Analysis of Tomato Agronomic Traits using Generation Mean
Received: 05-Jul-2022 / Manuscript No. JPGB-22-68631 / Editor assigned: 07-Jul-2022 / PreQC No. JPGB-22-68631(PQ) / Reviewed: 21-Jul-2022 / QC No. JPGB-22-68631 / Revised: 25-Jul-2022 / Manuscript No. JPGB-22-68631(R) / Accepted Date: 29-Jul-2022 / Published Date: 30-Jul-2022
Abstract
This study aimed at determining inheritance of agronomic traits viz. plant height, days to 50% flowering, inter truss spacing and number of trusses per plant in local and introduced tomato lines. Six generations; P₁, P₂, F₁, F₂, BC₁P₁ and BC₁P₂ were developed for each of four bi-parental crosses between five genetically diverse parental lines; AVTO1429, Roma VF, AVTO1424, AVTO1314 and Valoria. A split-plot design, crosses as main plots and generations as subplot with three replicates was used in two sites during 2019, long rain season. Cross Roma VF x AVTO1424 and Roma VF x AVTO1314 were the earliest to 50% flowering in 33 days while Roma VF x Valoria select was the latest in 35 days. F1 hybrid of Roma VF x AVTO1314 showed flowering within 32 days whereas 35 days in P1 (Roma VF). Mwea Station had the tallest mean plant heights of 62cm (at 50% flowering) compared to Kabete Station with 48cm in all crosses and generations. A significant increase (>10%) in plant height at 50% flowering in comparison to parental genotypes was registered in F1 generations. Final plant height across the environments ranged from 82cm for shorter parent (Roma VF) of Roma VF x AVTO1429 to 120 cm for taller offspring, BC₁P₁. Notably, both inter truss spacing and number of trusses per plant were not significantly different (P≤0.05) for crosses evaluated in both sites. Agronomic traits which showed significant genotype x environment interaction in Roma VF x AVTO1314 were days to 50% flowering, final plant height, and number of trusses per plant whereas, in Roma VF x AVTO1429 it was plant height at 50% flowering and number of trusses per plant. The importance of gene effects for agronomic trait inheritance was in additive and dominance-additive portions which implied that the traits were inherited.
Keywords
Generation mean analysis; inheritance; agronomic traits; additive; dominance
Introduction
Tomato constitutes 7% of total horticulture produce in Kenya and 14% of the entire vegetable produce [1]. The demand for fruit quality and diversity in agronomic traits attributes for diverse production systems by Kenyan consumers and growers continues to increase, hence the need to improve the existing cultivars to mitigate the gap [2]. Of concern, from 2016 to 2018 there has been 30% decrease in tomato production against increasing demand of 300,000 tonnes [3], despite the increase of more than 41.7% in tomato consumption per capita [4]. The consequence of decline in tomato productivity in Kenya was tomato prices became unaffordable, resulting to importation of over 27,000 tonnes from Ethiopia and Tanzania [3].
Generation means analysis had been widely adopted in assessing the estimation of main genetic effects such as additive, dominance and their digenic interactions as associated with the expression of quantitative traits [5]. Generation mean analysis had been used in the determination of yield, cold tolerance, vitamin C and total soluble solids and acyl sugar content in tomato [6-8]. Morphological and agronomic traits including leaf, floral, growth habit, crop yields and yield components provide valuable information for crop improvement programmes [9, 10] revealed that morphological and agronomic traits not only provide consumer satisfaction and quality raw materials for the processing industry but also enhance the competitiveness of tomato crop in horticultural sector. Knowledge on the relative contribution of various traits to yields can significantly facilitate identification of high yielding genotypes from a population of varying genotypes [11]. Study finding of Adelana, (1975) as reported by attributed poor tomato yields as a result of flowers not developing into fruits [12]. There are limited tomato technologies, innovations and management practices in Kenya and most African countries [1, 13]. Moreover, breeding programs in Kenya have only focused on cereals, pulses, root and beverage crops, hence no tomato breeding programme had been initiated by either public or private company [14].
Improved tomato cultivars especially hybrids are more productive because of the commonly reported fruit yield heterosis of 20 to 50%, which make farmers interested in growing F1 varieties. Besides the high yields, hybrids exhibit other advantages such as early maturity, resistance to pests and diseases, growth vigour that help overcome abiotic stresses like drought and big fruit size of high quality [15]. It is worth mention that, lack of varieties adapted to different agro-ecological zones across the country poses a challenge to farmers growing hybrid tomatoes [16]. This therefore means, demand-led breeding of locally adapted improved varieties especially hybrids with market preferred traits coupled with within seed production will ensure these varieties are easily accessible and affordable by farmers than the imported hybrid varieties [13]. The objective of this study was to determine the inheritance of growth attributes of tomato genotypes and identifying the cross family with great potential for further breeding.
