Research Article Open Access
Analysis of Soluble Microbial Products by Mass Spectrometry: Potential in Monitoring Bioprocesses of Wastewater Treatment
Huixing Feng1#, Yan Zhou1#, Guili Zhao1,3#, Jaslyn Lee1,3, Chenghong Guo1, Wun Jern Ng1, Wei Ning Chen2*
1Advanced Environmental Biotechnology Centre, Nanyang Environment and Water Research Institute, 1 Cleantech Loop, CleanTech One, #06-08 Singapore 637141
2School of Chemical and Biomedical Engineering, Nanyang Technological University, 62 Nanyang Drive, 637459, Singapore.
3Interdisciplinary Graduate School, Nanyang Technological University, Singapore
#These authors contribute equally to this work.
- *Corresponding Author:
- Wei Ning Chen
School of Chemical and Biomedical Engineering
Nanyang Technological University
62 Nanyang Drive, 637459, Singapore
Tel: +65 63162870
Fax: +65 62259865
E-Mail: WNChen@ntu.edu.sg
Received date: March 13, 2014; Accepted date: April 28, 2015; Published date: May 05, 2015
Citation: Feng H, Zhou Y, Zhao G, Lee J, Guo C et al. (2015) Analysis of Soluble Microbial Products by Mass Spectrometry: Potential in Monitoring Bioprocesses of Wastewater Treatment. J Anal Bioanal Tech 6:241. doi: 10.4172/2155-9872.1000241
Copyright: © 2015 Feng H. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
In biological wastewater treatment process, the analysis of metabolic compounds that are produced during the process is critical to monitor the performance of microorganisms. The soluble products present in the effluent directly affect the process efficiency and quality of the water after treatment, and it is also the major reason for fouling in membrane bioreactor. Currently, analytical methods are mainly restricted to the overall measurement of the total amount of polysaccharides, DNA and proteins without any specific identification of these compounds. Here we introduce an explorative mass spectrometry based strategy, for the analysis of soluble microbial products and other soluble impurities in the effluents of wastewater treatments using different digestion process. According to the results from this study, the two stage co digestion process indicated higher treatment efficiency compared with the single stage process, since fewer compounds were detected in the effluent. For the two-stage process, most of the fatty acids produced in the first stage of digestion by hydrolysis and acidogenesis, were digested in the second stage. The results also indicated that the digestion efficiency of the single stage process was lower than that of the separated two-stage process. This study is one of few exploration of analyzing and identifying unknown compounds using MS based technique from a metabolic analysis perspective. Our novel approach can be applied as an analytical platform to effectively monitor the biological processes and provides a different view point in wastewater treatment systems.
Keywords
Bioprocess; Wastewater treatment; Mass spectrometry; Soluble microbial product
Introduction
The routine parameters to analyze biological wastewater treatment systems are chemical oxygen demand (COD) and biological oxygen demand (BOC). The presence of organic compounds and biopolymers in the effluent are defined as soluble microbial products (SMP) when they are soluble, and defined as extracellular polymer substrates (EPS) when they are insoluble and eventually combine to form flocs or solid colloids together with other impurities [1]. The levels of SMP and EPS correspond directly to the levels of effluent COD and membrane fouling, which in turn affects effluent discharge levels, treatment efficiency and energy use. Moreover, SMP and EPS are believed to be the main causes of fouling in membrane bioreactors.
Generally, SMPs are produced during substrate metabolism or cell lysis and degradation. They are classified into two groups according to their origins; utilization associated products (UAP) and biomass associated products (BAP). UAP are usually composed of carbonaceous compounds, and are produced during cell growth and metabolism, and substrate utilization. The amount of UAP produced during substrate utilization is proportional to the amount of substrate utilized. BAP are produced from cell lysis, biomass degrade and endogenous decay. BAP are macromolecules produced from cell debris, and the major components are believed to be polysaccharides. The molecular weight of BAP is much larger than that of UAP. The boundary between BAP and EPS is not very clear but it is generally accepted that UAP are soluble hydrolyzed EPS [2].
SMP is critical because they are the main contributors of effluent chemical oxygen demand (COD) and biochemical oxygen demand (BOD). Together with EPS, they affect COD and the toxicology of the effluent and membrane fouling of the bioreactor systems [3]. Currently, the generation of EPS and SMP, factors affecting their composition and concentration, and fouling mechanism related with the properties of EPS/SMP are still unknown. Hence, intensive and accurate understanding of SMP and EPS is critical. Furthermore, the generation of EPS and SMP, and the complicated composition of them are the major causes of high COD and membrane fouling resulting in the low effluent quality. Therefore, it is essential to analyze SMP and EPS and identify their composition in the effluent, which can facilitate understanding of how to reduce membrane fouling and COD. Especially, based on the understanding of composition of SMP, the SMP generation resource can be further known and relevant effective strategies can be conducted and developed to reduce SMP resulting in reducing membrane fouling. Hence, analysis and identification of SMP composition is meaningful for monitoring bioprocess of wastewater treatment in membrane bioreactor system.
However, the identification of SMP and EPS is a challenge as it is a mixture of various unknown compounds. SMP and EPS have a wide range of molecular weights (MW) ranging from 0.5 kilo dalton (kDa) to 50 kDa [4]. The components are believed to include humic substances, proteins, DNAs, lipids, polysaccharides, carbohydrates and small molecules. Over the past years, several groups have been working on the characterization of SMP and other components in the effluents and sludge. Currently, there are many technologies and methods that have been applied to characterize SMP and EPS, and they are summarized and discussed in this paper.
