Analysis of the Molecular Diversity of Common Bacterial Blight (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscansi) Strains from Ethiopia Revealed by Rep-PCR Genomic Fingerprinting
Received: 09-Oct-2018 / Accepted Date: 24-Nov-2018 / Published Date: 27-Nov-2018 DOI: 10.4172/2155-952X.1000286
Abstract
Common bacterial blight (CBB) disease of the common bean (Phaseolus vulgaris) caused by Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans, is one of the most damaging foliar diseases of common bean production in Ethiopia. CBB causes economic losses due to reduction in seed quality and yield in common bean producing regions of Ethiopia. Currently, information on the genetic diversity of CBB strains in Ethiopia has been lacking. Here, for this specific study common bean bacterial blight strains were obtained from infected leaves collected from diverse bean growing areas. The collected strains of CBB were characterized to study the genetic diversity and relatedness of the CBB strains using repetitive extragenic elements polymerase chain reaction (rep-PCR) genomic fingerprinting technique. Analysis of Molecular Variance (AMOVA) revealed the existence of genetic diversity among bacterial strains and confirmed the presence of genetically distinct strains in Ethiopia. CBB pathogens are seed-borne so the lack of geographic differentiation among the six-different common bean growing localities could be the result of the distribution of one or some limited bacterial genotypes. Common bean improvement programs that develop CBB-resistant bean varieties for higher production should consider this information to determine the relevance and extent of resistance of improved bean cultivars.
Keywords: Rep-PCR fingerprinting; Genetic diversity; Common bacterial blight, Disease resistance
Introduction
Common bacterial blight (CBB) disease of the common bean (Phaseolus vulgaris) caused by Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans, is one of the most damaging foliar diseases of common bean production in Ethiopia. CBB causes economic losses due to reduction in seed quality and yield in common bean producing regions of Ethiopia. Currently, information on the genetic diversity of CBB strains in Ethiopia has been lacking. Here, for this specific study common bean bacterial blight strains were obtained from infected leaves collected from diverse bean growing areas. The collected strains of CBB were characterized to study the genetic diversity and relatedness of the CBB strains using repetitive extragenic elements polymerase chain reaction (rep-PCR) genomic fingerprinting technique. Analysis of Cluster and Molecular Variance (AMOVA) revealed the existence of genetic diversity among bacterial strains and confirmed the presence of genetically distinct strains in Ethiopia. CBB pathogens are seed-borne so the lack of geographic differentiation among the six-different common bean growing localities could be the result of the distribution of one or some limited bacterial genotypes. Common bean improvement programs that develop CBB-resistant bean varieties for higher production should consider this information to determine the relevance and extent of resistance of improved bean cultivars.
Materials and Methods
Collection of common bacterial blight strains
Strains of common bacterial blight were recovered from infected leaves of common bean which were collected from field surveys conducted during 2015 and 2016 main seasons from six geographical diverse major common-bean growing areas of Ethiopia (Wolaita, Gurage, Sidama, Gamogofa, Jimma, and Arisi) strains of CBB obtained from each location were considered as populations (Table 1).