Material and Methods
Experimental sites
Experiments were conducted at Kabete Field Station and Mwea Research Station (2018-2019) in Kiambu and Kirinyaga Counties, respectively. Kabete Field Station is located at 01°15’S; 036°44’E with an elevation of 1820m above sea level (ASL) which is at agro-ecological zone (AEZ) III. It has a bimodal rainfall of 1059 mm per year distributed in two seasons which are the long rains (March to May) and short rains (October to December). Temperature ranges from 12.3 to 22.5°C and soils are humic nitisols that are deep and well-drained with a pH of 5.0 to 5.4 [17].
Mwea Research Station is located at 0°41’S; 037°21’E with an elevation of 1247m ASL which is at agro-ecological zone II. The area has a bimodal rainfall regime of 973 mm annually with long rains (March to May) and short rains (October to December). Temperature ranges from 15.6 to 28.6°C and soils are Niti-rhodic ferrosols with a pH of about 5.1 [18].
Experimental design
The experiment involved development of study populations and field evaluations of progenies and their parents. Study populations were developed at Kabete Field Station (April and September 2018) using a randomized complete block design with three replicates. Hybridization of 5 parental lines in 10 x 10 half diallel mating design excluding reciprocals was carried out from April-August 2018 and backcrosses to both parents from September-December 2018 at Kabete Field Station following a modified protocol of [19].
Plant materials
The study used 5 tomato genotypes, i.e., 3 breeding genotypes from the World Vegetable Centre (AVRDC) in Taiwan namely; AVT01424, AVT01429 and AVT01314, a commercial cultivar from Continental Seeds Company Limited known as Roma VF, and Valoria selection from farmers in Kirinyaga County. Genotypes AVTO1424 and ATO1314 are semi-determinate and AVTO1429 is indeterminate that matures and flowers early suitable for open field cultivation. However, performances of these genotypes and productivity in terms of yields have not been determined in Kenya [13]. Commercial variety Roma VF is a determinate pure-line that flowers and matures early. Moreover, this variety is low yielding, require staking, lacks trait for resistance to bacterial wilt and insect pests [1]. Valoria selection is a determinate line preferred by farmers in Central Kenya and requires staking. Besides, the selection is low yielding, late flowering, late maturing, and their traits have not been validated.
Development of Study populations
Four bi-parental crosses were developed from Roma VF and AVTO1429, AVTAO1424, AVTO1314 and Valoria Select giving F1 hybrids. The F1s’ were backcrossed to both parents (BC₁P₁ and BC₁P₂) and also advanced to F2 at Kabete Field Station during September- December, 2018 following a protocol by [20]. Six generations were developed for each cross; P₁, P₂, F₁, F₂, BC₁P₁ and BC₁P₂. Field trial evaluations were carried out at Kabete Field Station and Mwea Research Station during the long rain season (April-August 2019).
Evaluation of study populations
Seedlings were raised in germination trays with 204-cells (3.5 cm deep and 2.5 cm wide) containing peat moss as planting media at Kabete Field Station on 6th March 2019. Trays were sown with one seed per cell and raised under a net-house. Seedlings were watered daily in hot weather and once on a two-day interval in cool weather to provide sufficient moisture for growth. Seedlings having 4 true leaves were hardened by reducing watering 25 days after sowing. Netting was removed to expose the seedling to sunlight to become stocky and sturdy. Seedlings were watered 12 hours before transplanting in the field. One-month-old seedlings having pencil thickness were then transplanted in open fields for evaluations at Kabete Field Station and Mwea Research Station on 8th April 2019. Transplanting was done early in the morning to reduce the transplanting shock and watered immediately as described in manual [21]. The land was prepared by deep (45cm) ploughing to improve the soil structure, water holding capacity and to achieve a fine tilth. Regular ridges of 30 cm high and 25 cm wide were made to raise the beds.
A split-plot design with four families as main plots and the six generations as subplot replicated three times was established. The main plot had a configuration of 36x54 meters with 18 subplots of 2x3 meters. Each subplot had four rows each having five plants. The number of plants per plot varied with generations. The segregating F2 and backcross populations were assigned more rows than the nonsegregating F1 and parental populations as follows; 40 rows with 200 plants for F2 generation, 20 rows with 100 plants for backcross generations and 4 rows with 20 plants for each non-segregating population (P1, P2 and F1) following a modified procedure of [22].
Crop Management
The crop was maintained weed-free by hand-weeding at a 2-3 weeks interval. The crop was mainly rain-fed and supplemented with drip irrigation. On planting, both Di-ammonium phosphate (DAP) fertilizer (18: 46: 0) and N: P: K (17: 17: 17) were each applied at the rate of 12g plant-1 during transplanting. The plants were top-dressed with calcium ammonium nitrate (CAN) at the rate of 100 kg/ha when plants were 25 cm high and 200kg/ha 55 days after transplanting. Fertilizer application was to ensure adequate nutrient levels for the crop to prevent deficiency disorders [21]. Metalaxyl-M and Propineb (700g/ kg) at the rate of 50g / 20 litres of water was applied at an interval of two weeks to manage against early and late blights. Imidacloprid (100g l-1) and betacyfluthrine (45g l-1) at the rate of 0.2 l ha-1 and Thiamethoxam at the rate of 8g / 20 litres water were used to control aphids, whiteflies, and leaf miners during the crop growth cycle.