Most of present studies focus on the overall measurement of SMP/EPS by measuring the COD and BOC amounts, and by the total quantification and size distribution of polysaccharides, proteins and biopolymers. Size distribution analysis is commonly used, by employing ultra-filtration (UF) or HPLC-SEC (high performance liquid chromatography-size exclusion chromatography) [5]. Some groups have employed different chemical methods in order to identify groups of compounds such as proteins and carbohydrates. It also is reported that excitation/emission matrix spectroscopy (EEM) has been applied as a substitute for chemical analysis. EEM is able to produce a fluorescence spectrum using many different excitation wavelengths, to form a unique spectral fingerprint of the sample, which then can be compared [6]. Resonance light-scattering (RLS) has been also used to determine the levels of proteins and carbohydrates [7]. For the direct analysis of SMP, FTIR (Fourier transform infrared spectrometry) can determine the functional groups in the membranes used in the wastewater treatment system, enabling the comparison of clean and SMP-fouled membranes. In Ni’s work, they employed EEM and FTIR together with mathematical modeling, and confirmed that the components of BAP are much larger than that of UAP [8]. The MWs of UAP are less than 290 kDa, while BAPs can reach a MW of nearly 5000 kDa. Considering the much larger size of BAP, it forms the majority of the soluble organic compounds in the effluent. Jarusutthirak et al. used HP-SEC and FTIR to analyze the sample, and they showed that high MW compounds from BAP play an essential role in creating a high resistance membrane, reducing permeate flux [9]. Sludge retention time (SRT), affects the characteristics and the amounts of SMP, based on analysis of sludge and effluent sample produced under different conditions. In Wang’s work, they analyzed SMP produced from activated sludge under stressful conditions such as high temperature, starvation, low pH, and heavy metals with the methods of SEC, HPLC and FEEM [6]. The results showed that stressful conditions rather than microbial species dominated the production of SMP composition and amounts [6]. High temperature and low pH showed strong effects of SMP, by stimulating the production of polysaccharides and polycarboxylate-type humic acid with high hydrophilicity (High Temperature) and hydrophobic humic-acid-like organics (low pH). The change of SMP under these stressful conditions could increase the formation of foulants and should be avoided during the process.
In recent several years, gas chromatography- mass spectrometry (GC-MS) has been applied for specific component identification of SMP, rather than the overall identification of a specific group of component [10-12]. GC-FID (flame ionization detector) was used to measure the volatile fatty acids in the effluents, while GC-TCD (thermal conductivity detector) was used to detect the presence of biogas. The volatile, non-polar compounds were extracted with SPE (solid phase extraction) column and analyzed using GC-MS. A number of compounds were identified in permeate by this method [12,13]. In Zhou’s work, GC-MS was used to analyze the effluents [11]. GC-MS results showed that the main SMP in the anaerobic effluent were long chain carbohydrates and esters, accounting for 55–65% of the total organic matter. Anaerobic SMP was more complex than the aerobic SMP. Soluble COD, protein and polysaccharides showed a clear decrease at the sludge layer from 10 to 15 m despite the low MLSS/MLVSS (mixed liquor suspended solids/ mixed liquor volatile suspended solids) content. Methanogens might be the main consumers of the SMP in anaerobic reactors. Aquino analyzed the effluents with UF and GC MS in 2002 [14]. In his work, it was found that the bulk of SMP are in the low MW range, although compounds with MW as high as 300 kDa were also present in all anaerobic effluents. Compound identification using GC-MS revealed the presence of long chain alkenes with carbon length C12-C24 and alkanes C12-C16, as well as some aromatic compounds. They concluded that these compounds, likely from the cell lysis and endogenous decay, may not be easily biodegradable, hence their presence in the effluent is likely to be the cause of residual COD.
Wastewater treatment is a biological process that involves the microorganisms in the sludge assimilating and degrading the organic impurities, to produce water (methane), carbon dioxide and biomass. Hence this bioprocess is essential the same as a community of bacteria. The bacteria thrive on the nutrients and the organic contaminants, to produce SMP, which includes the biomass and utilization byproducts of the microorganism such as metabolites, proteins and polysaccharides, which are exactly the same as the components in a microorganism's cell. Thus this inspired this study that the methods that are used for the analysis of metabolic molecules produced by microorganisms could also be applied for the analysis of wastewater bioprocess samples. In modern systematic biology research, MS based techniques have become the most convenient, reliable and widely used techniques for life science and biological research. Among above mentioned techniques, it is proposed that mass spectrometry as an analytical tool that can be applied to study SMP which produced during the metabolism and decay of a microorganism, and the rapidly developing field of mass spectrometry coupled with GC/LC could be used as a potential strategy to analyze wastewater bioprocess compounds. With this approach, the specific compound would be identified, rather than the overall levels of certain groups of compounds.
Analysis of the soluble compounds identification of the effluents from anaerobic co-digestion system was carried out in this study. This work established a pioneer application of qualitative analysis with time-of-flight (TOF) MS method in this area. A number of compounds were successfully identified and effluents from different bioprocess showed different compounds composition. This method could be expanded to soluble compound analysis in different bioprocess with various analytical purposes. It could greatly facilitate the exploration of unknown components analysis in the study of process efficiency and optimization, fouling mechanism, disinfection by-products and toxicity of the processed water.