| Code | Strain | Geographic Origin | Collection zone | Altitude m.a.s.l | Year of collection | Host |
|---|---|---|---|---|---|---|
| 1 | et xa 001 | Ethiopia | Arisi | 1785 | 2015 | Mesoamerican |
| 2 | et xa 002 | Ethiopia | Gurage | 1835 | 2015 | Andean |
| 3 | et xa 003 | Ethiopia | Arisi | 1999 | 2015 | Mesoamerican |
| 4 | et xa 004 | Ethiopia | Sidama | 1820 | 2015 | Mesoamerican |
| 5 | et xa 005 | Ethiopia | Jimma | 1216 | 2016 | Mesoamerica |
| 6 | et xa 006 | Ethiopia | Gurage | 1903 | 2015 | Mesoamerica |
| 7 | et xa 007 | Ethiopia | Sidama | 1823 | 2015 | Mesoamerican |
| 8 | et xa 008 | Ethiopia | Sidama | 1835 | 2015 | Mesoamerican |
| 9 | et xa 009 | Ethiopia | Jimma | 1713 | 2016 | Andean |
| 10 | et xa 010 | Ethiopia | Jimma | 1700 | 2016 | Andean |
| 11 | et xa 011 | Ethiopia | Jimma | 1216 | 2016 | Mesoamerica |
| 12 | et xa 012 | Ethiopia | Wolaita | 2058 | 2015 | Mesoamerica |
| 13 | et xa 013 | Ethiopia | Arisi | 1999 | 2015 | Mesoamerican |
| 14 | et xa 014 | Ethiopia | Arisi | 1835 | 2016 | Mesoamerica |
| 15 | et xa 015 | Ethiopia | Sidama | 1823 | 2015 | Mesoamerican |
| 16 | et xa 016 | Ethiopia | Wolaita | 1260 | 2015 | Mesoamerica |
| 17 | et xa 017 | Ethiopia | Jimma | 1383 | 2016 | Mesoamerica |
| 18 | et xa 018 | Ethiopia | Wolaita | 1606 | 2015 | Mesoamerica |
| 19 | et xa 019 | Ethiopia | Wolaita | 1257 | 2015 | Mesoamerica |
| 20 | et xa 020 | Ethiopia | Jimma | 1823 | 2016 | Mesoamerica |
| 21 | et xa 021 | Ethiopia | Wolaita | 1865 | 2015 | Mesoamerica |
| 22 | et xa 022 | Ethiopia | Jimma | 1920 | 2016 | Mesoamerica |
| 23 | et xa 023 | Ethiopia | Wolaita | 1262 | 2015 | Mesoamerica |
| 24 | et xa 024 | Ethiopia | Wolaita | 1909 | 2015 | Mesoamerica |
| 25 | et xa 025 | Ethiopia | Gurage | 1893 | 2015 | Mesoamerica |
| 26 | et xa 026 | Ethiopia | Gurage | 1835 | 2015 | Mesoamerica |
| 27 | et xa 027 | Ethiopia | Wolaita | 1835 | 2015 | Mesoamerica |
| 28 | et xa 028 | Ethiopia | Wolaita | 1835 | 2015 | Mesoamerica |
| 29 | et xa 029 | Ethiopia | Gurage | 1903 | 2015 | Andean |
| 30 | et xa 030 | Ethiopia | Gurage | 1903 | 2015 | Andean |
| 31 | et xa 031 | Ethiopia | Wolaita | 1262 | 2015 | Andean |
| 32 | et xa 032 | Ethiopia | Wolaita | 1709 | 2015 | Mesoamerica |
| 33 | et xa 033 | Ethiopia | Gamogofa | 1266 | 2015 | Mesoamerica |
| 34 | et xa 034 | Ethiopia | Gurage | 1855 | 2015 | Mesoamerica |
| 35 | et xa 035 | Ethiopia | Wolaita | 1862 | 2015 | Mesoamerican |
| 36 | et xa 036 | Ethiopia | Gamogofa | 1862 | 2015 | Mesoamerican |
| 37 | et xa 037 | Ethiopia | Wolaita | 1699 | 2015 | Mesoamerica |
| 38 | et xa 038 | Ethiopia | Wolaita | 1687 | 2015 | Mesoamerica |
| 39 | et xa 039 | Ethiopia | Gurage | 1861 | 2015 | Mesoamerica |
| 40 | et xa 040 | Ethiopia | Sidama | 1695 | 2015 | Mesoamerican |
Table 1: strains of common bacterial blight collected from diverse regions of Ethiopia.