Data collection
Data on 50 F1 plants, 50 plants of each parent in a cross, 300 plants of each backcross and 600 plants of the F2 generation was collected following protocol [20]. Assessed parameters in the field trials were plant height (cm), duration to 50% flowering and the number of trusses per plant. Plant height was determined from the soil base of the plant to the main stem using a metre rule at 50% flowering and physiological maturity stage. Duration to 50% flowering (days) was determined when half of plants population in a plot had one flower. A sample of 6 random plants per plot was assessed on the number of trusses from the main stem and averaged at the harvesting stage. Inter truss spacing was determined in centimetre using a metre rule as the distance between two trusses from a sample of 6 random plants per plot and averaged
Data analysis
Traits for each generation and crosses comparison data were subjected to analysis of variance (ANOVA) to determine their significant differences using GenStat software 15th edition. The variables for which ANOVA showed significant differences between the generations, their means were separated using Tukey’s procedure for multiple comparisons (P≤ 0.05) following a protocol of [22]. The significantly different variables showed by orthogonal contrasts between parents P1 and P2 were further subjected to generation mean analysis (GMA) to establish if the respective traits are quantitatively or qualitatively inherited using the methodology proposed by [22].
Segregation ratios were subjected to chi-square tests to establish goodness-of-fit for observed ratios. The outcome was compared with the observed results to determine whether the differences are because of chance or other traits hence:-
Chi-square= (Observed- Expected)2 / Expected
Hence, ᵡ2= Σ [(O-E)2 / E]
The calculated chi-square value was used to determine P (probability) value from the chi-square table. If P-value obtained <5%, the variation between the segregating ratios was influenced by other traits but if P-value >5% the variation was due to chance and within the acceptable deviation [23].
Generation mean analysis
Calculation of generation mean analysis followed the approach of [20] as follows: Development of generation means: - this was calculated by summing the number of observations for a trait in each generation and dividing by the total number (n) of sampled plant i.e. =T/n. Calculating the variance and mean-variance of each generation was: Variance for each generation =Σ SS/ (n-1) and Mean-variance for each generation was =V/n. Epistasis affects the estimation of additive and dominance components of variance. Scaling tests were used to determine epistatic effects for traits studied and appropriate model for genetic analysis. Four scales A, B, C, D were used to determine the presence of an additive, dominance, and the type of interaction effects. Computation of the scales was achieved as A= P1 +F1-2BC1, B=P2+F1- 2BC2, C=P1+P2+2 F1-4 F2 and D=2F2-BC1-BC2 Where: A= additive x dominance (P1), B= additive x dominance (P2); C= dominance x dominance; D=additive x additive. Test for significance of each scale was carried out using the equation t (A) = A/SE (A). Where: A= additive x dominance (P1) and SE= Standard error. This was done for each scaling test. Significance of even one of the 4 scales showed the presence of epistasis, therefore necessitated analysis of components of means. Analysis of components of means in crosses with epistasis was conducted using 6-parameters model since backcrosses were used following a procedure of [20].