Materials and Methods
Effluent samples
The effluents were obtained from anaerobic co-digestion system with feeding of municipal sludge and oil mixture. The feeding for anaerobic co-digestion consisted of waste sludge from a wastewater treatment plant mixed with 50% of cooked oil (based on volatile solids mass). The effluents treated from two independent anaerobic digestion systems were taken and analyzed. The first system was a single stage anaerobic reactor that was operated at hydraulic retention time (HRT) of 20 days at 35°C. The effluent from this single stage anaerobic digestion was named Sample SD. The second system used for treating the sludge mixture and cooking oil was a two-stage anaerobic digestion system. Sample TD1 was the effluent from stage 1 reactor, while sample TD2 was the effluent from stage 2 reactor. Stage 1 reactor was operated (3 days HRT and 35°C) for the hydrolysis and acidogenesis process, while stage 2 reactor was for the methanogenesis process (17 days HRT and 35°C). The feed for stage 2 reactor was the effluent from the stage 1 reactor. All samples were centrifuged and pre-filtered with 0.45 μm of nylon membrane after collection to remove any bulky impurities.
Sample preparation for soluble compounds analysis with LCMS/ MS
All of the collected effluents samples were filtered with a 0.22 μm membrane for detection of soluble compounds. The supernatant then was subjected to a modified Bligh and Dyer extraction method. Briefly, 800 μl methanol-chloroform with 3:5(v/v) was added to 300 μl wastewater sample. After vortexing, the mixture was centrifuged at 12000 rpm for 10 min. The supernatant aqueous phase containing the soluble compounds was transferred to a clean tube. The extracts were vacuum-dried, and the pellet dissolved with 60 μl H2O-methnol, 1:1 (v/v), and centrifuged at 12000 rpm for 2 min to remove the insoluble part. The supernatant was ready to be injected.
Compounds identification with LC-MS/MS
The supernatant fraction from sample preparation step was analyzed using Agilent 1200 HPLC system (Waldbronn, Germany) equipped with a 6530 Q-TOF mass detector managed by a MassHunter workstation. The column used for the separation was an Agilent rapid resolution HT Zorbax SB-C18 (0.5 × 50 mm, 1.8 mm; Agilent Technologies, Santa Clara, CA, USA). The gradient elution involved a mobile phase consisting of (A) 0.1% formic acid in H2O and (B) 0.1% formic acid in acetonitrile. The initial condition was set at 2% B. A linear gradient to 98% B was applied in 25 min, held for 2 min, then quickly returned to starting conditions for over 1 min and held for another 2 min. Flow rate was set at 20 μl/min, and 2 μl of sample was injected. The electrospray ionization mass spectra were acquired in positive ion mode. Mass data were collected between m/z 100 and 2000 at a rate of 3.35 spectra per second. The ion spray voltage was set at 3,500 V, and the heated capillary temperature was maintained at 350°C. The drying gas and nebulizer nitrogen gas flow rates were 9.0 L/ min and 45 psi, respectively. Two reference masses were continuously infused to the system to allow constant mass correction during the run: m/z 121.0509 (C5H4N4) and m/z 922.0098 (C18H18O6N3P3F24).
Compound identification by molecular features
Compounds chromatography was extracted from the raw data files using an unbiased, molecular feature extraction (MFE) algorithm in Agilent MassHunter Qualitative Analysis B.05.00 Software. Provisional compound identification was performed by matching accurate mass results to content from METLIN Personal Compound Database and Library (PCDL) (Agilent, Santa Clara, CA, USA).
Results and Discussion
In this article, we used effluents sample generated from anaerobic co-digestion as a model, and studied the SMP compounds produced using MS based tools. LC-MS/MS was used for the metabolic products analysis and identification of unknown soluble compounds in the effluents. In our anaerobic co-digestion system, the purpose of this process was to digest aerobic sludge with anaerobic sludge, and produce methane at the same time. The co-digestion process has been believed to be able to enhance biogas production and organic matter degradation [15,16]. The adding of cooking oil was used to boost the production of methaneas lipid rich waste with a high methane producing potential [17]. Here, cooking oil was added to mimic the lipid-rich municipal wastewater, and the digestion efficiency was observed. The process of anaerobic digestion goes through four steps, hydrolysis, acidogenesis, acetogenesis and methanogenesis. The effluent samples used for this study were obtained from two different anaerobic co-digestion systems, single stage and two-stage system. The efficiency of the single-stage and two-stage anaerobic co-digestion processes was then compared by analyzing the identities of the soluble compounds which were produced.
Soluble compounds identified with LC-MS/MS
Two systems were involved. The first was a single-stage digestion system, for the processes of hydrolysis, acidogenesis and methanogenesis to be carried out. The sample used for the single-stage anaerobic digestion was aerobic and anaerobic sludge obtained from wastewater treatment plant mixed with cooking oil, named Sample SD. The sludge and cooking oil mixture was also treated in a separate two step anaerobic digestion system, whereby the hydrolysis, acidogenesis and methanogenesis processes were performed in two separate stages. The effluent samples taken from stage 1 and stage 2 were named TD1 and TD2 respectively. As described, Sample TD1 was the effluent from stage 1 reactor, while sample TD2 was the effluent from stage 2 reactor. Stage 1 reactor was operated for 3 days HRT at 35°C, for hydrolysis and acidogenesis process to take place, while stage 2 reactor was for the methanogenesis process to take place (17 days HRT and 35°C). The feed for stage 2 reactor was the effluent from stage 1 reactor.