During surveys representative common bean field were selected and investigated for CBB symptoms. Leaves showing typical symptoms of CBB with water-soaked spots and irregular necrotic lesions with yellow border were collected and dried with paper bags. From the collected leaf sample, tissues were removed from the lesion margin, placed in a drop of distilled water on a microscope slide, and macerated. Loopful of macerate were streaked onto nutrient agar (NA) and the plates were incubated at 28°C for 24 h. Yellow, mucoid, xanthomonad-like colonies were selected from each leaf sample and sub cultured on NA.
DNA extraction
Genomic DNA was extracted using a protocol described by [18] with minor modification. Bacterial cells were harvested after 24 h of incubation on Yeast extract-dextrose-CaCO3 (YDC) medium, collected by centrifugation at 5000 × g for 5 min, and washed twice with 1 M NaCl and two times with sterile distilled water. The bacterial cell pellet was re-suspended in warm (55°C) extraction buffer (0.2 M Tris HCl pH 8.0; 10 mM EDTA, pH 8.0, 0.5 m NaCl, 1% SDS) containing Proteinase K (10 μg/ml). After 60 min at 55°C, 0.5 vol. of 7.5 M ammonium acetate were added, gently mixed and left to stand for 10 min at room temperature. Following centrifugation, the supernatant was transferred to a fresh tube and the DNA was precipitated by adding 1 vol. of ice-cold isopropanol. The pellet was washed with 70% ethanol, dried and re-suspended in 1X TE buffer (10 mM Tris–HCl, pH 8.0; 1 mM EDTA) containing 10 mg/ ml RNase A. Tubes were incubated at 37°C for an hour, and the DNA were precipitated with 1/10 vol. of 3 M NaAc, pH 5.2, and 2 vol. of 95% ethanol. The pellet was dried and finally suspended in 0.1X TE buffer. The quality of extracted DNA was determined by electrophoresis on 0.7% agarose gels.
Rep-PCR fingerprinting
Rep-PCR was done using the primer pairs REP1R-I and REP2-I and ERIC 1 and ERIC2 (Table 2). The reproducibility of rep-PCR was tested by amplifying DNA from three randomly chosen strains two times. Optimal PCR conditions for each of the primer sets were used as described by [15]. PCR amplifications were performed with applied bio system thermal cycler model ABI2720 using Taq DNA polymerase with 25 units/ml concentrations. The PCR products were electrophoresed in a 1.5% agarose gel for 1 h at a constant voltage of 55 V in 1×TAE buffer (40 mMTris–Acetate, 1 mM EDTA, pH 8.0). The rep-PCR profiles were visualized under UV light after staining of the gel with 0.5 μl/ml concentration of ethidium bromide, and digital image capturing was done using a Canon powers hot SX150 digital photo camera mounted on a hood.
| Genetic markers | SEQUENCES 5’ to 3’ | Ta 0C | GC % | Number of nucleotide |
|---|---|---|---|---|
| REP 1 | IIIICGICGICATCIGGC | 49 | 52.9 | 18 |
| REP 2 | IIICGNCGNCATCNGGC | 58 | 52.9 | 17 |
| ERIC 1 | ATGTAAGCTCCTGGGGATTCAC | 58 | 50 | 22 |
| ERIC 2 | AAGTAAGTGACTGGGGTGAGCG | 42 | 54.5 | 22 |
| BOX AIR | CTACGGCAAGGCGACGCTGACG | 50 | 68.2 | 22 |
Table 2: Molecular markers used to amplify PCR product of strains of common bacterial blight.