Results
Days to 50% flowering
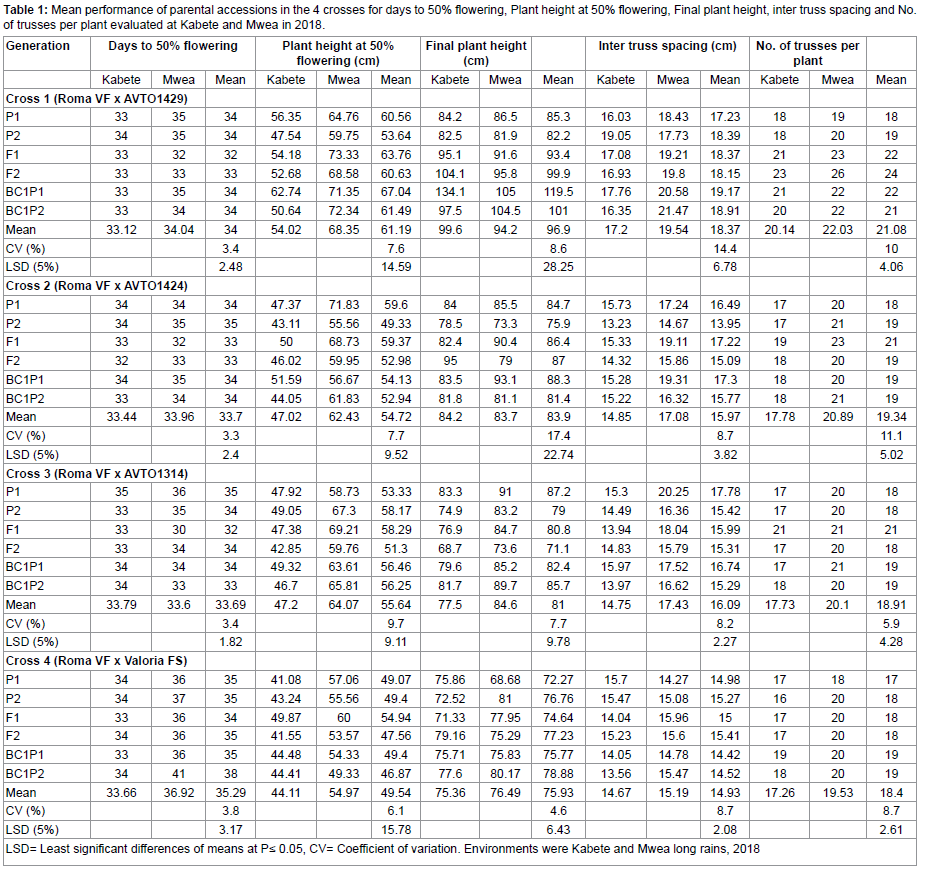
In the assessment of duration to 50% flowering, the six generations of cross Roma VF x AVTO1429 and Roma VF x AVTO1424 had no significant difference (P≤0.05) in all sites. Significant differences were observed in cross Roma VF x AVTO1314 and Roma VF x Valoria select at (P≤0.05). Based on the mean of days to 50% flowering, cross Roma VF x AVTO1424 and Roma VF x AVTO1314 had an equal mean of 33 days while Roma VF x AVTO1429 and Roma VF x Valoria select had a mean of 34 and 35, respectively. Despite the marginal differences in Roma VF x AVTO1314 six generations, the F1 hybrid showed flowering within 32 days whereas 35 days in P1 (Roma VF). Similarly, F1 hybrid in cross Roma VF x Valoria select flowered within 34 days which was significantly different from BC₁P₂ that flowered within 38 days (Table 1). Both Kabete Field Station and Mwea Research Station showed marginal differences in the days to 50% flowering in all the crosses (P≤0.05).
Interactions between the two environments (Kabete Field Station and Mwea Research Station) and the genotypes in cross Roma VF x AVTO1314 and Roma VF x Valoria select showed significant differences (P≤0.01) but none in cross Roma VF x AVTO1429 and Roma VF x AVTO1424, respectively. All the scaling tests showed significant differences (P≤0.01) in cross Roma VF x AVTO1429. From the scaling tests, the additive, dominance, additive x additive interaction and additive x dominance effects were -0.08**, -0.63**, 1.76** and -2.04**, respectively (** means significant at 1 % probability levels) (Table 2). Therefore, further analysis using a 6-parameter model was carried out since backcrosses were used. Roma VF x AVTO1429 showed presence of epistasis. Results showed that a combined gene effect of 3.6 was higher than the interaction components of 2.29 put together (Table 3).
| Scales | Days to 50% flowering | Plant height at 50% flowering (cm) | Final plant height (cm) | Inter truss spacing (cm) | No. of trusses per plant |
|---|---|---|---|---|---|
| A= (P͞1 +F͞1-2B͞C1) | -0.08** | -9.76** | -60.3ns | -2.74ns | -2.61** |
| B= (P͞2+F͞1-2B͞C2) | -0.63** | -5.58** | -26.4ns | -1.06ns | -1.66ns |
| C= (P͞1+P͞2+2 F͞1-4 F͞2) | 1.76** | -0.8** | -45.3ns | -0.24ns | -16.21ns |
| D= (2F͞2-B͞C1-B͞C2) | -2.04** | -7.27ns | -20.7ns | -1.78** | 5.97ns |
| Gene effects / Components | Final plant height (cm) | Plant height at 50% flowering (cm) | Days to 50% flowering | Number of trusses per plant | Inter truss spacing (cm) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| at df. | Expectation / Estimate | Standard error | t (gene effect/SE) | Expectation / Estimate | Standard error | t (gene effect/SE) | Expectation / Estimate | Standard error | t (gene effect/SE) | Expectation / Estimate | Standard error | t (gene effect/SE) | Expectation / Estimate | Standard error | t (gene effect/SE) | |
| Mean | 629 | 99.9 | 1.43 | 69.69** | 60.63 | 0.64 | 94.49** | 32.87 | 0.13 | 247.74** | 24.41 | 0.34 | 70.83** | 18.15 | 0.24 | 76.054** |
| additive effect | 502 | 18.5 | 3.15 | 5.87** | 5.55 | 1.38 | 4.03** | 0.26 | 0.28 | 0.91ns | 0.27 | 0.6 | 0.45ns | 0.26 | 0.58 | 0.45ns |
| dominance effect | 1,506.00 | 51.05 | 6.34 | 8.06** | 21.2 | 3.11 | 6.82** | 2.28 | 0.85 | 2.69** | -8.28 | 1.96 | -4.21** | 4.12 | 1.61 | 2.55** |
