Research Article Open Access
Allelic Variants of KLK2 Gene Predict Presence of Prostate Cancer at Biopsy
Emmanuel Okechukwu Nna*, Sam Tothill and Tracey Bailey
Safety Molecular Pathology Laboratory, Enugu, Nigeria
- *Corresponding Author:
- Mbamalu ON
Discipline of Pharmacology, School of Pharmacy
University of the Western Cape
Bellville 7535, South Africa
Tel: +27219593229/2190
Fax: +278615107002
E-mail: ombamalu@uwc.ac.za
Received Date: March 14, 2016; Accepted Date: April 19, 2016; Published Date: April 22, 2016
Citation: Nna EO, Tothill S, Bailey T (2016) Allelic Variants of KLK2 Gene Predict Presence of Prostate Cancer at Biopsy. J Cancer Diagn 1:105. doi: 10.4172/2476-2253.1000105
Copyright: © 2016 2016 Nna EO, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Cancer Diagnosis
Abstract
Objective: Several single nucleotide polymorphisms associated with prostate cancer risk have been reported in recent years. We evaluated polymorphisms in the human glandular kallikrein 2 (KLK2) genes because the protein product of this gene is known to be increased in prostate cancer.
Materials and methods: Blood samples were collected from sixty patients who underwent prostate biopsy sectioning, and from their genomic DNA the SNPs in KLK2 gene were investigated by direct DNA sequencing. Another 138 archived prostate tissue sections were also evaluated using the TaqMan SNP genotyping assay.
Results: Eighteen known SNPs were identified in the KLK2 gene. The SNPs were located in introns, coding exons and untranslated regions of the gene. Further analysis showed that two of the SNPs were associated with prostate disease. The T/T allele of rs198977 was significantly predictive of the presence of prostate cancer at biopsy and was also associated with high tumour grade. The A/A allele of rs2664155 was also significantly associated with the presence of benign hyperplasia at biopsy.
Conclusion: Our results support previous reports of association of the rs198977 SNP with prostate cancer risk and also indicated a link with the disease phenotype. However, the second SNP (rs2664155) was more associated with benign hyperplasia than prostate cancer risk. The method of TaqMan SNP genotyping could be clinically useful in genetic screening and risk stratification of patients for prostate diseases.
Keywords
Allelic variants; SNP; KLK2 gene; Prostate cancer risk; Prostate
Introduction
Single nucleotide polymorphisms (SNPs) are the most abundant form of human genetic variations and a resource for mapping complex genetic traits [1,2]. Several efforts, for example the HapMap project, have been made to document the frequencies of various SNPs in different ethnic groups and human races and to evaluate their associations with diseases [3]. Currently, there are no clinical tests to evaluate genetic predisposition to prostate cancer risk in men with or without elevated prostate specific antigen (PSA), abnormal digital rectal examination (DRE), or both. Reports by Nam et al. showed that two SNPs in KLK2 gene: rs2664155 (AA or AG variants) and rs198977 (TT or TC variants) were strongly associated with the presence of prostate cancer at biopsy [4]. Genome-wide association studies have also identified several groups of SNPs (haplotypes) in different genes that are linked to prostate cancer risk [5,6]. However, the SNPs in KLK2 gene warrant further investigation because the protein product of KLK2 gene (hK2) is only secreted by the prostate and its serum levels correlate with prostate cancer development [3]. Biologically, the hK2 activates the PSA, which is involved in the liquefaction of the seminal fluid thereby aiding sperm motility [7].
Objective
The objective of this study was to find out if any of the SNPs in the KLK2 gene could predict the presence of prostate cancer at biopsy. The second objective was to find out if any associated SNP has a correlation with the disease phenotype (tumour grade and/ or pathological grade). The third objective was to evaluate the performance of a TaqMan SNP genotyping assay for any of the associated SNP and to use the assay for genotyping DNA from archived formalin fixed, paraffin-embedded (FFPE) tissue sections.
Materials and Methods
Subjects
Sixty consecutive patients who consented to the study (after favourable ethical approval by the Bedfordshire Research Ethics Committee) were sampled for 5 ml of peripheral blood before they had transrectal ultrasound (TRUS) guided prostate biopsy. Most of the patients had elevated PSA level (≥ 4.0 ng/ml) and/ or abnormal DRE at the time of referral for biopsy. Some patients were referred to the clinic due to persistent urinary symptoms. No patient was a known case of prostate cancer. The patients were Caucasian whites (British). The blood samples were centrifuged at 1840 g for 5 minutes and plasma removed for PSA testing. The corpuscular components were lysed in a cold red cell lysis buffer (1.55 M Ammonium chloride, 0.01 M EDTA and 0.1 M Potassium bicarbonate; adjusted pH 7.4 using 10 M HCl). After two washes in the lysis buffer at 10 minutes intervals, white cell pellets were finally washed in phosphate buffered saline (PBS) before lysis in 1 ml of guanidine isothiocyanate (GITC) buffer. Genomic DNA was extracted from 200 μl of the lysates using QiaAmp DNA kit and the Qiacube automated extraction system (Qiagen UK).