Data Analysis and Interpretation
Analysis of molecular variance (AMOVA) was used to partition the genetic diversity among and within the bacterial strain populations and tested whether there is a hierarchy of rep-PCR sequence variation among individuals. The genetic structure of common bacterial blight isolates was obtained from the infected common bean leaves by the DNA finger printing using rep-PCR. Each unit of band pattern generated by rep-PCR (ERIC, BOX and REP PCR primers) and the fingerprints were decoded into a binary matrix (1,0) where 1 represents presence of band and 0 absence. The genetic relationships between strains were evaluated using a matrix of genetic distances constructed using the complement of the Jaccard similarity coefficient (CSJ), which does not consider negative similarities and the absence of the product. The binary matrix was used to drive a distance matrix using Jaccard’s matrix from which an average linkage (UPGMA or unweighted pair group method with arithmetic averages) dendrogram was derived. Sources of genetic differentiation were analyzed using Analysis of Molecular Variance (AMOVA) which were performed using GenAlEx6.1to assess genotypic variations across all the populations studied (Tables 3 and 4) [19,20]. The analysis included partitioning of total genetic variation into within-groups and among-groups variance components; hence it provided a measure of intergroup genetic distance as proportion of the total variation residing between populations. The significance of analysis was tested using 999 random permutations. Principal coordinate analysis (PCA) was computed with individual isolates using GenAlEx.
| Source | df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Among Pops | 5 | 33.782 | 6.756 | 0.338 | 7% |
| Within Pops | 34 | 158.068 | 4.649 | 4.649 | 93% |
| Total | 39 | 191.850 | 4.987 | 100% | |
| Stat | Value | P (rand > = data) | |||
| PhiPT | 0.068 | 0.020 | |||
