will be called ‘‘indicator’’ strains. Briefly, 107 cells of each producer strain were spotted onto BHI-agar and incubated at 25°C until the colony diameter reached 8 mm. In parallel, each indicator strain was grown in liquid medium at 25°C for 24-48 h. Subsequently, 105 cells of the indicator culture were mixed with 3 ml of BHI soft agar and poured over the plates. Plates were incubated at 25°C for 24 h and the diameter of the inhibition zone around the spotted strain was measured. An indicator strain was considered sensitive to the activity of the producer strain when it exhibited a clear inhibition zone with a diameter ≥ 8 mm. When the inhibition zone was <8 mm or when bacterial colonies grew inside the inhibition zone, the indicator strain was considered resistant [18].
A total of 3,080 tests, i.e. a 56 × 55 array, were performed for assessing antagonistic interactions among the bacterial strains. This means that each strain was tested against the 55 other strains for cross-inhibition. In order to calculate the relative frequencies of antagonistic interactions, the number of antagonistic interactions observed for a given strain (absolute frequency) was divided by the total number of interactions performed with this strain.
Bacterial isolates were then operationally grouped into three different interactivity clusters: (I) active, if they were able to inhibit the growth of at least one indicator strain; (II) sensitive, if their growth was inhibited by at least one producer strain; and (III) resistant, if their growth was not inhibited by any producer strain. It must be noted that an individual strain could be simultaneously included in the interactivity clusters I and II (active and sensitive), or I and III (active and resistant), but never in II and III [10].
The antagonistic relationships were also plotted in network graphs using the program Cytoscape 3.1.0 (http://www.cytoscape.org).
Results
Phylogenetic affiliation of isolates
Fifty-six bacterial strains were isolated from the 13 sampled sponge species. Based on 16S rRNA gene sequence analysis (targeting the V3-V5 region), they represented four classes: Firmicutes (52%), Gammaproteobacteria (39%), Actinobacteria (7%) and Alphaproteobacteria (2%). The most frequently isolated genus was Bacillus (17 isolates), followed by Pseudomonas (10 isolates), Citrobacter (5 isolates), Enterococcus (5 isolates) and Lactococcus (4 isolates). Other genera isolated in smaller numbers included Brachybacterium, Psychrobacter and Shigella (2 isolates each), and Acinetobacter, Brevibacillus, Klebsiella, Kocuria, Lysinibacillus, Pseudovibrio, Rhodococcus, Serratia and Staphylococcus (1 isolate each) (Table 1).
| Sponge species | Voucher MMBLa | Colletionsiteb | Marine strainsc | Bacterial genera | Phylum/Class |
| Clathrinaaurea | 42006Ca | PV | Ca31 | Pseudovibrio sp. | Proteobacteria /Alphaproteobacteria |
| Cliona aff. celata | 92010Cc | Us | Cc81 | Pseudomonas sp. | Proteobacteria /Gammaproteobacteria |
| Cliona aff. celata | 92010Cc | Us | Cc82 | Citrobacter sp. | Proteobacteria /Gammaproteobacteria |
| Cliona aff. celata | 82011Cc | Us | Cc92 | Citrobacter sp. | Proteobacteria /Gammaproteobacteria |
| Cliona aff. celata | 82011Cc | Us | Cc93 | Rhodococcus sp. | Actinobacteria |
| Cliona aff. celata | 82011Cc | Us | Cc94 | Pseudomonas sp. | Proteobacteria /Gammaproteobacteria |
| Dragmacidon reticulatum | 42006Dr | PV | Dr32 | Lactococcus sp. | Firmicutes / Bacilli |
| Dragmacidon reticulatum | 42006Dr | PV | Dr34 | Psychrobacter sp. | Proteobacteria /Gammaproteobacteria |
| Dragmacidon reticulatum | 42006Dr | PV | Dr35 | Psychrobacter sp. | Proteobacteria /Gammaproteobacteria |
| Dragmacidon reticulatum | 42006Dr | PV | Dr36 | Brachybacterium sp. | Actinobacteria |
| Dragmacidon reticulatum | 42006Dr | PV | Dr37 | Brachybacterium sp. | Actinobacteria |
| Dragmacidon reticulatum | 32007Dr | PV | Dr5 | Enterococcus sp. | Firmicutes / Bacilli |
| Dragmacidon reticulatum | 62009Dr | CA | Dr72 | Kokuria sp. | Actinobacteria |
| Geodia corticostylifera | 32007Gc | CA | Gc51 | Serratia sp. | Proteobacteria /Gammaproteobacteria |
| Geodia corticostylifera | 32007Gc | CA | Gc54 | Lactococcus sp. | Firmicutes / Bacilli |
| Haliclona vansoesti | 12007Hv | CA | Hv40 | Pseudomonas sp. | Proteobacteria /Gammaproteobacteria |
| Haliclona vansoesti | 12007Hv | CA | Hv41 | Pseudomonas sp. | Proteobacteria /Gammaproteobacteria |
| Haliclona fugidia | 32007Hf | PV | Hf51 | Pseudomonas sp. | Proteobacteria /Gammaproteobacteria |
| Haliclona fugidia | 32007Hf | PV | Hf52 | Enterococcus sp. | Firmicutes / Bacilli |
| Hymeniacidon heliophila | 32007Hh | PV | Hh5 | Acinetobacter sp. | Proteobacteria /Gammaproteobacteria |
| Hymeniacidon heliophila | 92010Hh | Us | Hh81 | Bacillus sp. | Firmicutes / Bacilli |
| Hymeniacidon heliophila | 92010Hh | Us | Hh82 | Bacillus sp. | Firmicutes / Bacilli |
| Hymeniacidon heliophila | 82011Hh | Us | Hh91 | Pseudomonas sp. | Proteobacteria /Gammaproteobacteria |
| Hymeniacidon heliophila | 82011Hh | Us | Hh92 | Pseudomonas sp. | Proteobacteria /Gammaproteobacteria |
| Hymeniacidon heliophila | 82011Hh | Us | Hh93 | Pseudomonas sp. | Proteobacteria /Gammaproteobacteria |
| Hymeniacidon heliophila | 82011Hh | Us | Hh94 | Bacillus sp. | Firmicutes / Bacilli |
| Mycale microsigmatosa | 52005Mm | PV | Mm1 | Bacillus sp. | Firmicutes / Bacilli |
| Mycale microsigmatosa | 52005Mm | PV | Mm3 | Pseudomonas sp. | Proteobacteria /Gammaproteobacteria |
| Mycale microsigmatosa | 42006Mm | PV | Mm31 | Bacillussp. | Firmicutes / Bacilli |
| Mycale microsigmatosa | 42006Mm | PV | Mm32 | Shigellasp. | Proteobacteria /Gammaproteobacteria |
| Mycale microsigmatosa | 42006Mm | PV | Mm33 | Bacillussp. | Firmicutes / Bacilli |
| Mycale microsigmatosa | 42006Mm | PV | Mm35 | Bacillussp. | Firmicutes / Bacilli |
| Mycale microsigmatosa | 32007Mm | CA | Mm51a | Bacillus sp. | Firmicutes / Bacilli |
| Mycale microsigmatosa | 32007Mm | CA | Mm51b | Lactococcussp. | Firmicutes / Bacilli |
| Mycale microsigmatosa | 92010Mm | Us | Mm81 | Pseudomonassp. | Proteobacteria /Gammaproteobacteria |
| Mycale microsigmatosa | 92010Mm | Us | Mm82 | Bacillus sp. | Firmicutes / Bacilli |
| Mycale microsigmatosa | 92010Mm | Us | Mm84 | Bacillus sp. | Firmicutes / Bacilli |
| Mycale microsigmatosa | 82011Mm | Us | Mm91 | Enterococcus sp. | Firmicutes / Bacilli |
| Pachychalina alcaloidifera | 32007Pa | CA | Pa51 | Bacillussp. | Firmicutes / Bacilli |
| Pachychalina alcaloidifera | 32007Pa | CA | Pa52 | Enterococcus sp. | Firmicutes / Bacilli |
| Pachychalina alcaloidifera | 32007Pa | CA | Pa53 | Bacillussp. | Firmicutes / Bacilli |
| Petromica citrina | 42006Pc | CA | Pc31 | Bacillussp. | Firmicutes / Bacilli |
| Petromica citrina | 42006Pc | CA | Pc32 | Bacillussp. | Firmicutes / Bacilli |
| Petromica citrina | 32007Pc | CA | Pc5a | Shigellasp. | Proteobacteria /Gammaproteobacteria |
| Petromica citrina | 32007Pc | CA | Pc5b | Enterococcussp. | Firmicutes / Bacilli |
| Polymastia janeirensis | 52005Pj | PV | Pj1 | Bacillussp. | Firmicutes / Bacilli |
| Polymastia janeirensis | 52005Pj | PV | Pj2 | Bacillus sp. | Firmicutes / Bacilli |
| Polymastia janeirensis | 42006Pj | PV | Pj32 | Lactococcussp. | Firmicutes / Bacilli |
| Polymastia janeirensis | 42006Pj | PV | Pj33 | Klebsiellasp. | Proteobacteria /Gammaproteobacteria |
| Polymastia janeirensis | 32007Pj | PV | Pj52 | Lysinibacillussp. | Firmicutes / Bacilli |
| Paraleucilla magna | 12007Pm | CA | Pm42 | Citrobactersp. | Proteobacteria /Gammaproteobacteria |
| Paraleucilla magna | 32007Pm | CA | Pm52 | Bacillussp. | Firmicutes / Bacilli |
| Tedania ignis | 12007Ti | CA | Ti41 | Brevibacillus sp. | Firmicutes / Bacilli |
| Tedania ignis | 32007Ti | CA | Ti54 | Staphylococcussp. | Firmicutes / Bacilli |
| Tedania ignis | 32007Ti | CA | Ti55 | Citrobactersp. | Proteobacteria /Gammaproteobacteria |
| Tedania ignis | 32007Ti | CA | Ti56 | Citrobactersp. | Proteobacteria /Gammaproteobacteria |