Sample SD contained very high amounts of soluble COD (sCOD), where 70%-80% of the sCOD was found to be volatile fatty acids (VFAs) (C2-C5) and 20-30% sCOD was found to be other components (data not shown here). In the identified compounds lists, we can find organic acids, lipids, cellular metabolites, antibiotics, antiparasitics, plasticizer, fragrance, pharmaceutical products and various other unknown molecules. Table 1 showed that 29 compounds were identified from sample SD. The results indicated that the organic acids and lipids are main components of SMP in this effluent sample, especially some long chain fatty acid, which probably were derived from the degradation of feed wastewater by microorganisms and produced by microorganisms. These results obtained here were similar to the results in the previous work [12-14]. Additionally, these compounds in the samples may be cause membrane fouling and low effluent quality, thus some effective strategies can be taken to reduce these particular compounds. For example, in Antoine’s work [12], powdered activated carbon (PAC) were added into the membrane bioreactor system adsorb the soluble compounds in wastewater. As a result, less amount of compounds was detected in the permeate samples. Specifically, removal of the phenanthrenecarboxylic acids could reach to 100% and some other compounds also were removed with high removal efficiency. However, the different removal rate of these compounds was associated with the surface hydrophobicity of both compounds and PAC. Thus it provides us inspiration that the appropriate adsorbents can be used to remove particular compounds according to their characteristic. Moreover, change of treatment temperature also could affect the removal efficiency of different compounds in Antoine’s work. These results prompt researchers to study different effective strategies to control and reduce known compounds.
| Name | Formula | m/z | Score (DB) | |
|---|---|---|---|---|
| 1 | Undecanoic acid, 3-hydroxy-, (S)- | C11H22O3 | 225.146 | 86.55 |
| 2 | Sugetriol | C15H24O3 | 253.1808 | 81.89 |
| 3 | Sapelin A | C30H50O4 | 475.3792 | 76.7 |
| 4 | Proclavaminic acid | C8H14N2O4 | 203.1029 | 78.55 |
| 5 | Phosphatidyl glycerol | C6H15O8P | 269.0385 | 81.53 |
| 6 | PGH2-EA | C23H39NO4 | 416.2772 | 89.45 |
| 7 | Lauryl hydrogen sulfate | C12H26O4S | 267.1628 | 75.88 |
| 8 | Ivermectin B1b | C47H72O14 | 861.5023 | 90.92 |
| 9 | Dihydrojasmonic Acid, Methyl Ester | C13H22O3 | 227.1631 | 82.7 |
| 10 | dexpanthenol | C9H19NO4 | 223.1647 | 78.87 |
| 11 | Decanoic acid, 9-hydroxy-, (R)-; (-)-(R)-9-Hydroxydecanoic acid; D-9-Hydroxydecanoic acid | C10H20O3 | 211.1296 | 86.46 |
| 12 | Cycluron | C11H22 N2O | 221.1626 | 87.43 |
| 13 | Chrysanthetriol | C15H26O3 | 255.1958 | 82.66 |
| 14 | Arachidic acid(d3) | C20H37D3O2 | 316.3285 | 92.13 |
| 15 | AAPH | C8H18N6 | 199.1671 | 90.99 |
| 16 | 9-Tridecynoic acid | C13H22O2 | 211.169 | 85.89 |
| 17 | 9-Methyl-undecanoic acid | C12H24O2 | 223.1679 | 82.59 |
| 18 | 8Z-Undecenyl acetate | C13H24O2 | 213.1847 | 99.55 |
| 19 | 8-oxo-3,7-Dimethyl-6E-octenyl acetate | C12H20O3 | 213.1483 | 76.71 |
| 20 | 7-oxo-11E,13-Tetradecadienoic acid | C14H22O3 | 239.1636 | 75.14 |
| 21 | 4-Imidazolone-5-acetate | C5H6N2O3 | 165.0265 | 79.57 |
| 22 | 4-Heptyloxyphenol | C13H20O2 | 209.1532 | 80.4 |
| 23 | 4,4',alpha-Trihydroxy-2'-methoxydihydrochalcone | C16H16O5 | 311.0888 | 80.34 |
| 24 | 4-(Glutamylamino) butanoate | C9H16N2O5 | 233.1135 | 98.78 |
| 25 | 2-Nor-1,3-seco-1α,25-dihydroxyvitamin D3 | C26H44O3 | 427.3178 | 88.2 |
| 26 | 23,23-difluoro-25-hydroxyvitamin D3 / 23,23-difluoro-25-hydroxycholecalciferol | C27H42F2O2 | 454.3498 | 83.38 |
| 27 | (6S,9R)-Vomifoliol | C13H20O3 | 225.1474 | 81.21 |
| 28 | (3S,7R)-iso-jasmonic acid | C12H18O3 | 211.1321 | 83.5 |
| 29 | (+)-trans-alpha-Irone | C14H22O | 207.1734 | 83.18 |
Table 1: Identified compounds from sample SD.
The formulas listed here were provided by searching against the METLIN database. The scores were obtained by compare the molecular features of the compounds in the sample against those in the database. The higher score represents the higher possibility of the match with the mass feature of the compound in the database.