Table 3: Analysis of molecular variance (AMOVA) within and among bacterial strain populations collected from Ethiopia.
| Axis | 1 | 2 | 3 |
|---|---|---|---|
| % | 24.44 | 13.89 | 10.71 |
| Cumulative | 38,33 | 49.04 |
Table 4: Percentage of variation explained by the first 3 axes.
Results
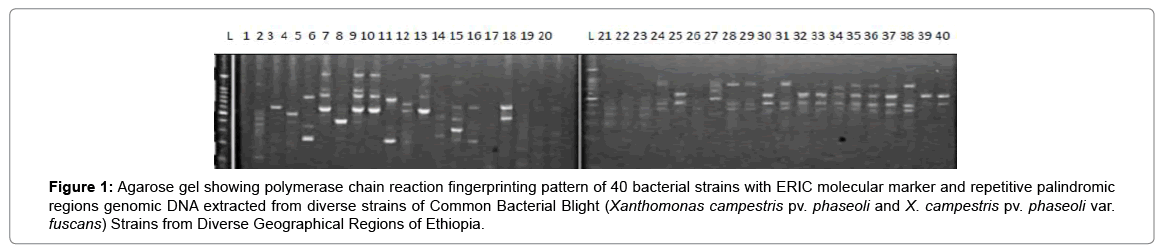
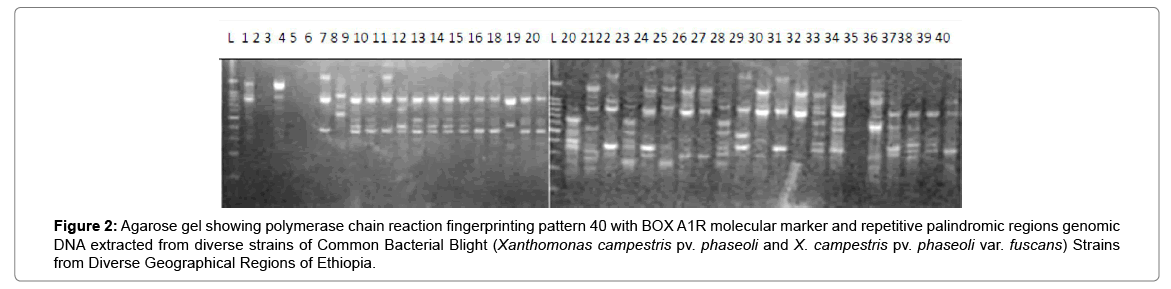
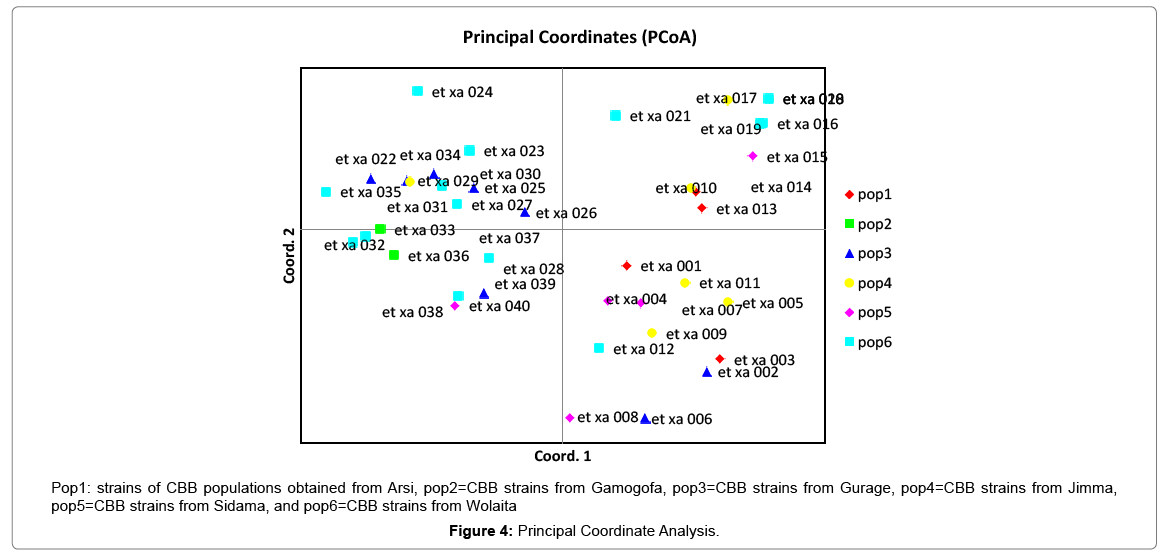
Results of molecular characterization with DNA fingerprinting techniques (Figures 1 and 2) indicated that genetic diversity exists among isolates of common bacterial blight collected from diverse bean growing areas (Wolaita, Gurage, Sidama, Gamogofa, Jimma, and Arisi) of Ethiopia (Table 1). AMOVA was used to partition the genetic diversity among the populations and tested whether there is any hierarchy of rep-PCR fingerprinting variation among individuals. The analysis of 40 CBB isolates revealed that 7% of the genetic variation were distributed among populations while 93% of the genetic variations were within the groups (Table 3). The common bacterial blight strains showed no geographic differentiation. The first and the second principal coordinates account for 24.44% and 13.89% of the variations respectively. The result indicates that gene flow is common at the group level.
Figure 1: Agarose gel showing polymerase chain reaction fingerprinting pattern of 40 bacterial strains with ERIC molecular marker and repetitive palindromic regions genomic DNA extracted from diverse strains of Common Bacterial Blight (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans) Strains from Diverse Geographical Regions of Ethiopia.
Figure 2: Agarose gel showing polymerase chain reaction fingerprinting pattern 40 with BOX A1R molecular marker and repetitive palindromic regions genomic DNA extracted from diverse strains of Common Bacterial Blight (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans) Strains from Diverse Geographical Regions of Ethiopia.
