An Evaluation Model of Enhancing and Releasing Effect of the Blue Swimming Crab, Portunus trituberculatus, Based on Molecular Marker Technique
Received: 22-Aug-2018 / Accepted Date: 10-Sep-2018 / Published Date: 18-Sep-2018 DOI: 10.4172/2332-2608.1000279
Keywords: Portunus trituberculatus; mtSNP; Stock enhancement; Recapture rate; Evaluation model
Introduction
As a kind of large economic crabs, the blue swimming crab, Portunus trituberculatus, widely distributed in the Yellow Sea, Bohai Sea and the west coast of the Pacific, is an important capture target in China, the North Korea, the South Korea, Japan, the Philippines and Vietnam, etc. [1-3]. It is also the most important species for marine culture in China [4]. Over the past several decades, the number of P. trituberculatus, living in the natural sea area, has dramatically dropped as a result of overfishing and the change in natural ecological environment. In order to make up for the shortage of natural fishing resources, the Chinese and Japanese governments have begun to implement the stock enhancement plan for P. trituberculatus since 1985. The development of enhancing and releasing activities significantly increases the wildcaught catches of P. trituberculatus. For example, only 349,000 tons of P. trituberculatus were caught in 2010 off the coast of China [5], compared with 540,000 tons in 2017, an increase of 54.7% [6].
In spite of the remarkable economic benefits from enhancing and releasing, there has been no evaluation method for the effect all the time. In order to evaluate the effect of enhancing and releasing, the marking releasing must be carried out in the estimation of the recapture rate and multiplication effect in natural population (proportions) [7]. As a kind of crustaceans, the conventional physical and chemical marker techniques cannot be applied to the enhancing and releasing marking of P. trituberculatus, since on the one hand, this kind of species has to experience multiple shelling to grow [8] and on the other hand, the larvae are vulnerable to enhancing and releasing and any marker technique that damages the body can result in a high mortality rate, which further leads to the inaccurate recapture rate. In this regard, molecular marker technique, which does not cause any damage to the released individuals, is an ideal means for evaluating the stock enhancement effect of P. trituberculatus. However, few studies about the evaluation model of enhancement and release effect based on molecular marker technique have been conducted yet. In view of the current development of SNP in mitochondrial genome of P. trituberculatus, the present paper attempts to propose a model for evaluating the enhancing and releasing effect based on this marker technique, with an aim to provide theoretical guidance for future researches.
The establishment of mathematical model
As the survey of germplasm resources indicate, the blue swimming crab does not take long-distance migration, with the maximum of 25 km [9]. On this basis, assumptions can be made that after artificial release, the P. trituberculatus will distribute in the water area of 25 km around the releasing point, that is, the total area S=πr2≈1962.5 km2. In an attempt to investigate the recapture rate of enhancing and releasing individuals, samples are required within the area. Suppose ten points are randomly selected with the total area of S1. The number of P. trituberculatus collected in S1 is N1 and the individuals selected by SNP marker identification technology is N1'. In addition, assuming that the probability of “actual identification rate of molecular marker technique” is Pm, then the actual N1 sample should contain the releasing individuals, N2=N1'x Pm On this basis, it can be concluded that the total number of released individuals N'2 in the total area S (1962.5 km2) is 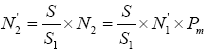 .
.
Based on the mitochondrial genomic SNP markers, it can be found that different from the previous physical markers [10], this method can be used to label all the releasing individuals, not only part of them. Therefore, the number of released individuals and their corresponding ratios can be obtained directly from the total number of catches in any batch. Moreover, the probability distribution law of individuals released with molecular markers can be regarded as normal distribution, compared with Poisson distribution, the distribution pattern only for marking thousands to tens of thousands of individuals [10]. Therefore, we further propose the following mathematical models for the total number of enhancing and releasing individuals from the population.
Assuming that the P. trituberculatus collected at a certain time is N1 with the released individuals of N2, then the probability (ri) of released individuals at any time is presented as follows:
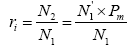 (1)
(1)
Where N1' is the released individuals identified by molecular marker technique and Pm is the molecular marker recognition rate.
According to the probability of normal distribution, the average expected value (μ) of individuals released at a certain time point in the sea area is:
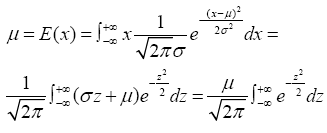 (2)
(2)
Where the x refers to the number of released individuals from a single sampling and σ is the overall standard deviation. Therefore, supposing that the released individuals per sampling is μi, the surface sea area is S1 and
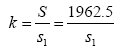
Where S refers to the total distribution of released individuals, then the total number of enhancing and releasing individuals can be estimated by the following formula, which is 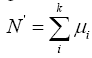 , where i=1,2,3…k.
, where i=1,2,3…k.