| Add. x Add. Interaction | 1,131.00 | 41.4 | 8.52 | 4.86** | 14.54 | 3.76 | 3.86** | 4.08 | 0.78 | 5.24** | -11.94 | 1.83 | -6.52** | 3.56 | 1.5 | 2.38ns |
| Add. Dom. Interaction | 752 | 33.9 | 6.78 | 5.00** | 4.18 | 3.12 | 1.34ns | 1.06 | 0.69 | 1.54ns | 0.95 | 1.39 | 0.68ns | 1.68 | 1.36 | 1.23ns |
| Dom. x Dom. Interaction | 1,381.00 | -128.1 | 14.52 | -8.82** | -29.88 | 6.88 | -4.35** | -6.4 | 1.43 | -4.49** | 7.67 | 3.12 | 2.46ns | -7.36 | 2.77 | 2.65** |
Source |
Mean squares for plant height at 50% flowering | ||||
|---|---|---|---|---|---|
| Df | Cross 1 (Roma VF x AVTO1429) | Cross 2 (Roma VF x AVTO1424) | Cross 3 (Roma VF x AVTO1314) | Cross 4 (Roma VF x Valoria select) | |
| Replication | 2 | 826.97 | 446.12 | 200.62 | 639.67 |
| Environmentsß | 1 | 1848.12 | 2136.13* | 2560.76** | 1062.71 |
| Residual | 2 | 179.77 | 70.07 | 27.69 | 183.44 |
| Generations | 5 | 118.35** | 97.08** | 46.41 | 48.52 |
| Environment. Generations | 5 | 45.67 | 64.82* | 22.46 | 20.00** |
| Residual | 35 | 21.41 | 17.64 | 28.95 | 9.08 |
| ßEnvironments were Kabete and Mwea long seasons, 2018. *, ** Significant at 5 and 1 % probability levels, respectively. | |||||
Plant height at 50% flowering (cm)
Plant height at 50% flowering varied significantly (P≤0.01) in the six generations for crosses Roma VF x AVTO1429, Roma VF x AVTO1424 and Roma VF x Valoria select but not in the six-generation of cross Roma VF x AVTO1314 (Table 4). Plant height also showed highly significant differences between Kabete Field and Mwea Research Stations in cross Roma VF x AVTO1424 at (P≤0.05) and Roma VF x AVTO1314 at (P≤0.01). With exception of cross Roma VF x AVTO1424 having significant variations at (P≤0.05) for interactions between the two environments x genotypes, all the other crosses were not significantly different at (P≤0.01). Results showed that; for all generation in the 4 crosses, Mwea Research Station had the tallest mean plant heights at 50% flowering as compared to Kabete Field Station. Except for the six generations of cross Roma VF x AVTO1424, the rest of the crosses had no significant difference (P≤0.05) for plant height at 50% flowering. Both P2 in Roma VF x AVTO1429 and Roma VF x AVTO1424 had short plant height at 50% flowering whereas the tallest were BC₁P₁ and P1 with 67.04cm and 59.60 cm, respectively (Table 1). Besides, a significant increase (>10%) in plant height at 50% flowering in comparison to parental genotypes was registered in F1 generation for all crosses and Roma VF x AVTO1429 had the highest plant height of 63.76cm. F2 hybrids in all the crosses had shorter plant height at Kabete Field Station ranging from 41.55 to 52.68cm than at Mwea Research Station which ranged from 53.37 to 68.58cm.
The scaling tests showed significant differences at (P≤0.01) in cross Roma VF x AVTO1429. The scaling tests showed additive effects of -9.76**; dominance effect of -5.58**; and additive x additive interaction effects of -0.8** (Table 2). Therefore, further analysis using a 6-parameter model was carried out since backcrosses were used. Roma VF x AVTO1429 showed presence of epistasis. Results showed that a combined gene effect of 10.85 was higher than the interaction components of 0.85 put together (Table 3).