Discussion
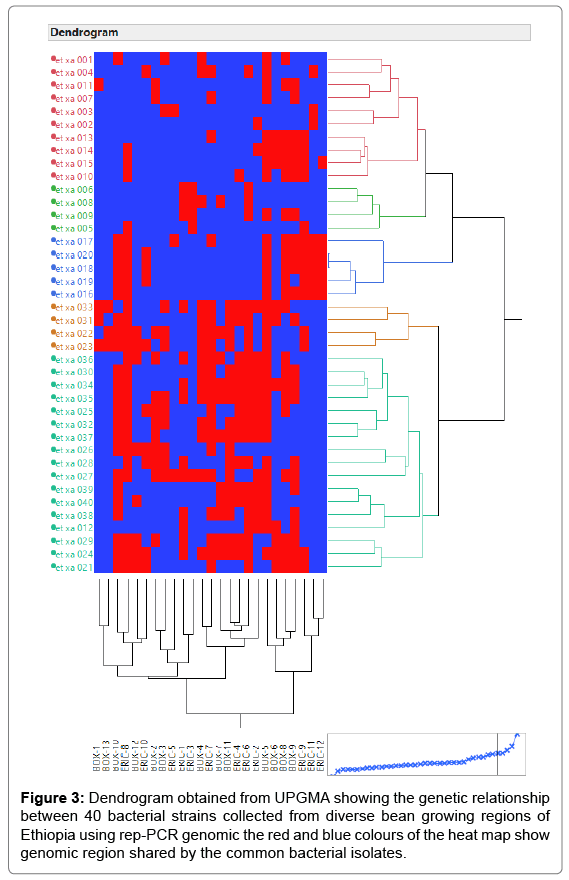
The principal mechanism of gene flow was the informal bean seed system operating in the areas and the predominant seed born nature of the pathogen [21,22]. Gene flow or migration refers the exchange of genetic information among geographic population through the movement of gamete [23]. This is consistent with the fact that several genomic regions were shared by samples of CBB isolates from different populations (Figure 3) which was indicated by the heat map of the red color and the blue. This indicated that gene flow occurred between populations. Genetic differences among common bacterial blight strains within populations might be the result of gene flow between populations and this were indicated by the principal component analysis (Figure 4) where individuals within the same populations marked with the same symbols. The first and the second principal coordinates account for 24.44% and 13.89% of the variations respectively. Out of one hundred forty-three local collocations genetic variation of common bacterial blight strains were also reported by [24]. The same authors reported that both RAPD and AFLP analyses revealed high frequency of DNA polymorphism among isolates and could distinguish between Xap, Xapf and a non-pathogenic isolate. Differences between Xap and Xapf isolates demonstrated the existence of two distinct groups of bacteria [25]. Rep-PCR genomic fingerprinting has proven to have high discrimination power and reproducibility for bacterial diversity study (Figures 1 and 2). It generated estimation of genetic relatedness among the common bean bacterial blight strains. The rep-PCR genomic fingerprinting has been shown to be very valuable tool in molecular biology in studying the classifications and diversity of microbial isolates. The result from our genetic study based on the combined ERIC and BOX-PCR fingerprinting data with computer-based clustering analysis method, revealed the existence of genetic diversity between the bacterial strains. This confirmed the presence of genetically dissimilar strains of common bean bacterial blight in Ethiopia. The resulted differences might be between Xap and Xapf isolates representing the existence of two distinct groups of bacteria. Similar distinction between these two groups was also reported by [26], using RFLP’s. Non-pathogenic Xanthomonas commonly associated with beans could be distinguished from Xap and Xapf using both RAPD and AFLP techniques. Knowledge on the existence of variability in CBB isolates populations is important for plant breeding and program. Hence the common bean improvement program that designs developing disease resistance common bean varieties for wider production should consider this information (presence of diverse strains) during evaluation process for better achievements. Rep-PCR to be very useful for studying plant pathogen population structure and a powerful tool for the molecular genetic analysis of bacteria. In other reports with rep-PCR techniques, strains of different Xanthomonas species were differentiated. The potential of rep-PCR in discrimination the strains and the potential of rep-PCR patterns obtained with ERIC, BOX and REP primers showed polymorphism among Brazilian X. campestris pv. strains [27]. The rep-PCR is an effective method in determining genetic diversity among populations of many bacterial pathogenic genera, including Xanthomonas and Pseudomonas [28,29]. In the present study, the genetic diversity of CBB strains collected from bean growing areas of Ethiopia was determined. With combined analysis of rep-PCR patterns obtained showed the existence of genetic diversity among the Ethiopian CBB strains. Although pathogenicity test was under investigation the molecular characterization revealed the existence of different genotypes of common bean strains in Ethiopia. X. axonopodis pv. phaseoli is genetically diverse. X. axonopodis pv. phaseoli strains were grouped in at least 4 genetic lineages spanning over two different homology groups defined by [7]. Genetic heterogeneity within non-fuscous strains of X. axonopodis pv. phaseoli was also revealed by [16] Moreover, our cluster analysis of the pairwise similarity values performed using UPGMA confirmed differences in fingerprint patterns. CBB pathogens are seed-borne so the lack of geographic differentiation among the six-different common bean growing localities could be the result of the distribution of one or some limited bacterial genotypes. In current study, CBB isolates obtained from the same common bean growing area present different rep-PCR pattern and clustered differently. The existence of two distinct clusters among CBB isolates in Ethiopia could suggest existence of both CBB strains.