bAll the specimens of sponges were collected by scuba diving at depths of 4-2 m, at 18-25°C, in the Cagarras Archipelago (CA), Praia Vermelha beach (PV) and Urca square (Us), Rio de Janeiro, Brazil.
cThe strains belong to the sponge-associated bacteria collection of the Molecular and Marine Bacteriology Laboratory (MMBL) of the Microbiology Institute, UFRJ, Brazil.
Table 1: Sponges, collection sites and isolated bacteria.
Antagonistic interactions among bacterial isolates
The 56 isolated bacterial strains were screened against each other in 3,080 cross-tests (56 × 55 tests) for antagonistic interactions by a protocol to test for antimicrobial substance production (Table 2).
| Indicatora | Pa51 | Pa52 | Pa53 Ca31 Cc81 | Cc82 | Cc92 | Cc93 | Cc94 | Dr32 | Dr34 | Dr35 | Dr36 | Dr37 | Dr5 | Dr72 | ||
| Producera | ||||||||||||||||
| Pa51 | 0 | 0 | 0 | 0 | 0 | 10b | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Pa52 | 0 | 0 | 0 | 0 | 10 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Pa53 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Ca31 | 0 | 0 | 0 | 0 | 0 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cc81 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cc82 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cc92 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cc93 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 10 | 10 | |
| Cc94 | 25 | 23 | 28 | 0 | 0 | 0 | 0 | 0 | 26 | 24 | 0 | 26 | 22 | 25 | 22 | |
| Dr32 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Dr34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Dr35 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Dr36 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Dr37 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | |
| Dr5 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Dr72 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | |
| G51 | 0 | 0 | 0 | 0 | 10 | 10 | 10 | 24 | 0 | 0 | 36 | 32 | 0 | 0 | 10 | 0 |
| G54 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 10 | 0 |
| Hv40 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hv41 | 18 | 22 | 25 | 22 | 0 | 0 | 0 | 10 | 0 | 22 | 25 | 28 | 28 | 22 | 22 | 35 |
| Hf51 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hf52 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hh5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hh81 | 25 | 0 | 25 | 20 | 10 | 0 | 0 | 0 | 0 | 24 | 20 | 0 | 0 | 0 | 0 | 0 |
| Hh82 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hh91 | 0 | 0 | 0 | 0 | 0 | 0 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hh92 | 0 | 0 | 0 | 0 | 0 | 0 | 22 | 0 | 0 | 0 | 10 | 10 | 0 | 0 | 0 | 0 |
| Hh93 | 30 | 35 | 33 | 30 | 0 | 0 | 20 | 0 | 0 | 25 | 42 | 15 | 34 | 0 | 10 | 0 |
| Hh94 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mm1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mm3 | 0 | 0 | 0 | 0 | 0 | 0 | 20 | 10 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 0 |
| Mm31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mm32 | 0 | 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mm33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mm35 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mm51a | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 |
| Mm51b | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 |
| Mm81 | 12 | 0 | 13 | 12 | 0 | 0 | 0 | 0 | 0 | 12 | 16 | 12 | 0 | 0 | 0 | 0 |
| Mm82 | 13 | 0 | 13 | 12 | 0 | 0 | 0 | 0 | 0 | 12 | 16 | 12 | 0 | 0 | 0 | 0 |
| Mm84 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mm91 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pc31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pc32 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pc5a | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pc5b | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pj1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pj2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pj32 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pj33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pj52 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pm42 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pm52 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ti41 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 10 | 0 | 0 | 0 | 0 |
| Ti54 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ti55 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ti56 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Indicator | G51 | G54 | Hv40 | Hv41 | Hf51 | Hf52 | Hh5 | Hh81 | Hh82 | Hh91 | Hh92 | Hh93 | Hh94 | Mm1 | Mm3 | |
| Producer | ||||||||||||||||
| Pa51 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |
| Pa52 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |
| Pa53 | 0 | 38 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |
| Ca31 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |
| Cc81 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cc82 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | |
| Cc92 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cc93 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cc94 | 10 | 0 | 0 | 25 | 0 | 26 | 28 | 0 | 0 | 18 | 0 | 0 | 15 | 18 | 0 | |
| Dr32 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |
| Dr34 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |
| Dr35 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Dr36 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |
| Dr37 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |
| Dr5 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |
| Dr72 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| G51 | 12 | 12 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 14 | 0 | ||
| G54 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | ||
| Hv40 | 0 | 0 | 0 | 0 | 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Hv41 | 30 | 24 | 0 | 30 | 12 | 22 | 0 | 0 | 0 | 0 | 10 | 23 | 25 | 20 | ||
| Hf51 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Hf52 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Hh5 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | ||
| Hh81 | 0 | 20 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 10 | 0 | ||
| Hh82 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Hh91 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Hh92 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Hh93 | 33 | 0 | 0 | 28 | 0 | 30 | 31 | 0 | 0 | 28 | 0 | 15 | 20 | 0 | ||
| Hh94 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Mm1 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | ||
| Mm3 | 0 | 10 | 0 | 0 | 0 | 10 | 25 | 0 | 0 | 0 | 10 | 0 | 0 | 15 | ||
| Mm31 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |
| Mm32 | 10 | 10 | 0 | 10 | 0 | 10 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |
| Mm33 | 0 | 12 | 0 | 10 | 0 | 10 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |
| Mm35 | 0 | 10 | 0 | 12 | 0 | 10 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 12 | 0 | |
| Mm51a | 0 | 12 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 12 | 0 | 10 | 0 | 0 | |
| Mm51b | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |
| Mm81 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Mm82 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Mm84 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Mm91 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | |
| Pc31 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 20 | |
| Pc32 | 0 | 25 | 0 | 0 | 0 | 12 | 15 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 15 | |
| Pc5a | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Pc5b | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Pj1 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Pj2 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Pj32 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Pj33 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Pj52 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Pm42 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Pm52 | 15 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Ti41 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | |
| Ti54 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Ti55 | 10 | 0 | 0 | 10 | 0 | 10 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 10 | 0 | |
| Ti56 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Indicator | Mm32 | Mm33 | Mm35 | Mm51a | Mm51b | Mm81 | Mm82 | Mm84 | Mm91 | Pc31 | Pc32 | Pc5a | Pc5b | |||
| Producer | ||||||||||||||||
| Pa51 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 10 | |||
| Pa52 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 10 | |||
| Pa53 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 10 | |||
| Ca31 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 10 | |||
| Cc81 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |||
| Cc82 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Cc92 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | |||
| Cc93 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Cc94 | 18 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 20 | 20 | 20 | 0 | |||
| Dr32 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 10 | |||
| Dr34 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 12 | 0 | 10 | |||
| Dr35 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 12 | 0 | 10 | 0 | 10 | |||
| Dr36 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 21 | 0 | 10 | 0 | 10 | |||
| Dr37 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 10 | |||
| Dr5 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 10 | |||
| Dr72 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |||
| G51 | 0 | 0 | 0 | 0 | 10 | 10 | 10 | 30 | 20 | 0 | 15 | 0 | 10 | |||
| G54 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 10 | 10 | 0 | 0 | 0 | |||
| Hv40 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |||
| Hv41 | 30 | 20 | 20 | 28 | 12 | 0 | 0 | 0 | 18 | 28 | 24 | 28 | 0 | |||
| Hf51 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Hf52 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | |||
| Hh5 | 0 | 0 | 0 | 10 | 10 | 0 | 0 | 0 | 15 | 0 | 10 | 12 | 10 | |||
| Hh81 | 0 | 0 | 18 | 0 | 0 | 10 | 0 | 0 | 25 | 0 | 0 | 0 | 0 | |||
| Hh82 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |||
| Hh91 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | |||
| Hh92 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 10 | 0 | 0 | |||
| Hh93 | 0 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 25 | 20 | 0 | |||
| Hh94 | 0 | 0 | 0 | 0 | 10 | 10 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | |||
| Mm1 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | |||
| Mm3 | 15 | 13 | 0 | 21 | 15 | 0 | 0 | 0 | 11 | 22 | 0 | 28 | 25 | |||
| Mm31 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | |||
| Mm32 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | ||||
| Mm33 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | ||||
| Mm35 | 23 | 20 | 21 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 10 | ||||
| Mm51a | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 10 | ||||
| Mm51b | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 10 | ||||
| Mm81 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| Mm82 | 0 | 0 | 0 | 0 | 10 | 15 | 12 | 12 | 0 | 0 | 0 | 0 | ||||
| Mm84 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 0 | 0 | 0 | ||||
| Mm91 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| Pc31 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 25 | 10 | ||||
| Pc32 | 0 | 0 | 0 | 15 | 20 | 0 | 0 | 0 | 10 | 12 | 0 | 11 | ||||
| Pc5a | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | ||||
| Pc5b | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 11 | 0 | ||||
| Pj1 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 10 | |||
| Pj2 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 10 | |||
| Pj32 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 10 | |||
| Pj33 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | |||
| Pj52 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | |||
| Pm42 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Pm52 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | |||
| Ti41 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 10 | 0 | 0 | 10 | 0 | 0 | |||
| Ti54 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 15 | 0 | 0 | 0 | 0 | 0 | |||
| Ti55 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | |||
| Ti56 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | |||
| Indicator | Pj1 | Pj2 | Pj32 | Pj33 | Pj52 | Pm42 | Pm52 | Ti41 | Ti54 | Ti55 | Ti56 | |||||
| Producer | ||||||||||||||||
| Pa51 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | |||||
| Pa52 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | |||||
| Pa53 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | |||||
| Ca31 | 10 | 10 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | |||||
| Cc81 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Cc82 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Cc92 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | |||||
| Cc93 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Cc94 | 0 | 0 | 10 | 10 | 10 | 0 | 10 | 10 | 25 | 0 | 0 | |||||
| Dr32 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | |||||
| Dr34 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 10 | |||||
| Dr35 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18 | 0 | 0 | 10 | |||||
| Dr36 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 10 | |||||
| Dr37 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 18 | 0 | 10 | 13 | |||||
| Dr5 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 27 | 0 | 10 | 0 | |||||
| Dr72 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | |||||
| G51 | 10 | 10 | 0 | 0 | 10 | 0 | 0 | 10 | 10 | 20 | 0 | |||||
| G54 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | |||||
| Hv40 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | |||||
| Hv41 | 0 | 0 | 0 | 24 | 0 | 10 | 0 | 10 | 10 | 0 | 20 | |||||
| Hf51 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Hf52 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 10 | |||||
| Hh5 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | |||||
| Hh81 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | |||||
| Hh82 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Hh91 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Hh92 | 20 | 11 | 20 | 0 | 0 | 18 | 0 | 0 | 0 | 15 | 0 | |||||
| Hh93 | 0 | 0 | 10 | 10 | 10 | 25 | 0 | 15 | 26 | 10 | 0 | |||||
| Hh94 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Mm1 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 10 | 0 | |||||
| Mm3 | 16 | 10 | 0 | 14 | 0 | 15 | 0 | 10 | 0 | 21 | 0 | |||||
| Mm31 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Mm32 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Mm33 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Mm35 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Mm51a | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Mm51b | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Mm81 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Mm82 | 0 | 12 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | |||||
| Mm84 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Mm91 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Pc31 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | |||||
| Pc32 | 22 | 10 | 12 | 0 | 17 | 0 | 0 | 10 | 0 | 0 | 0 | |||||
| Pc5a | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 10 | |||||
| Pc5b | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 10 | |||||
| Pj1 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Pj2 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 10 | ||||||
| Pj32 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Pj33 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Pj52 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Pm42 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | ||||||
| Pm52 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Ti41 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Ti54 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Ti55 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Ti56 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
bDiameter of the inhibition zones in millimeters of each interaction.