Final plant height (cm)
The final plant height demonstrated significant differences (P≤0.01) across the six generations of crosses Roma VF x AVTO1429 and Roma VF x AVTO1314. These variations (P≤0.01) were also noted across Kabete Field Station and Mwea Research Station. Interactions between the two stations (Kabete Field Station and Mwea Research Station) and the genotypes for crosses Roma VF x AVTO1429 (P≤0.05) and Roma VF x Valoria select (P≤0.01) were observed. Results showed that the final plant height for parents in all the crosses ranged from 72.27 to 91.00cm with Roma VF as the shortest parent recording ≤83cm in all cross as shown in table 1. Similarly, the F1 hybrids in all the crosses had final plant height range of 71.33 to 95.10cm at both study stations. Also, the F2 hybrids had similar final plant height that ranged from 71.10 to 104.10cm in all cross at both study stations. Results for F1 and F2 hybrids were consistent across the crosses. The final plant height across the two environments ranged from 82.20 cm for shorter parent (Roma VF) of cross Roma VF x AVTO1429 to 119.50 cm for taller offspring, BC₁P₁ (Table 1).
Due to insignificant difference (P≤0.01) on additive, dominance, additive x additive interaction, and additive x dominance interaction effects (Table 2), further analysis was not necessary since not even one cross showed significant differences in the scaling tests. However, a 6-parameter model was carried out since backcrosses were used. Results showed a combined gene effect of 13.93 was higher than the interaction components of 1.04 put together (Table 3).
Inter truss spacing (cm)
The inter truss spacing across the two study stations ranged from 17.23cm for parent P₁ (AVTO1429) of cross Roma VF x AVTO1429 to 19.17cm offspring F₁ x BC₁P₁ (Table 1). For cross Roma VF x AVTO1424, the range was from 13.95cm for parent P₂ (Roma VF) to 17.30cm offspring, F₁ x BC₁P₁ while cross Roma VF x AVTO1314 ranged from 15.29cm for offspring, BC₁P₂ to 17.78cm for parent (AVTO1314). The inter truss spacing across the two study stations for cross Roma VF x Valoria select ranged from 14.42cm for offspring, BC₁P₁ to 15.41cm offspring, BC₁P₂. Results showed that all the scaling tests had no significant differences at (P≤0.01) for all the crosses. However, for cross Roma VF x AVTO1429, the scaling test for additive x dominance interaction effects (D) showed significant differences at (P≤0.01). Scaling test showed presence of additive x additive interaction represented by -1.78** (Table 2). Therefore, further analysis using a 6-parameter model was carried out since backcrosses were used. Cross Roma VF x AVTO1429 showed epistasis. Results showed that the combined gene effects (3.0) were lower than the interaction components (6.26) put together (Table 3).
Number of trusses per plant
There was a highly significant difference across the six generations of cross Roma VF x AVTO1429 and Roma VF x AVTO1314 for the number of trusses per plant at (P≤0.01). However, the number of trusses per plant across the two study stations (Kabete Field Station and Mwea Research Station) in all crosses were not significantly different (P≤0.01). Interactions between the two study stations and the genotypes in all the crosses except Roma VF x Valoria select were significantly different (P≤0.05). The number of trusses per plant across the two study stations ranged from 18 trusses for parent P₁ (AVTO1429) of cross Roma VF x AVTO1429 to 24 trusses in offspring F₂ and from 18 trusses for parent P₁ (AVTO1424) of cross Roma VF x AVTO1424 to 21 trusses in offspring F1 (Table 1). Similarly, number of trusses per plant across the two environments ranged from 18 trusses for parent P₂ (Roma VF) of cross Roma VF x AVTO1314 to 21 trusses in offspring F1 and from 17 trusses for parent P₁ (Valoria select) of cross Roma VF x Valoria select to 19 trusses in offspring BC₁P₁.
Results showed that all the scaling tests had no significant differences at (P≤0.01) for all the crosses. However, for cross Roma VF x AVTO1429 the scaling test for additive effects (A) was significant at (P≤0.01). Scaling test also showed additive x dominance interaction of -2.61** (Table 2). Therefore, further analysis using a 6-parameter model was carried out since backcrosses were used. Roma VF x AVTO1429 showed epistasis. Results showed that the combined gene effects of -3.76 were lower than the interaction components of -3.16 put together (Table 3).
Discussion
A six-parameter model was identified as most suitable for data analysis for P1, P2, F1, F2 and backcrosses of the four crosses studied. The crosses were Roma VF x AVTO1424, Roma VF x AVTO1429, Roma VF x AVTO1314 and Roma VF x Valoria select. This model was adopted because one cross (Roma VF x AVTO1429) showed epistasis and backcrosses were used. The six parameters in this model were; Mean (ḿ), additive effect (d̂), dominance effect (ĥ), Additive x Additive interaction (î), Additive x Dominance interaction (ĵ) and Dominance x Dominance interaction (l̂). The parents in each cross were contrasting for all the traits evaluated. The offspring derived from the cross-combination Roma VF x AVTO1429 were earlier flowering and maturing, taller, had higher inter truss spacing and number of trusses per plant compared to other crosses and the better parent AVTO1429. Earliest flowering and maturing offspring were the F₁ hybrid Roma VF x AVTO1429 while tallest offspring was the backcross F1 x AVTO1429 BC₁P₁. Offspring with the smallest inter truss spacing was the F₂, followed closely by F₁. Offspring with the smallest number of trusses per plant were backcross of cross F₁ x Roma VF BC₁P₂, followed closely BC₁P₂. Differences among the generations for each trait were significant.