Conclusion
These findings can help to provide better understanding of the CBB strains for disease resistance in the common bean breeding program. The authors also suggest the information obtained could be complimented with pathotype characterization so that the bean breeding program should use the most virulence pathotypes during developing and screening of resistance bean lines.
Acknowledgement
The authors acknowledge the support and provision of lab facilities by the Kirkhouse Trust. The contributions of Molecular Biotech Lab Technicians Especially Bethel Mulugeta and Mihiret Tadesse was highly appreciated. We also thank the Southern Agricultural Research Institute (SARI) and the ABC partner countries.
References
- Asfaw A, Blair MW, Almekinders C (2009) Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) landraces from the East Africa highlands. Theor Appl Genet 120: 1-12.
- Jones LA (1999) Phaseolus Bean: Post-Harvest Operations. Int Cent Trop Agric 3: av015e.
- PABRA (2014)Phase Report and Partnership in Research for Impact: Case of Common Beans in Ethiopia. Nairobi, Kenya. Website: www.pabra-africa.org
- Belachew K, Gebremariam M, Alemu K (2015) Integrated Management of Common Bacterial Blight (Xanthomonas axonopodis pv. Phaseoli) of Common Bean (Phaseolus vulgaries) in Kaffa, Southwest Ethiopia. Malays J Med Boil Res 2: 149-154.
- Belete T, Bastas KK (2017) Common Bacterial Blight (Xanthomonas axonopodis pv. phaseoli) of Beans with Special Focus on Ethiopian Condition. J Plant Pathol Microbiol 8: 403.
- Lopez R, Asensio C, Gilbertson RL (2006) Phenotypic and Genetic Diversity in Strains of Common Blight Bacteria (Xanthomonas campestris pv . phaseoli and X . campestris pv . phaseoli var . fuscans) in a Secondary Center of Diversity of the Common Bean Host Suggests Multiple Introduction Events. Phytopathol 53: 1204-1213.
- Mkandawire AB, Mabagala RB, Guzman P, Gepts P, Gilbertson RL (2004) Genetic Diversity and Pathogenic Variation of Common Blight Bacteria (Xanthomonas campestris pv. phaseoli and X . campestris pv . phaseoli var . fuscans) Suggests Pathogen Coevolution with the Common Bean. Phytopathol 94: 593-603.
- Mahuku GS, Jara C, Henriquez MA, Castellanos G, Cuasquer J (2006) Genotypic Characterization of the Common Bean Bacterial Blight Pathogens, Xanthomonas axonopodis pv. phaseoli and Xanthomonas axonopodis pv. phaseoli var. fuscans by repâ€PCR and PCR–RFLP of the Ribosomal Genes. J Phytopathol 154: 35-44.
- Wortmann CS (1998) Atlas of common bean (Phaseolus vulgaris L.) production in Africa (No. 297). CIAT.
- Katungi E, Farrow A, Chianu J, Sperling L, Beebe S (2009) Common bean in Eastern and Southern Africa: a situation and outlook analysis. International Centre for Tropical Agriculture p: 61.