Table 2: Antagonistic interactions among sponge-associated bacteria from the Brazilian coast.
General cross-inhibition among bacteria isolated from different species of sponges collected of the different sites between 2005 and 2011
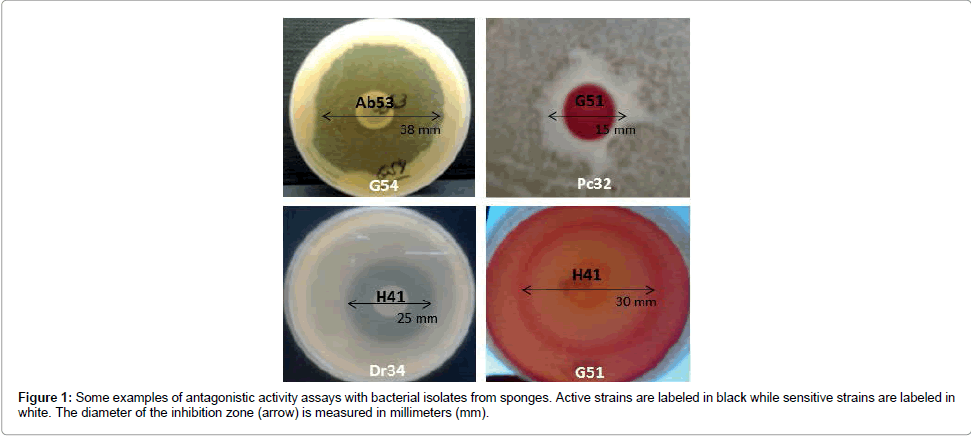
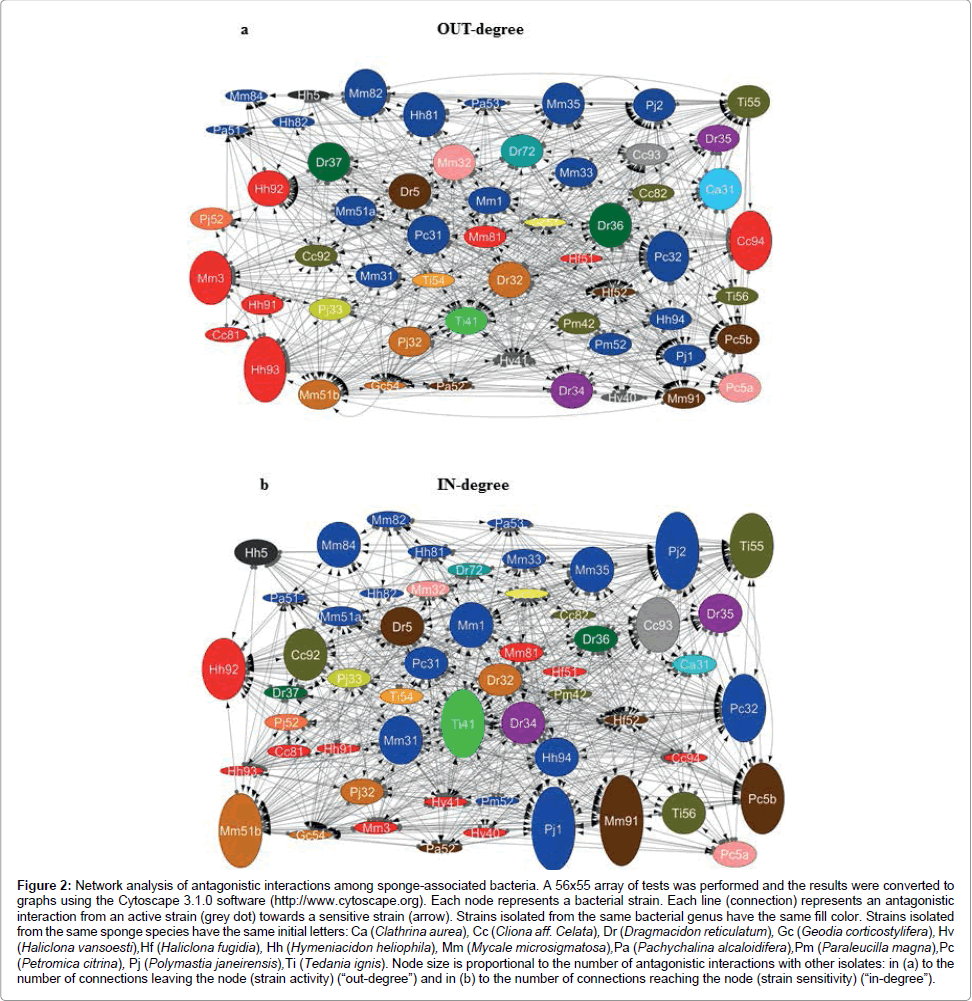
General cross-inhibition among bacteria isolated from all sponge samples showed a relative frequency of 18% (555 positive tests out of 3,080). The diameter of the inhibition zone varied from 10 to 38 mm (Table 2 and Figure 1). In total, 55 of out 56 isolates (98.2%) were active, i.e. producing antimicrobial substances against at least one indicator strain (Figure 2a). In these cross-inhibition tests, several active strains also proved to be sensitive to the effects of other tested isolates (Figure 2b). Actually, all isolates were sensitive, as their growth was inhibited by at least one strain used as a producer.