FFPE Prostate tissue sections
Archived FFPE prostate tissue blocks (n=138) were retrieved from the Royal Gloucestershire tissue store following favourable ethical approved by the Royal Gloucestershire Research Ethics Committee. Two 25 micrometre thick sections were asceptically cut from each tissue block and picked into a 2 ml tube. The tubes were briefly centrifuged, deparaffinised in two washes of 1 ml xylene for 10 minutes each; the xylene decanted and the tissue rehydrated by two washes in 1 ml of 100% ethanol before allowing the pellets to dry for 5 minutes on a dry heat block kept at 37°C. The tissue pellets were digested overnight in 540 μl of ATL tissue lysis buffer and 60 μl of proteinase K (Qiagen, UK). The digest was centrifuged at 800 g for 5 minutes and the supernatants (containing nucleic acids) were collected. A 200 μl of the supernatant was used for DNA extraction. Another 300 μl of the supernatant was used for RNA extraction; and subsequent complementary DNA synthesis using random hexamer priming and the Molony murine leukaemic virus reverse transcriptase (M-MLV RT) in a final 50 μl reaction volume according to manufacturer’s recommendation (Promega, UK). The RNA elute was 30 μl and the quality was checked by automated electrophoresis using BioRad Experion system. Quality of gDNA was checked by the amplification of a 150 bp G6PD from each sample using 2 μl of the genomic DNA, 300 nM each of forward and reverse primers in a final 20 μl reaction volume using a standard polymerase chain reaction (PCR). The primer sequences and thermal protocol are available on request.
Direct sequencing
The KLK2 gene was sequenced in four parts covering nearly the entire KLK2 gene. The Big Dye X terminator purification system and the ABI 3130 genetic analyzer (Applied biosystems) were used for the sequencing according the manufacturer’s instructions. The nucleotide sequences for primers have been described in details elsewhere [4]. The sequences were aligned using the SeqMan component of the DNA star software (DNASTAR Inc, USA). The sequences were correlated with canonical sequences from the UCSC genome browser (http://genome. ucsc.edu/) and Entrez SNP database (http://www.ncbi.nlm.nih.gov/).
Allelic discrimination assays
Using the same genomic DNA from blood samples, predesigned SNP genotyping assays, based on TaqMan VIC- and FAMfluorescent labelled minor groove binding (MGB) probes for the two alleles and specific primers (Applied biosystems, UK) were used in genotyping SNPs rs198972 and rs198977: the assay identities were C_8705643 and C_736084 respectively. The ABI 7900HT sequence detection system (Applied biosystems, UK) was used according to manufacturer’s instructions. Genomic DNA from the FFPE tissue materials was also genotyped by this method. For each reaction, 50 to 100 ng (1-2 μl) of sample gDNA was used in setting up the real time PCR reaction, for allelic discrimination, in a final 25 μl reaction volume using thermal protocol as recommended by the manufacturer (Applied Biosystems).
Relative Quantification of KLK2 transcript
The KLK2, G6PD and ABL1 transcripts in the samples were measured by a triplex real time PCR in a 20 μl reaction volume using 2 μl of cDNA, TaqMan gene expression master mix and TaqMan probes (FAM for ABL1 and TET for KLK2 using a black hole quencher BHQ and CY5 for G6PD with a BHQ). Primer and probe sequences are shown in Table 1. The thermal profile was 50°C for 2 minutes, initial denaturation at 95°C for 10 minutes; cycle denaturation of 95°C for 15 seconds, annealing and extension at 60oC for 1 minute for 40 cycles. Data was acquired at all stages. The geometric mean of G6PD and ABL1 genes were used in normalizing the gene expression of KLK2. The normalized relative quantity (NRQ) of KLK2 was calculated using the formula NRQ=ECt KLK2/EGm of Ct ABL and G6PD where E is the PCR efficiency for each target, Gm is the geometric mean of Ct values of ABL1 and G6PD, the endogenous control genes [8].
| Gene | Sequence(5’-3’) | Chromosome location | Amplicon size |
|---|---|---|---|
| Abelson | Forward primer:GATACGAAGGGAGGGTGTACCA | 9q34 | 94bp |
| (ABL1)2 | Reverse primer:CTCGGCCAGGGTGTTGAA | ||
| Probe: FAM-GCTTCTGATGGCAAGCTCTACGTCTCCT-TAMRA. | |||
| G6PD | Forward primer: GGCGATGCCTTCCATCAG | Xq28 | 63bp |
| Reverse primer: CCAGGTCACCCGATGCA | |||
| Probe: CY5-CGGATACACACATATTC-BHQ | |||
| KLK2 | Forward primer:TGCCCATTGCCTAAAGAAGAATAG | 19q13 | 72bp |
| Reverse primer: CCTGTGTCTTCAGGCTCAAACA | |||
| Probe:Tet-CTGGGTCGGCACAAC-BHQ |
Table 1: Primer and Probe sets for RQ-PCR (qPCR).