For the two-stage system, the results of LC-MS analysis showed that 42 and 25 compounds were identified in sample TD1 (the first stage) and TD2 (the second stage) respectively (Tables 2 and 3). The results indicated that much less compounds were detected in the stage 2 compared with the stage 1, which is due to the fact that these compounds were degraded during the second stage. From the two tables, it can be seen that some of the compounds identified in TD1 were also found in TD2, which indicated that those compounds were not degraded during the stage 1 and stage 2. . This might due to the fact that those compounds also could not be degraded in stage 2 and the systems might require an acclimation period to receive the co-substrates for higher efficiency. Furthermore, the main compounds detected in TD1 and TD2 were also long chain fatty acid or their ester, similar to the results obtained in SD. Additionally, compared the SD and TD2, it might be impossible to remove some compounds by one process, and further purification methods might be required depending on the standard of the discharge or the usage of the purified water.
| Name | Formula | m/z | Score (DB) | |
|---|---|---|---|---|
| 1 | Undecanoic acid, 3-hydroxy-, (S)- | C11H22O3 | 203.1631 | 81.91 |
| 2 | U-51605 | C20H32N2O2 | 350.2804 | 84.99 |
| 3 | Thermospermine | C10H26N4 | 241.1787 | 94.04 |
| 4 | Strigolactone ABC-rings | C14H18O3 | 235.1331 | 80.05 |
| 5 | SerSer | C6H12N2O5 | 215.0629 | 92.44 |
| 6 | Sapelin A | C30H50O4 | 475.3802 | 82.46 |
| 7 | Proclavaminic acid | C8H14N2O4 | 203.1031 | 79.08 |
| 8 | Phosphatidyl glycerol | C6H15O8P | 269.0392 | 84.1 |
| 9 | N-palmitoyl glycine | C18H35NO3 | 331.2964 | 76.67 |
| 10 | N-oleoyl glutamine | C23H42N2O4 | 411.3214 | 85.78 |
| 11 | N-3-oxo-hexadec-11(Z)-enoyl-L-Homoserine lactone | C20H33NO4 | 369.2749 | 84.25 |
| 12 | Lubimin | C15H24O4 | 237.186 | 77.23 |
| 13 | Lauryl hydrogen sulfate | C12H26O4S | 267.163 | 75.07 |
| 14 | EPA (d5) | C20H25D5O2 | 308.2642 | 76.49 |
| 15 | Dihydrojasmonic Acid, Methyl Ester | C13 H22 O3 | 227.165 | 86.4 |
| 16 | Dihydrojasmonic Acid, Methyl Ester | C13H22O3 | 227.1633 | 83.26 |
| 17 | Dihydroflavokawain C | C17H18O5 | 325.1062 | 75.16 |
| 18 | D-Fucosamine | C6H13NO4 | 181.1185 | 75.01 |
| 19 | Decanoic acid, 9-hydroxy-, (R)-; D-9-Hydroxydecanoic acid | C10H20O3 | 211.1301 | 86.32 |
| 20 | Corey Lactone Aldehyde Benzoate | C15H14O5 | 297.0728 | 78.23 |
| 21 | Cnidilide | C12H18O2 | 195.1385 | 75.9 |
| 22 | Chrysanthetriol | C15H26O3 | 255.1966 | 79.66 |
| 23 | Arachidic acid(d3) | C20H37D3O2 | 316.3295 | 99.17 |
| 24 | altretamine | C9H18N6 | 211.1669 | 83.48 |
| 25 | 9Z-Dodecen-7-ynyl acetate | C14H22O2 | 223.1687 | 85.6 |
| 26 | 8Z-Undecenyl acetate | C13H24O2 | 213.1849 | 97.69 |
| 27 | 8-Methyl-undecanoic acid | C12H24O2 | 223.1662 | 82.83 |
| 28 | 7-oxo-11E,13-Tetradecadienoic acid | C14H22O3 | 239.1644 | 79.01 |
| 29 | 7-Aminomethyl-7-carbaguanine | C7 H9N5O | 197.1154 | 84.15 |
| 30 | 5Z-Decenyl acetate | C12H22O2 | 199.1684 | 96.53 |
| 31 | 4-Heptyloxyphenol | C13H20O2 | 209.1533 | 85.25 |
| 32 | 4,4',alpha-Trihydroxy-2'-methoxydihydrochalcone | C16H16O5 | 311.0896 | 77.25 |
| 33 | 23,23-difluoro-25-hydroxyvitamin D3 / 23,23-difluoro-25-hydroxycholecalciferol | C27H42F2O2 | 454.3496 | 76.03 |
| 34 | 1β,3α,4α-p-menthane-3,8-diol | C10H20O2 | 195.1349 | 85.45 |
| 35 | 1-tetradecanyl-2-(8-[16]-ladderane-octanyl)-sn-glycerophosphoethanolamine | C39H74NO6P | 701.5625 | 83.23 |
| 36 | 1-amino-3,3-diethoxypropane | C7H17NO2 | 165.1591 | 78.08 |
| 37 | 1-amino-3,3-diethoxypropane | C7H17NO2 | 165.1592 | 79.41 |
| 38 | (6S,9R)-Vomifoliol | C13H20O3 | 225.1491 | 82.66 |
| 39 | (3S,7R)-iso-jasmonic acid | C12H18O3 | 211.133 | 83.47 |
| 40 | (25S)-3-oxo-cholest-1,4-dien-26-oic acid | C28H42O3 | 427.3202 | 78.51 |
| 41 | (±)-Ibuprofen | C13H18O2 | 207.1378 | 82.36 |
| 42 | (+)-trans-alpha-Irone | C14H22O | 207.174 | 86.74 |
Table 2: Identified compounds from sample TD1.