Figure 2: Network analysis of antagonistic interactions among sponge-associated bacteria. A 56x55 array of tests was performed and the results were converted to graphs using the Cytoscape 3.1.0 software (http://www.cytoscape.org). Each node represents a bacterial strain. Each line (connection) represents an antagonistic interaction from an active strain (grey dot) towards a sensitive strain (arrow). Strains isolated from the same bacterial genus have the same fill color. Strains isolated from the same sponge species have the same initial letters: Ca (Clathrina aurea), Cc (Cliona aff. Celata), Dr (Dragmacidon reticulatum), Gc (Geodia corticostylifera), Hv (Haliclona vansoesti),Hf (Haliclona fugidia), Hh (Hymeniacidon heliophila), Mm (Mycale microsigmatosa),Pa (Pachychalina alcaloidifera),Pm (Paraleucilla magna),Pc (Petromica citrina), Pj (Polymastia janeirensis),Ti (Tedania ignis). Node size is proportional to the number of antagonistic interactions with other isolates: in (a) to the number of connections leaving the node (strain activity) (“out-degree”) and in (b) to the number of connections reaching the node (strain sensitivity) (“in-degree”).
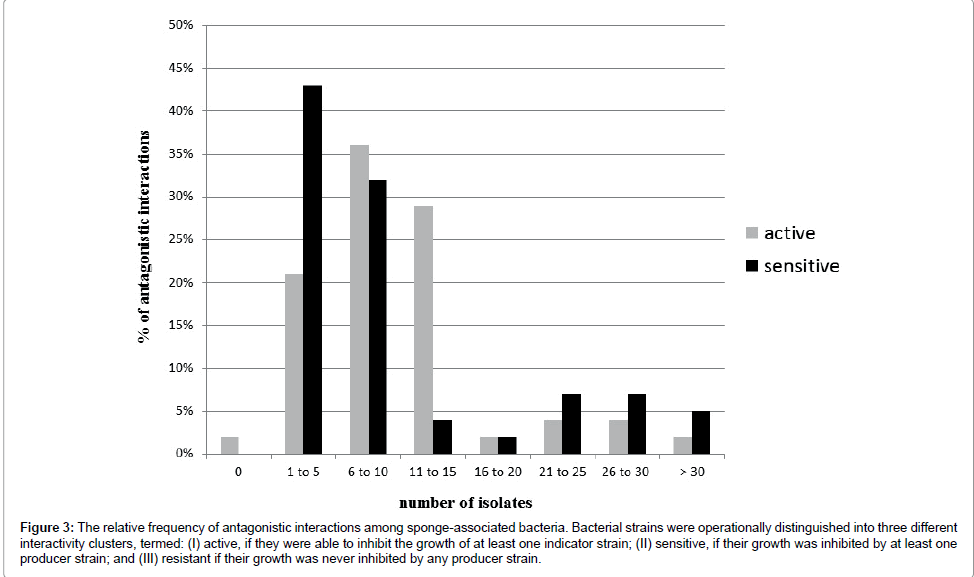
Each producer strains inhibited a mean of 10 indicator strains. While the majority of producers inhibited between 6 and 12 indicator strains, eight producer strains inhibited 15 or more indicator strains (Figure 3).
Figure 3: The relative frequency of antagonistic interactions among sponge-associated bacteria. Bacterial strains were operationally distinguished into three different interactivity clusters, termed: (I) active, if they were able to inhibit the growth of at least one indicator strain; (II) sensitive, if their growth was inhibited by at least one producer strain; and (III) resistant if their growth was never inhibited by any producer strain.
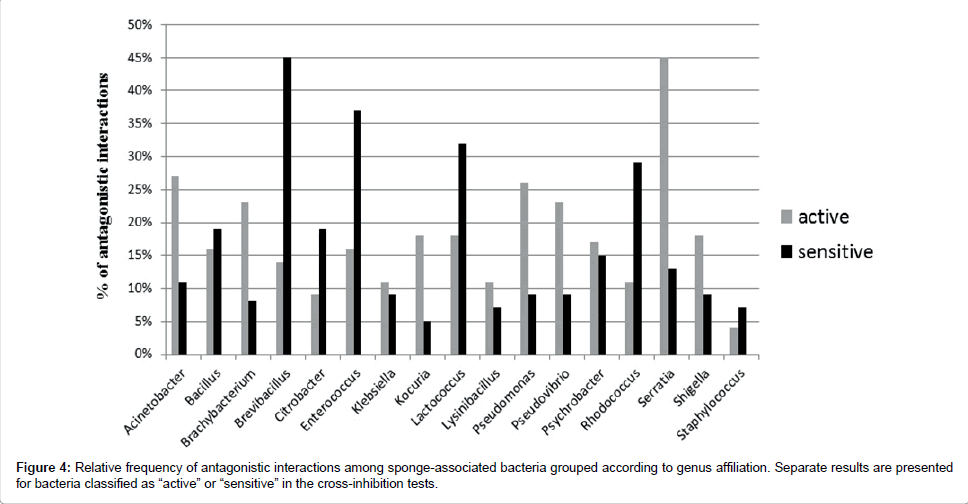
Bacteria belonging to all 17 cultured genera showed inhibitory activity. Bacillus, Klebsiella and Psychrobacter were involved in antagonistic interactions as both, active and sensitive strains in a similar proportion. However, some bacterial genera included a higher number of active than sensitive strains (like Brachybacterium, Kocuria, Pseudomonas, Pseudovibrio and Serratia), while others included more sensitive than active strains (Brevibacillus, Enterococcus, Lactococcusand Rhodococcus) (Figure 4).
