Annotsv and Knotannotsv: Human Structural Variations Annotations, Ranking and Analysis
Received: 01-Sep-2022 / Manuscript No. jpgb-22-129 / Editor assigned: 05-Sep-2022 / PreQC No. jpgb-22-129(PQ) / Reviewed: 19-Sep-2022 / QC No. jpgb-22-129 / Revised: 26-Sep-2022 / Manuscript No. jpgb-22-129 (R) / Published Date: 30-Sep-2022
Abstract
With the dramatic increase of pangenomic analysis, Human geneticists generate large amount of genomic data including millions of small variants (SNV/Indel) but also thousands of structural variations (SV) mainly from nextgeneration sequencing and array-based techniques. To help identifying human pathogenic SV, we have developed a webserver dedicated to their annotation and ranking (AnnotSV, PMID: 29669011, PMID: 34023905) as well as their visualization and interpretation (knotAnnotSV, PMID: 34023905) at the following address: https://www.lbgi.fr/ AnnotSV/.
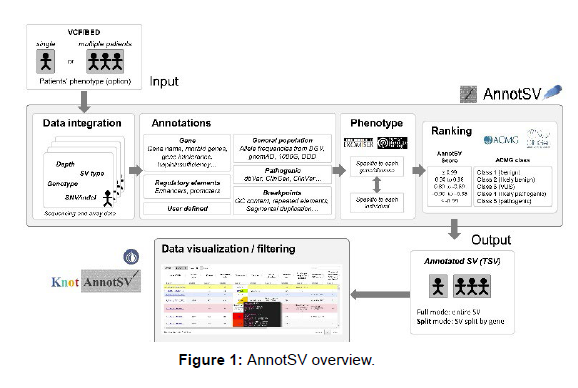
First, the available data sources in our annotation engine (AnnotSV) includes, among others, databases such as DGV, gnomAD, DDD, OMIM, intolerance score and known pathogenic SV (dbVar, ClinVar and ClinGen) as well as users own annotations (e.g. patient’s SNV/indel...). Second, a phenotype driven analysis based on HPO and Exomiser has been implemented. Third, an automatic SV classification based on the latest ACMG recommendations (PMID:31690835) is available. Finally, knotAnnotSV displays the annotated SV in an interactive way including popups, filtering options, advanced colouring to highlight pathogenic SV and hyperlinks to the UCSC genome browser or other public databases. The annotation is available for the SV as a whole (full/compact mode) or divided for each overlapping gene (split/expanded mode). Output can be either visualized in a web browser directly or using a specific link, or downloaded as a tab separated file. To our knowledge, this makes our webserver the most comprehensive online SV annotation and interpretation tool.
This new version of the AnnotSV web server can be accessed at the following address: https://lbgi.fr/AnnotSV/. The underlying annotation engine (AnnotSV) has been upgraded to version 3 and is getting more and more citations since its publication in 2018 (80 in total, decomposed in 2 in 2018, 8 in 2019, 17 in 2020 and already 53 in 2021)
First, the available data sources in our annotation engine (AnnotSV) includes, among others, databases such as DGV, gnomAD, DDD, OMIM, intolerance score and known pathogenic SV (dbVar, ClinVar and ClinGen) as well as users own annotations (e.g. patient’s SNV/indel...). Second, a phenotype driven analysis based on HPO and Exomiser has been implemented. Third, an automatic SV classification based on the latest ACMG recommendations (PMID:31690835) is available. Finally, knotAnnotSV displays the annotated SV in an interactive way including popups, filtering options, advanced colouring to highlight pathogenic SV and hyperlinks to the UCSC genome browser or other public databases. The annotation is available for the SV as a whole (full/compact mode) or divided for each overlapping gene (split/expanded mode). Output can be either visualized in a web browser directly or using a specific link, or downloaded as a tab separated file. To our knowledge, this makes our webserver the most comprehensive online SV annotation and interpretation tool.
This new version of the AnnotSV web server can be accessed at the following address: https://lbgi.fr/AnnotSV/. The underlying annotation engine (AnnotSV) has been upgraded to version 3 and is getting more and more citations since its publication in 2018 (80 in total, decomposed in 2 in 2018, 8 in 2019, 17 in 2020 and already 53 in 2021)
Recent Publications
1. Geoffroy et al. (2018) AnnotSV: An integrated tool for Structural Variations annotation. Bioinformatics.
2. Geoffroy et al. (2021) AnnotSV and knotAnnotSV: a web server for human structural variations annotations, ranking and analysis.
3. Riggs et al. (2020) Technical Standards for the Interpretation and Reporting of Constitutional Copy-Number Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genetics in Medicine.
4. Ann M. Mc Cartney et al. (2021) An international virtual hackathon to build tools for the analysis of structural variants within species ranging from coronaviruses to vertebrates. F1000Research. 5. Surajit Bhattacharya et al. (2019) Guidelines for
Bioinformatics and the Statistical Analysis of Omic Data. Omics Approaches to Understanding Muscle Biology 45-75.
Biography
Véronique Geoffroy professional interests are focused on Next Generation Sequencing data, especially the use of computational methods in human genetics and genomics. She strongly supports open sciences and she has developed under the GNU-GPL open source license, AnnotSV, a program designed for annotating and ranking Structural Variations.
Citation: Veronique G (2022) Annotsv and Knotannotsv: Human Structural Variations Annotations, Ranking and Analysis. J Plant Genet Breed 6: 129.
Copyright: © 2022 Veronique G. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.