| Name | Formula | m/z | Score (DB) | |
|---|---|---|---|---|
| 1 | Undecanoic acid, 3-hydroxy-, (S)- | C11H22O3 | 225.1473 | 79.49 |
| 2 | U-51605 | C20H32N2O2 | 350.2801 | 80.54 |
| 3 | Thermospermine | C10H26N4 | 241.1786 | 97.17 |
| 4 | Proclavaminic acid | C8H14N2O4 | 203.1028 | 79.77 |
| 5 | Phosphatidyl glycerol | C6H15O8P | 269.0392 | 85.35 |
| 6 | Oleic acid (d5) | C18H29D5O2 | 288.2953 | 77.99 |
| 7 | N-3-oxo-hexadec-11(Z)-enoyl-L-Homoserine lactone | C20H33NO4 | 369.2752 | 83.5 |
| 8 | Lauryl hydrogen sulfate | C12H26O4S | 267.1629 | 76.2 |
| 9 | Dodecanoic acid, 11-hydroxy-, (R)- | C12H24O3 | 217.1802 | 86.47 |
| 10 | Dihydrojasmonic Acid, Methyl Ester | C13H22O3 | 227.1634 | 84.38 |
| 11 | Corey Lactone Aldehyde Benzoate | C15H14O5 | 297.0727 | 76.35 |
| 12 | Arachidic acid(d3) | C20H37D3O2 | 316.3288 | 97.73 |
| 13 | 9Z-Dodecen-7-ynyl acetate | C14H22O2 | 223.1684 | 82.52 |
| 14 | 9-Tridecynoic acid | C13H22O2 | 211.1692 | 86.71 |
| 15 | 8Z-Undecenyl acetate | C13H24O2 | 213.185 | 99.6 |
| 16 | 8-oxo-3,7-Dimethyl-6E-octenyl acetate | C12H20O3 | 213.1485 | 75.49 |
| 17 | 8-Methyl-undecanoic acid | C12H24O2 | 223.1671 | 86.46 |
| 18 | 7-oxo-11E,13-Tetradecadienoic acid | C14H22O3 | 239.164 | 85.28 |
| 19 | 4,4',alpha-Trihydroxy-2'-methoxydihydrochalcone | C16H16O5 | 311.0894 | 75.69 |
| 20 | 2-Nor-1,3-seco-1α,25-dihydroxyvitamin D3 | C26H44O3 | 427.3189 | 82.42 |
| 21 | 23,23-difluoro-25-hydroxyvitamin D3 / 23,23-difluoro-25-hydroxycholecalciferol | C27H42F2O2 | 454.3497 | 91.75 |
| 22 | 1-Phenylbiguanide | C8H11N5 | 195.1345 | 77.8 |
| 23 | (6S,9R)-Vomifoliol | C13H20O3 | 225.149 | 81.38 |
| 24 | (3S,7R)-iso-jasmonic acid | C12H18O3 | 211.1322 | 82.44 |
| 25 | (+)-trans-alpha-Irone | C14H22O | 207.1736 | 84.93 |
Table 3: Identified compounds from sample TD2.
Compared with the compounds detected in TD1, some organic acids and their derivatives, amino acids and their derivatives, and antibiotics were degraded and removed completely in sample TD2 during stage 2, for example, phosphatidyl glycerol, N-palmitoyl glycine, N-oleoyl glutamine, decanoic acid, altretamine, 5Z-decenyl acetate, 4-heptyloxyphenol and so on. For the two-stage digestion, stage 1 reactor carried out the process of hydrolysis and acidogenesis. The hydrolytic bacteria transformed the particulate organic substrate into liquefied monomers and polymers i.e. proteins, carbohydrates and fats into amino acids, mono saccharides and fatty acids, respectively. Following this, the acidogenic bacteria transformed the products of hydrolysis into short chain volatile acids, ketones, alcohols, hydrogen and carbon dioxide. The products of this acidogenesis process could be utilized directly by the methanogenic bacteria in the second stage to produce CH4 for the methanogenesis process. Therefore, the compounds existed in TD1 but removed from TD2 probably were directly degraded by methanogenic bacteria. Similarly, compared with compounds identified in SD, some compounds such as sugetriol, decanoic acid, 4-imidazolone-5-acetate, 4-heptyloxyphenol could be removed from TD2 sample during two-stage digestion process. The single step process combined all the four processes of hydrolysis, acidogenesis and methanogenesis in one single reactor, and there were still more compounds left after the four steps compared with two-stage process. On the other hand, comparing table 1 and table 3, there are also some compounds degraded in single-stage system but still existed in TD2 during co-digestion process, which might due to the fact that the period of stage 1 was short so that many compounds could not be degraded completely by hydrolytic and acidogenic bacteria, leading to exist in the final effluent of two-stage digestion process. Therefore, the stage and stage 2 should be arranged appropriately in order to obtain high digestion efficiency in the future study.