Cross-inhibition among isolates associated with different species of sponges collected at the same site and on the same date
Under this condition, relative frequencies of antagonistic interactions among bacteria ranged from 5.5 to 50.0% (Table 3). The weighted mean of all frequencies was 17.7%, a value very close to that (18%) observed among the 56 isolates. Therefore, for a given site at a given sampling date, bacteria isolated from different sponge species cross-inhibited their growth at a frequency that was comparable to the situation in which all strains were tested.
| Site / year | Sponges | Frequencies | Interactivity clusters | ||
| absolute (relative) | I | II | III | ||
| PV/2005 | Mm, Pj | 4 (50.0%) | 2 | 2 | 2 |
| PV/2006 | Ca, Dr, Mm, Pj | 3 (5.5%) | 3 | 2 | 10 |
| PV/2007 | Dr, Hf, Hh, Pj | 2 (11.1%) | 2 | 1 | 4 |
| CA/2007 | Gc, Mm, Pa, Pc, Pm, Ti | 31 (22.5%) | 12 | 7 | 6 |
| CA/2008 | Hv, Pm, Ti | 3 (30.0%) | 2 | 2 | 2 |
| Us/ 2010 | CC, Hh, Mm | 4 (12.5%) | 2 | 4 | 3 |
| Us/2011 | CC, Hh, Mm | 6 (15.8%) | 5 | 3 | 5 |
| Weighted mean 17.7% | |||||
Table 3: Cross-inhibition among bacteria isolates from different sponge species collected from the same site and date.
When isolates were classified into three interactivity clusters, 60% of the strains were resistant in their relationships, followed by 40% of the sensitive strains and finally 32% of the active strains. The resistant cluster includes strains that are either exclusively resistant or both, resistant and active; the sensitive cluster includes strains that are either exclusively sensitive, or sensitive and active; and the active cluster includes strains which proved to be either active and sensitive or active and resistant.
Cross-inhibition among isolates associated with the same sponge species collected at different sites and on different dates
The frequency of antagonistic interactions was also analyzed among isolates from the same sponge species which were spatially separated by about 10 km along a pollution gradient. In this condition, 17.5% of the interactions were inhibitory. This percentage is similar to those measured in the aforementioned analyses. The majority of isolates were classified as resistant, except for isolates from the sponge M. microsigmatosa (Mm), where the active cluster was predominant (Table 4).
| Voucher | Sponge | Site / year | Frequencies | Interactivity clusters | ||
| absolute (relative) | I | II | III | |||
| 62009Dr | Dr | Ca/2009 | 1 (8.3%) | 1 | 1 | 6 |
| 42006Dr | PV/2006 | |||||
| 32007Dr | PV/2007 | |||||
| 12007Hv | H | Ca/2007 | 3 (37.5%) | 2 | 2 | 2 |
| 32007Hf | PV/2007 | |||||
| 32007Hh | Hh | PV/2007 | 2 (16.6%) | 2 | 2 | 5 |
| 92010Hh | Us/2010 | |||||
| 82011Hh | Us/2011 | |||||
| 82005Mm | Mm | Pv/2005 | 15 (17.0%) | 10 | 4 | 8 |
| 42006Mm | Pv/2006 | |||||
| 92010Mm | Us/2010 | |||||
| 82011Mm | Us/2011 | |||||
| Weighted mean | 17.50% | |||||
Table 4: Cross-inhibition among bacteria isolated from the same sponge collected from different sites and dates.
Cross-inhibition among isolates associated with the same species of sponge collected at the same site but in different years
Relationships among bacteria obtained from the same sponge species collected at the same site over 6 years (2005-2011) showed a relative frequency of antagonistic interactions ranging from 0.0 to 50.0% (Table 5). The results were very different among sponge-associated bacteria, and the lowest percentage was observed among the isolates from site Us.
| Sponge | Site / years | Frequencies | Interactivity clusters | ||
| absolute (relative) | I | II | III | ||
| Pc | CA/ 2006, 2007 | 4 (50.0%) | 3 | 3 | 1 |
| Pm | CA/ 2007, 2008 | 0 (0.0%) | 0 | 0 | 2 |
| Ti | CA/ 2007,2008 | 0 (0.0%) | 0 | 0 | 4 |
| Dr | Pv/ 2006, 2007, 2009 | 1 (10.0%) | 1 | 1 | 5 |
| Mm | Pv/ 2005, 2006 | 4 (25.0%) | 2 | 4 | 2 |
| Pj | Pv/ 2005, 2006, 2007 | 5 (31.2%) | 3 | 2 | 3 |
| Cc | Us/ 2010, 2011 | 0 (0.0%) | 0 | 0 | 0 |
| Hh | Us/ 2010, 2011 | 1 (6.2%) | 1 | 1 | 5 |
| Mm | Us/ 2010, 2011 | 1 (16.7%) | 1 | 1 | 3 |
Table 5: Cross-inhibition among bacteria isolates from the same sponge species collected from the same sites and in different years.
Cross-inhibition among isolates associated with the same sponge specimen
Few bacteria inhibited isolates from the same sponge specimen, especially among those from sponges collected at the CA site (Table 6). Relative frequencies of antagonism were closer between the bacteria from Us and PV sites. In addition, the majority of isolates from CA site was classified in the resistant interactivity cluster, (i.e. resistant and active strains, as well as exclusively resistant strains). However, the distribution of the isolates from PV and Us was similar among the three interactivity clusters.
| Voucher number | Sponges | Site / year | Number of bacteria | Frequencies absolute (relative) | Interactivity clusters | ||
| I | II | III | |||||
| 32007Gc | Gc | CA / 2007 | 2 | 1 (50.0%) | 1 | 1 | 1 |
| 12007Hv | Hv | CA / 2007 | 2 | 0 (0.0%) | 0 | 0 | 2 |
| 32007Mm | Mm | CA / 2007 | 2 | 0 (0.0%) | 0 | 0 | 2 |
| 32007Pa | Pa | CA / 2007 | 3 | 0 (0.0%) | 0 | 0 | 3 |
| 42006Pc | Pc | CA / 2006 | 2 | 1 (50.0%) | 1 | 1 | 1 |
| 32007Pc | Pc | CA / 2007 | 2 | 0 (0.0%) | 0 | 0 | 2 |
| 32007Ti | Ti | CA / 2007 | 3 | 0 (0.0%) | 0 | 0 | 3 |
| 42006Dr | Dr | Pv / 2006 | 5 | 0 (0.0%) | 0 | 0 | 0 |
| 42006Hf | Hf | Pv / 2007 | 2 | 1 (50.0%) | 1 | 1 | 1 |
| 82005Mm | Mm | Pv / 2005 | 4 | 2 (16.7%) | 1 | 2 | 2 |
| 42006Mm | Mm | Pv / 2006 | 2 | 1 (50.0%) | 1 | 1 | 1 |
| 52005Pj | Pj | PV / 2005 | 2 | 2 (100.0%) | 2 | 2 | 0 |
| 42006Pj | Pj | PV / 2006 | 2 | 0 (0.0%) | 0 | 0 | 2 |
| 92010Cc | Cc | Us / 2010 | 2 | 0 (0.0%) | 0 | 0 | 0 |
| 82011Cc | Cc | Us / 2011 | 3 | 0 (0.0%) | 0 | 0 | 0 |
| 92010Hh | Hh | Us / 2010 | 2 | 0 (0.0%) | 0 | 0 | 2 |
| 82011Hh | Hh | Us / 2011 | 4 | 2 (16.7%) | 1 | 2 | 2 |
| 92010Mm | Mm | Us / 2010 | 3 | 2 (33.3%) | 1 | 2 | 1 |
Table 6: Cross-inhibition among bacteria isolated from the same sponge specimen.