- Louws FJ, Fulbright DW, Stephens CT, De Bruijn FJ (1994) Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl Envirl Microbiol 60: 2286-2295.
- Barak JD, Gilbertson RL (2003) Genetic diversity of Xanthomonas campestris pv. vitians, the causal agent of bacterial leafspot of lettuce. Phytopathol 93: 596-603.
- Rademaker JLW, De Bruijn FJ, Caetano-Anolles G, Gresshoff PM (1997) DNA markers: protocols, applications and overviews.
- Gilson E, Saurin W, Perrin D, Bachellier S, Hofnung M (1991) The BIME family of bacterial highly repetitive sequences. Res Microbiol 142: 217-222.
- Versalovic J, Koeuth T, Lupski R (1991) Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucl Acids Res 19: 6823-6831.
- Arnaud-Santana E, Coyne DP, Eskridge KM, Vidaver AK (1994) Inheritance; low correlations of leaf, pod, and seed reactions to common blight disease in common beans; and implications for selection. J American Society Horticul Sci 119: 116-121.
- Mahuku GS (2004) A simple extraction method suitable for PCR-based analysis of plant, fungal, and bacterial DNA. Plant Molec Biol Reporter 22: 71-81.
- Gevers D, Huys G, Swings J (2001) Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiology Letters 205: 31-36.
- Peakall ROD, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molec Ecol Reso 6: 288-295.
- Rubyogo JC, Sperling L, Nasirumbi L, Kasambala S (2007) Developing seed systems with and for the marginalized: case of common beans (Phaseolus vulgaris L.) in East, Central and Southern Africa. In Proceedings of Farmer First Revisited Conference, Sussex, UK pp: 393-407.
- Webster DM, Atkin JD, Cross JE (1983) Bacterial Blights of Snap Beans. Plant Disease 67: 935.
- Hutchison DW, Templeton AR (1999) Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution 53: 1898-1914.
- Zhu W, Zhan JS (2012) Population genetics of plant pathogens. Yi chuan Hereditas 34: 157-166.
- Fourie D (2002) Distribution and severity of bacterial diseases on dry beans (Phaseolus vulgaris L.) in South Africa. J Phytopathol 150: 220-226.
- Gilbertson RL, Otoya MM, Pastor-Corrales MA, Maxwell DP (1991) Genetic diversity in common blight bacteria is revealed by cloned repetitive DNA sequences.
- Gilbertson RL, Rojas MR, Russell DR, Maxwell DP (1991) Use of the asymmetric polymerase chain reaction and DNA sequencing to determine genetic variability of bean golden mosaic geminin virus in the Dominican Republic. J Gen Virol 72: 2843-2848.
- Trindade PVO, Sobral LG, Rizzo ACL, Leite SGF, Soriano AU (2005) Bioremediation of weathered and recently oil-contaminated soils from Brazil: a comparison study. Chemosphere 58: 515-522.
- Weingart H, Völksch B (1997) Genetic fingerprinting of Pseudomonas syringae pathovars using ERICâ€, REPâ€, and IS50â€PCR. J Phytopathol 145: 339-345.
- Mondal KK, Mani C (2009) ERIC-PCR-generated genomic fingerprints and their relationship with pathogenic variability of Xanthomonas campestris pv. punicae, the incident of bacterial blight of pomegranate. Curr Microbiology 59: 616.
Citation: Rezene Y, Mitiku M, Tesfaye K, Male A, Gepts P (2018) Analysis of the Molecular Diversity of Common Bacterial Blight (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans) Strains from Ethiopia Revealed by Rep-PCR Genomic Fingerprinting. J Biotechnol Biomater 8:286. DOI: 10.4172/2155-952X.1000286
Copyright: © 2018 Rezene Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 3669
- [From(publication date): 0-2018 - Mar 31, 2025]
- Breakdown by view type
- HTML page views: 2840
- PDF downloads: 829