Compounds identification and analysis method
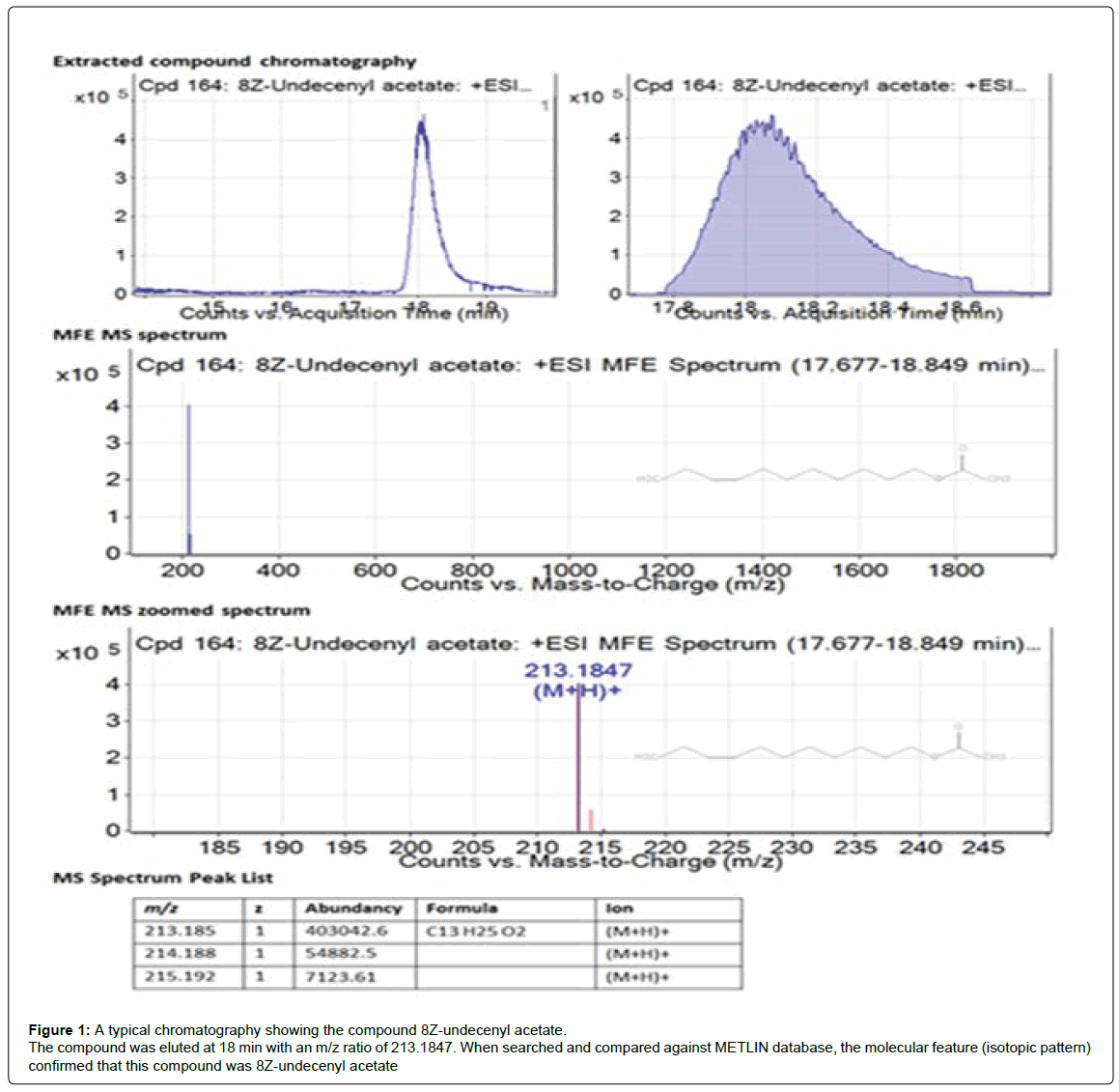
In this method and database analysis with LC-MS, the compounds eluted and their molecular features were used for identification. Figure 1 showed a typical extracted compound chromatography of a certain compound (undecenyl acetate) as an example. As shown in the MFE MS zoomed spectrum, three isotopes were detected (red lines). The m/z value of the main isotope together with and the ratio among the isotopes, combined the established isotopic patterns in the database, provided sufficient information that this compound could be identified as undecenyl acetate. Based on the identification methods, the compounds of the three samples SD, TD1 and TD2 are identified and list in the Tables 1-3 respectively.
From the Tables 1-3, it can be seen that the largest number of compounds were detected in TD1, and the number reached only 42 and some compounds had low match relationship with the mass feature of compounds in the database. In fact, the fragmentation pattern of a compound by MS/MS analysis can further help to validate or negate the search hits. According to preliminary results obtained, fragmentation pattern analysis using MS/MS could be carried out in the next step in order to confirm the compounds with low match in the database. Moreover, in some studies using developed MS method to analyze metabolic molecular and compounds produced by microorganisms, more compounds can be detected and identified with the help of metabolic profile analysis. In fact, this metabolic analysis method also can be used to monitor the activity of functional microorganisms in the wastewater so that it can monitor the operation process of membrane bioreactor system and wastewater treatment. These microorganisms play an essential role in compounds degradation process duding wastewater treatment, and complicated compounds are produced by the mixture of microorganisms and from decay of microorganisms, such as SMP.
However, compared with the number of compounds listed in Tables 1-3 (29, 42 and 25 respectively), it was clear that only a small portion of the compounds managed to be identified from the database. The main reason for this is despite recent developments, the MS database for small molecules are still far from comprehensive. Limited entries of molecular information are present in the database. Other possible reason could lie in the running condition, sample preparation and low abundance of some compounds, therefore insufficient information could be obtained during the analysis. For example, in this work, we used HPLC for the online separation of the compounds but the separation efficiency is limited. An ultra-performance liquid chromatography (UPLC) could obtain higher separation efficiency due to the small particle size of the packing material. Therefore, ultraperformance liquid chromatography could address the problem and might be able to boost the analysis result, and it can be tried in the future.