Interestingly, sponge-associated bacteria which were active against numerous strains when all were cross-tested (see previous results) and showed no inhibitory activity against bacterial isolates from the same sponge. This was the case for strains H41 and Hh93 which inhibited 64% and 50% of all 55 tested bacteria, respectively (Table 2).
Discussion
Bacteria from a wide range of marine environments, including sediments, seawater, biofilms, and tissues/surfaces of invertebrate and algae, have been shown to possess antagonistic activities. In most cases, these bacteria are members of complex communities in which competition for limited space and resources can be intense [19]. Antagonistic interactions may play an important role in structuring these communities, where the evolutionary advantages afforded by an effective chemical defense may be crucial for survival.
The bacterial isolates belonged to the Firmicutes, Gammaproteobacteria, Actinobacteria and Alphaproteobacteria classes. These bacterial groups represented the predominant cultivable sponge-associated bacteria reported in other studies [2,3,20,21]. It is known that many bacterial inhabitants in sponges appear to be recalcitrant to cultivation on laboratory media, probably reflecting their evolutionary adaptation to the conditions provided by the host [2]. Therefore, the classical culture dramatically underestimates microbial numbers and diversity found in the samples under study. However, the major advantage of this approach over modern molecular techniques is that it provides the researcher with live microbes, which can be used in further studies [22], such as the present one.
This study is the first to analyze the antagonistic interactions among 56 sponge-associated bacteria. Bacteria belonging to all 17 cultured genera showed inhibitory activity. Our data demonstrate that the antagonistic interaction is present among the isolates and expression of this phenotype depends on both the identity of the producer strain and that of the indicator strain. In contrast to terrestrial environments, which are essentially static, the marine environment also involves dispersion and movement of communities driven by hydrography, thus complicating the interpretation of results [23].
All strains used in this study were previously analyzed for their production of antimicrobial substances against bacteria of medical importance [15,18]. Overall, the percentage of active bacteria found in this study (98.2%) was much higher than those (22%) reported for the same isolates against clinical pathogenic bacteria. We suggest that the enhanced production of bioactive compounds may occur to inhibit other sponge bacteria, i.e. potential in situ competitors for nutrients and space [24]. Clinical pathogenic bacteria are not natural competitors for sponge-associated bacteria and therefore lack the secondary metabolites that would induce the antimicrobial activity [25].
It is relevant to note that secondary metabolites from marine bacteria are involved in a variety of processes, including nutrient acquisition [26] and chemical communication [27]. Mechanisms responsible for antagonistic effects can vary widely, ranging from direct cell killing, as in the case of an antibiotic, to the removal of an essential nutrient, as in the case of an iron chelating siderophore. Antagonism also can result from the production of small organic acids or other compounds that render the environment unsuitable for growth of competing bacteria [28]. These compounds likely play important ecological roles that ultimately affect ecosystem structure and function; however, much remains to be discovered before these processes can be fully appreciated [29].
Temporal and spatial variability of sponge bacterial communities has been discussed and some researchers have shown that there are differences among bacterial communities across sponge species or even specimens [30,31]. Indeed marine sponges are well known for their associations with highly diverse, yet very specific and often highly similar microbiota [2]. Furthermore, sponges filter large amounts of water and can collect contaminants from both dissolved and particulate phases [32]. The nature of accumulation between different sponge species may be related to composition of symbiotic microorganism communities, skeletal composition, histology, and life cycle [14]. This can be seen with the data obtained in this study, where no antagonistic activity was observed among isolates from C. celata (Us site, 2010-2011), P. magna (CA site, 2007-2008) and T. ignis (CA site, 2007-2008) sponges, whereas isolates from P. citrina (CA site, 2006/2007) were the most active, including among themselves. This data was not surprising, since bacterial communities are dynamic in respect of responding to environmental conditions. Moreover, seasonal changes in the production of bioactive compounds by sponges may be considered [33]. Sponges therefore contain a uniform, sponge-specific bacterial community, although each sponge species contains different bacterial species [34]. Recent advances in studying the dynamics of marine bacterial communities have shown that the composition of these communities follows predictable patterns and involves complex network interactions, which shed light on the underlying processes regulating these globally important organisms [35].
It is important to point out that the Guanabara Bay, in the Rio de Janeiro state, Southeast Brazil, is one of the largest and most polluted estuaries on the Brazilian coast [36]. CA integrates a protected marine area situated approximately 8 km southwest from the entrance of the Guanabara Bay. These islands are important areas for fishery and tourism and receive alternating influence from clean waters from oceanic currents and polluted waters from coastal discharges. The polluted waters from Guanabara Bay also have some influence in this area, which are, however, much less polluted than the center of the bay [14]. The collection sites are therefore located along a marked environmental gradient, from highly polluted (Us) to moderately (PV) and slightly polluted sites (CA).
Our study traced the broad profile of antagonistic interactions among sponge-associated bacteria isolated from the same or different sponge species, sites and years. However, in order to establish a connection between an antagonistic activity observed in the laboratory and an ecologically meaningful effect, many factors must be considered, such as the biogeography, seasonal variation and environmental factors. Future studies will be conducted with greater control over these variables. Overall, our study demonstrates that antagonism could be a structuring force in sponge-associated microbial communities.
Acknowledgements
This work was supported by grants from CAPES, CNPq and FAPERJ to M.S. Laport, and by a grant from FRS-FNRS to I. George. We are also grateful to Science without Borders Program / CNPq for the post doctorate scholarship to M.S. Laport.
References
- Wilkinson CR, Garrone R, Vacelet J (1984) Marine sponges discriminate between food bacteria and bacterial symbionts: electron microscope radioautography and in situevidence.Proc R Soc Lond B Biol Sci 220: 519-528.
- Taylor MW, Radax R, Steger D, Wagner M (2007) Sponge associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev 71: 295-347.
- Webster NS, Taylor MW (2012) Marine sponges and their microbial symbionts: love and other relationships. Environ Microbiol 14: 335-346.
- Vacelet J (1975) Étude en microscopie e´lectronique de l’association entre bactéries et spongiaires du genre Verongia (Dictyoceratida). J De Microscopie Et De BiologieCellulaire 23: 271-288.