From the 6-parameter model, the combined gene effects (3.6) were higher than the interaction components (2.29) put together. Duration to flowering and maturity was controlled by dominance gene effects (2.69**) and the interactions of additive x additive effects (5.24**) and dominance x dominance interactions (-4.49**). These results were in agreement with the findings of after crossing a 9 x 9 half diallel in Gazipur, Bangladesh [16]. They recorded that non-allelic gene interaction or epistasis was observed on days to 50% flowering and P6 had most of the dominant genes for both numbers of flowers/ cluster and number of locules. Similarly, plant height was controlled by main gene effects which are the additive and dominance effects and the interactions between the two. Similar findings were confirmed by [16]. Findings of also reported significant fully adequate additive-dominance model for plant height and number of fruits per plant [24]. However, studies by using a 7 x 7 half diallel mating design failed to establish the additive-dominance gene effects in their experiment in Ethiopia [25]. From their experiment additive-dominance, gene interactions were evident in tomato fruit shape index and acidity that can be titrated. The reason for the observed additive and dominance gene effects in most traits studied was a failure to identify parents with far contrasting traits. The parents used were Marglobe, Roma VF and Esthete. Evaluation of genetic inheritance in Shaanix, China by using six generations namely; P1, P2, F1, F2, BC1P1 and BC1P2 showed significant gene effects of all plant characters that included plant height, number of branches per plant, days to 50% flowering, number of flowers per cluster, number of fruits per cluster and average fruit weight [26].
Inter truss spacing was controlled by the dominance gene effects and the interaction components of the dominance x dominance effects. Similar studies were conducted by to determine internode length using multi-generation joint analysis of major genes plus the polygene model [27]. Results demonstrated that internode length is controlled by the major gene and needs early selection in the pedigree selection. The number of trusses per plant was controlled by major dominance gene and the additive x additive interaction of polygenes. Similar studies were conducted by to determine the inheritance of yield and yield components in tomato [16]. The deduction was that additive and dominant components of genes are important in the plant characters.
Conclusion
Desirable agronomic traits of tomato despite being influenced by many genes, environment contribute to their expression significantly. For the four crosses, significant traits such as days to 50% flowering, plant height at 50% flowering, final height, inter truss spacing and the number of trusses per plant that contribute to desirable yields are influenced by the environment. Earliest flowering and maturing offspring were the F₁ hybrid from cross Roma VF x AVTO1429. Tallest offspring was the backcross of cross F1 x AVTO1429 BC₁P₁. The offspring’s derived from the cross-combination Roma VF x AVTO1429 had highest inter truss spacing and number of trusses per plant. F₁, F₂ and BC₁P performed better in all traits evaluated than the better parent. This study also found importance of gene effects for agronomic trait inheritance was in additive and dominance-additive portions which implied that the traits were inherited. Contribution of both parents in the subsequent generations (offspring) is vital in developing a breeding program for a particular trait
Acknowledgement
The authors are grateful to Continental Seeds Company for providing financial support for this work and the University of Nairobi for providing physical facilities to conduct various experiments.
Author’s Contribution
All the authors jointly designed the experiments. FKK conducted the experiment, performed data analysis, and drafted the manuscript with inputs from all the authors. PMK, RDN and LMK supervised the experiments and reviewed the manuscript. All authors read and approved the final manuscript.
Funding
Continental Seeds Company funded this research. The funder had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Abbreviations
AVRDC: World Vegetable Centre; g: grams; l: litres; ha: Hectares; t: tonnes; AEZ: Agro-ecological zones; ASL: Above Sea Level; rpm: revolution per minute; m: meters
Availability of Data and Materials
The data used and analysed during this study is available from the corresponding author on request.
Ethics Approval and Consent to Participate
Not applicable.
Consent For Publication
Not applicable
Competing Interests
The authors declare that they have no competing interests.
References
- Ochilo WN, Nyamasyo GN, Kilalo D, Otieno W, Otipa M, et al. (2019) Characterization and production constraints of smallholder tomato production in Kenya. Sci Afr 2: e00014.
- Agong SG, Schittenhelm S, Freidt W (2001) Genotypic variation of Kenya tomato (Lycopersicum esculentum L.) germplasm. J Food Technol Afr 6: 13-17.
- Mwangi TM, Ndirangu SN, Isaboke HN (2020) Technical efficiency in tomato production among smallholder farmers in Kirinyaga County, Kenya. Afr J Agric Res 16: 667-677.
- Kenya’s Facts and Figures (2019) Kenya National Bureau of Statistics.
- Mather K, Jinks IL (1982) Biometrical Genetics. 3rd Edn, Champan and Hall, London, UK.