Long chain fatty acids analysis with GC/MS
Based on the results obtained at present, in each of the samples, many compounds identified with LC-MS were long chain fatty acids (LCFAs) and their derivatives, so LCFAs could be detected with GCMS in future and the levels of LCFAs may be quantified to analyze change tendency between different samples. According to previous study, LCFAs were specifically analyzed using a GC machine coupled with MS, and the samples were subject to derivatization of their fatty acids using BF3-methanol derivatization method for the analysis of non-volatile LCFA. From the Tables 1-3, it can be seen that after the process of single digestion and two-stage digestion, there was still LCFAs present in the effluent but the less type of LCFAs was left in TD2 compared with TD. This indicate that during the digestion in a single-stage system, many fatty acids could not be acidized by bacteria. Comparatively, it was found in the two stage process, that the type of LCFA in sample TD1 was even more than the effluent in the single step process. However, after treatment from stage two, many types of LCFA were degraded by microorganisms. In the future, the amount of the same type LCFA in different samples was quantified with GS-MS which can assist us to analyze different efficiency between different treatment processes. Generally, during the methanogenesis stage, most of the fatty acid may be degraded into methane. However, probably there is still amount of fatty acids left in the effluent due to the inhibition of oil and unknown compounds in the sludge. Quantification of LCFAs also can monitor wastewater treatment and boost generation of modification method of the process system to obtain a higher level of digestion.
Conclusions
LC-MS/MS based metabolomics analysis technique was successfully used for the analysis of untargeted soluble compounds in the effluent obtained from anaerobic co-digestion system. Different profiles of compounds were identified. In the data from this study, the two stage co digestion process showed higher digestion efficiency as compared with the single stage process, as fewer compounds were found in the effluent. In the two-stage process, most of the fatty produced in the first stage of digestion by hydrolysis and acidogenesis, were digested in the second stage. Although the single stage process combined all the steps, the digestion efficiency was lower than the separated twostage process. Our work is one of few exploration of analyzing and identifying unknown compounds using MS based technique from a metabolic analysis perspective. It could provide a brand new analytical platform and a different view point in various wastewater bioprocess treatment analysis. Its ability of unknown compounds identification could be used as a strong tool in the study of bioprocess efficiency and parameters optimization, fouling mechanism, disinfection byproducts, and toxicity of the processed water. In addition to analysis of unknown soluble compounds using LC-MS/MS, the amount of LCFA can be analyzed by GC-MS in the future.
Acknowledgements
The authors acknowledge funding support from Environment and Water Industry Programme Office (EWI), Singapore. We also thank the support from Nanyang Environment and Water Research Institute, Nanyang Technological University.
References
- Laspidou CS, Rittmann BE (2002) A unified theory for extracellular polymeric substances, soluble microbial products, and active inert biomass. Water Res 36: 2711-2720.
- Ni BJ, Rittmann BE, Yu HQ (2011) Soluble microbial products and their implications in mixed culture biotechnology.Trends Biotechnol 29: 454-463.
- Silva DGV, Rittmann BE (2000) Nonsteady-state modeling of multispecies activated-sludge process. Water Environ Res 72: 554-565.
- Barkers DJ, Stuckey DC (1999) A review of soluble microbial products (SMP) in wastewater treatment system. Water Research 33: 3063-3082.
- Jiang T, Kennedy MD, De Schepper V, Nam SN, Nopens I, et al. (2010) Characterization of soluble microbial products and their fouling impacts in membrane bioreactors.Environ SciTechnol 44: 6642-6648.
- Wang ZP, Zhang T (2010) Characterization of soluble microbial products (SMP) under stressful conditions.Water Res 44: 5499-5509.
- Zhang ML, Sheng GP, Yu HQ (2008) Determination of proteins and carbohydrates in the effluents from wastewater treatment bioreactors using resonance light-scattering method.Water Res 42: 3464-3472.
- Ni BJ, Zeng RJ, Fang F, Xie WM, Sheng GP, et al. (2010) Fractionating soluble microbial products in the activated sludge process.Water Res 44: 2292-2302.
- Jarusutthirak C, Amy G (2006) Role of soluble microbial products (SMP) in membrane fouling and flux decline.Environ SciTechnol 40: 969-974.
- Aquino SF, Stuckey DC (2006) Chromatographic characterization of dissolved organics in effluents from two anaerobic reactors treating synthetic wastewater.Water SciTechnol 54: 193-198.
- Zhou W, Wu B, She Q, Chi L, Zhang Z (2009) Investigation of soluble microbial products in a full-scale UASB reactor running at low organic loading rate.BioresourTechnol 100: 3471-3476.
- Trzcinski AP, Stuckey DC (2010) Treatment of municipal solid waste leachate using a submerged anaerobic membrane bioreactor at mesophilic and psychrophilic temperatures: analysis of recalcitrants in the permeate using GC-MS. Water Res 44: 671-680.
- Trzcinski AP, Stuckey DC (2009) Continuous treatment of the organic fraction of municipal solid waste in an anaerobic two-stage membrane process with liquid recycle.Water Res 43: 2449-2462.
- Aquino SF, Stuckey DC (2002) Characterization of soluble microbial products (SMP) in effluents from anaerobic reactors.Water SciTechnol 45: 127-132.
- Mata-Alvarez J, Mace S, Llabres P (2000) Anaerobic digestion of organic solid wastes. An overview of research achievements and perspectives. BioresourceTechnol 74: 3-16.
- Luostarinen S, Luste S, Sillanpää M (2009) Increased biogas production at wastewater treatment plants through co-digestion of sewage sludge with grease trap sludge from a meat processing plant.BioresourTechnol 100: 79-85.
- Long JH, Aziz TN, Reyes FL, Ducoste JJ (2012) Anaerobic co-digestion of fat, oil, and grease (FOG): A review of gas production and process limitations. Process Saf Environ 90: 231-245.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 15687
- [From(publication date):
June-2015 - Apr 02, 2025] - Breakdown by view type
- HTML page views : 11017
- PDF downloads : 4670