- Santos-Gandelman JF, Giambiagi-deMarval M, Oelemann WMR, Laport MS (2014) Biotechnological potential of sponge-associated bacteria. Curr Pharm Biotechnol 15: 143-155.
- Van Soest RWM, Boury-Esnault N, Hooper JNA, Rützler K, de Voogd NJ, et al. (2016) World Porifera database.
- Muricy G (2015) Porifera Brasil.
- Rypien KL, Ward JR, Azam F (2010) Antagonistic interactions among coral-associated bacteria. Environ Microbiol 12: 28-39.
- Shnit-Orland M, Sivan A, Kushmaro A (2012) Antibacterial activity of Pseudoalteromonas in the coral holobiont. Microb Ecol 64: 851-859.
- Mangano S, Michaud L, Caruso C, Brilli M, Bruni V, et al. (2009) Antagonistic interactions between psychrotrophic cultivable bacteria isolated from Antarctic sponges: a preliminary analysis. Res Microbiol 160: 27-37.
- Strahl ED, Dobson WE, Lundie LL Jr (2002) Isolation and screening of brittlestar-associated bacteria for antibacterial activity. Curr Microbiol 44: 450-459.
- Grossart HP, Schlingloff A, Bernhard M, Simon M, Brinkhoff T (2004) Antagonistic activity of bacteria isolated from organic aggregates of the German Wadden Sea. FEMS Microbiol Ecol47: 387-396.
- Santos-Gandelman JF, Santos OC, Pontes PV, Andrade CL, Korenblum E, et al. (2013) Characterization of cultivable bacteria from Brazilian sponges. Mar Biotechnol 15: 668-676.
- Batista D, Muricy G, Rocha RC, Miekeley NF (2014) Marine sponges with contrasting life histories can be complementary biomonitors of heavy metal pollution in coastal ecosystems. Environ Sci Pollut Res Int 21: 5785-5794.
- Santos OCS, Pontes PVML, Santos JFM, Muricy G, Giambiagi-deMarval M, et al. (2010) Isolation, characterization and phylogeny of sponge-associated bacteria with antimicrobial activities from Brazil. Res Microbiol 161: 604-612.
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol173: 697-703.
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, et al. (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188-7196.
- Marinho PR, Moreira AP, Pellegrino FL, Muricy G, Bastos MC, et al. (2009) Marine Pseudomonasputida: a potential source of antimicrobial substances against antibiotic-resistant bacteria. Mem Inst Oswaldo Cruz 104: 678-682.
- Hibbing ME, Fuqua C, Parsek MR, Peterson SB (2010) Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8: 15-25.
- Sipkema D, Schippers K, Maalcke WJ, Yang Y, Salim S, et al. (2011) Multiple approaches to enhance the cultivability of bacteria associated with the marine sponge Haliclona (gellius) sp. Appl Environ Microbiol 77: 2130-2140.
- Montalvo NF, Davis J, Vicente J, Pittiglio R, Ravel J, et al. (2014) Integration of culture-based and molecular analysis of a complex sponge-associated bacterial community. PLoS One 9: e90517.
- Al-Awadhi H, Dashti N, Khanafer M, Al-Mailem D, Ali N, et al. (2013) Bias problems in culture-independent analysis of environmental bacterial communities: a representative study on hydrocarbonoclastic bacteria. SpringerPlus 2: 369.
- Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, et al. (2012) Defining seasonal marine microbial community dynamics. ISME J 6: 298-308.
- Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, et al. (1994) Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad SciUSA 91: 197-201.
- Kanagasabhapathy M, Nagata S (2008) Cross-species induction of antibacterial activity produced by epibiotic bacteria isolated from Indian marine spongePseudocratina purpurea. World J Microbiol Biotechnol 24: 687-691.
- Hider RC, Kong X (2010) Chemistry and biology of siderophores. Nat Prod Rep 27: 637-657.
- Straight PD, Kolter R (2009) Interspecies chemical communication in bacterial development. Annu Rev Microbiol 63: 99-118.
- Schnurer J, Magnusson J (2005) Antifungal lactic acid bacteria as bio-preservatives. Trends Food Sci Technol 16: 70-78.
- Wietz M, Duncan K, Patin NV, Jensen PR (2013) Antagonistic interactions mediated by marine bacteria: the role of small molecules. J Chem Ecol 39: 879-891.
- Kvennefors ECE, Sampayo E, Ridgway T, Barnes AC, Hoegh-Guldberg O (2010) Bacterial communities of two ubiquitous Great Barrier Reef corals reveals both site-and species-specificity of common bacterial associates. PLoS ONE 5: e10401.
- Hardoim CC, Esteves AI, Pires FR, Gonçalves JM, Cox CJ, et al. (2012) Phylogenetically and spatially close marine sponges harbour divergent bacterial communities. PLoS One 7: e53029.
- Pérez T, Longet D, Schembri T, Rebouillon P, Vacelet J (2005) Effects of 12 years’ operation of a sewage treatment plant on heavy metal occurrence within a Mediterranean commercial sponge (Spongia officinalis, Demospongiae). Mar Pollut Bull 50: 301-309.
- Sacristán-Soriano O, Banaigs B, Becerro MA (2012) Temporal Trends in the Secondary Metabolite Production of the Sponge Aplysina aerophoba. Mar Drugs 10: 677-693.
- Schmitt S, Tsai P, Bell J, Fromont J, Ilan M, et al. (2012) Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J 6: 564-576.
- Fuhrman JA, Cram JA, Needham DM (2015) Marine microbial community dynamics and their ecological interpretation. Nat Rev Microbiol 13: 133-146.
- Kjerfve B, Ribeiro CA, Dias GTM, Filippo A, Quaresma VS (1997) Oceanographic characteristics of an impacted coastal bay: Baía de Guanabara, Rio de Janeiro. Coast Shelf Res 17: 1609-1643.
Relevant Topics
- Algal Blooms
- Blue Carbon Sequestration
- Brackish Water
- Catfish
- Coral Bleaching
- Coral Reefs
- Deep Sea Fish
- Deep Sea Mining
- Ichthyoplankton
- Mangrove Ecosystem
- Marine Engineering
- Marine Fisheries
- Marine Mammal Research
- Marine Microbiome Analysis
- Marine Pollution
- Marine Reptiles
- Marine Science
- Ocean Currents
- Photoendosymbiosis
- Reef Biology
- Sea Food
- Sea Grass
- Sea Transportation
- Seaweed
Recommended Journals
Article Tools
Article Usage
- Total views: 11594
- [From(publication date):
April-2016 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 10710
- PDF downloads : 884