- Bhatt RP, Biswas BR, Kumar N (2001) Heterosis, combining ability and genetics for vitamin C, total soluble solids and yield in tomato (Lycopersicon esculentum) at 1700 m altitude. J Agric Sci 137: 71-75.
- Foolad MR, Lin GY (2001) Genetic analysis of cold tolerance during vegetative growth in tomato, Lycopersicon esculentum Mill. Euphytica 122: 105-111.
- Resende JTV, Maluf WR, Das Graças CM, Nelson DL, Faria MV (2002) Inheritance of acyl sugar contents in tomatoes derived from an interspecific cross with the wild tomato, Lycopersicon pennellii and their effect on spider mite repellence. Genet Mol Res 1: 106-116.
- Valls JFM (2007) Characterization of Plant Genetic Resources. Plant genetic resources. Brasilia: Embrapa Genetic Resources and Biotechnology 1: 283-305.
- Huang XH, Zhao Y, Wei XH, Li CY, Wang A, et al. (2012) Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nature Review Genetics Journal 44: U32–U53.
- Singh NB, Paul A, Wani SH, Laishram JM (2012) Heterosis studies for yield and its components in tomato (Solanum lycopersicum Mill.) under valley conditions of Manipur. Int J Life Sci 1: 224-232.
- Olainyi JO, Akanbi WB, Adejumo TA, Ak OG (2010) Growth, fruit yield and nutritional quality of tomato varieties. Afr J Food Sci 4: 398-402.
- Fufa F, Hanson P, Dagnoko S, Dhaliwal MS (2011) AVRDC - the world vegetable centre tomato breeding in sub- Saharan Africa: lessons from the past, present work and future prospects. In I All African Horticulture Congress 911: 87-98.
- Kenneth TO (2016) Agro-morphological and nutritional characterization of tomato landraces (Lycopersicon species) in Africa. Masters of Science thesis, University of Nairobi, Kenya.
- Kumar P, Singh N, Singh PK (2017) A study on Heterosis in Tomato (Solanum lycopersicum L.) for Yield and its Component Traits. Int J Curr Microbiol Appl Sci 6: 1318-1325.
- Goffar MA, Ahmed A, Halim GMA (2016) Inheritance mechanism of yield and yield components in tomato Bangladesh. J Agric Res 41: 335-344.
- Lengai WM (2016) Efficacy of plant extracts and antagonistic fungi as alternatives to synthetic pesticides in management of tomato pests and diseases.
- Waiganjo MM, Wabule NM, Nyongesa D, Kibaki JM, Onyango I, et al. (2006) Tomato production in Kirinyaga district, Kenya, a baseline survey report. Kenya Agricultural Research Institute, Nairobi, Kenya 1-43.
- Griffing B (1956) Concept of general and specific combining ability in relation to diallel system. Aust J Biol Sci 9: 463-493.
- Sharma JR (2001) Statistical and Biometrical Techniques in Plant Breeding. Indian J Genet Plant Breed 61: 391–392.
- Kenya Agricultural and Livestock Research Organization (KALRO) (2016) Tomato Production Manual, Fruits and vegetables technical handbook. Retrieved from http://www.kalro.org.
- Checa O, Ceballos H, Blair MW (2006) Generation mean analysis of climbing ability in common beans (Phaseolus vulgaris L.). J Hered 97: 456-465.
- Squillance AE, Squillance DJ (1970) A graphic chi-square test for two-class genetic segregation ratios. South Eastern Forest Experiment Station, U.S. Dept. of Agriculture, Forest Service.
- Gul R, Rahman HU, Khalil IH, Shah SMA, Ghafoor A (2011) Estimate or heterosis in tomato (Solanum lycopersicum L.). Bangladesh J Agric Res 36: 521-527.
- Tasisa J, Mohammed W, Hussien S, Kumar V (2017) Heterosis for some physiological traits in tomato (Solanum lycopersicum L.) hybrids in East Hararghe, Ethiopia. World Appl Sci J 35: 2300-2307.
- Thainukul N, Sakdarueakrot S, Yan L (2017) The evaluation of genetic inheritance, heritability and correlations coefficient of mutant tomato. School J Agric Vet Sci 4: 209-213.
- Sun XR, Liu L, Zhi XN, Bai JR, Cui YN, et al. (2019) Genetic analysis of tomato internode length via mixed major gene plus polygene inheritance model. Sci Hortic 246: 759-764.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: Kathimba FK, Kimani PM, Narla RD, Kiriika LM (2022) Analysis of Tomato Agronomic Traits using Generation Mean. J Plant Genet Breed 6: 126.
Copyright: © 2022 Kathimba FK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Usage
- Total views: 1771
- [From(publication date): 0-2022 - Feb 22, 2025]
- Breakdown by view type
- HTML page views: 1561
- PDF downloads: 210