The verification of mathematical model
Sample marking: In order to verify the applicability of the above model, the present study employs a circular aquaculture pond with a radius of 65 m (the total area is about 13,266.5 m2 with 140,000 P. trituberculatus in total), and takes the offspring of 60 parent P. trituberculatus as the research objects. 50 juvenile crabs in the second phase are selected from each family. The physical marks of individuals are obtained by injecting different colors of fluorescent markers (Visible Implant Fluorescent tag, North West Marine Technology Inc.) into the fourth foot base. After 24-36 hours of short-term feeding observation, 30 crabs with the best preserved markers and the strongest vitality are selected from each family and put into the aquaculture pond. Altogether 1,800 crabs with marks are put into ponds.
The detection and recognition of marked individuals: After 72 hours of artificial marking, the P. trituberculatus in the pond are randomly sampled and examined. In the aquaculture pond above, 10 sampling points are evenly selected and one crab traps are placed at each sampling point. After one night, the number and ratio of artificially marked individuals can be statistically and calculated and collected. Furthermore, 18 mtSNPs markers can be used for the identification of the captured individuals [11]. The results show that the recognition rate of molecular markers is 95.5%. On this basis, Pm value of the molecular marker recognition rate in the above model can be set to 0.955.
The number of marked individuals (enhancing and releasing individuals) obtained from the 10 sampling are 9, 11, 6, 8, 9, 11, 8, 9, 13, 7, respectively. The average value is 9.1 and the sampling area is about 0.122 m2. According to the model above, it is estimated that the number of marked individuals in the pond is 1776. Chi- square test shows that there is no significant difference from the actual marked individuals (1800). The results indicate the high reliability of this model.
Acknowledgment
This project was supported by the National Natural Science Foundation of China (31472282), the Priority Academic Program Development of Jiangsu Higher Education Institutions PAPD, Agricultural science and technology support Project of Jiangsu (BE2009339) and the projects of "Six Talent Summit" team of Jiangsu (2016-HYGC-CXTD-004).
References
- Cui Z, Liu Y, Luan W, Li Q, Wu D, et al. (2010) Molecular cloning and characterization of a heat shock protein 70 gene in swimming crab (Portunus trituberculatus). Fish Shellfish Immunol 28: 56-64.
- Lv J, Liu P, Wang Y, Gao B, Chen P, et al. (2013) Transcriptome analysis of Portunus trituberculatus in response to salinity stress provides insights into the molecular basis of osmoregulation. PLoS One 8: e82155.
- Imai H, Fujii Y, Karakawa J, Yamamoto S, Numachi K (1999) Analysis of the population structure of the swimming crab, Portunus trituberculatus in the coastal waters of Okayama Prefecture, by RFLPs in the whole region of mitochondrial DNA. Fisheries Sci 65: 655-656
- Huo YW, Jin M, Zhou PP, Li M, Mai KS, et al. (2014) Effects of dietary protein and lipid levels on growth, feed utilization and body composition of juvenile swimming crab, Portunus trituberculatus. Aquaculture 434: 151-158.
- Tringali MD, Leber KM, Halstead WG, Mcmichael R, O'hop J, et al. (2008) Marine stock enhancement in Florida: a multi-disciplinary, stakeholder-supported, accountability-based
- He J, Gao Y, Xu W, Yu F, Su Z, et al. (2017) Effects of different shelters on the molting, growth and culture performance of Portunus trituberculatus. Aquaculture 481: 133-139
- Okamoto K (2004) Juvenile release and market size recapture of the swimming crab Portunus trituberculatus (Miers) marked with coded wire tags. In: Leber KM, Kitada S, Blankenship HL, Svasand T (eds) Stock enhancement and sea ranching: development, pitfalls and opportunities (2nd Edition). Oxford, pp: 181-186.
- Tuck GN, De La Mare WK, Hearn WS, Williams R, Smith ADM, et al. (2003) An exact time of release and recapture stock assessment model with an application to Macquarie Island Patagonian toothfish (Dissostichus eleginoides). Fish Res 63: 179-191.
- Zhao L, Li ZH, Zhang P, Xue B, Lai XF, et al. (2018) Analysis of single nucleotide polymorphism(SNP)in mitochondrial genome of Portunus trituberculatus. J Fisheries of China 42: 196-203.
Citation: Yan B, Xu W, Sun J, Gao H (2018) An Evaluation Model of Enhancing and Releasing Effect of the Blue Swimming Crab, Portunus trituberculatus, Based on Molecular Marker Technique. J Fisheries Livest Prod 6: 279. DOI: 10.4172/2332-2608.1000279
Copyright: © 2018 Yan B, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 2762
- [From(publication date): 0-2018 - Apr 19, 2025]
- Breakdown by view type
- HTML page views: 1995
- PDF downloads: 767