Results
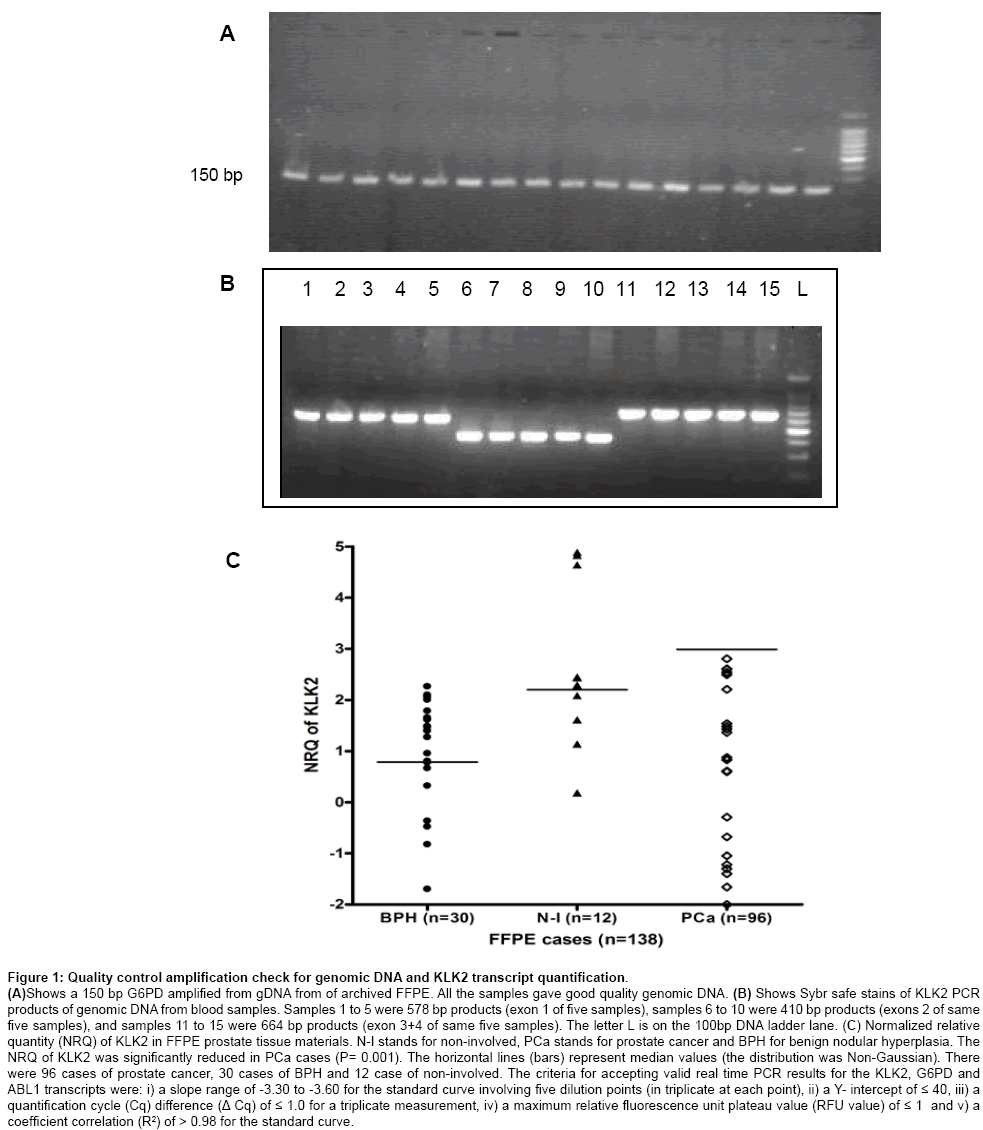
Figure 1a shows a representative SYBR safe stain of 2% agarose gel of 150 bp G6PD quality control check on gDNA from FFPE tissue materials. Figure 1b shows a representative SYBR safe stain of PCR products prior to setting up Big Dye terminator reaction. Single intense bands yielded good electropherograms during sequencing. Figure 1c shows the normalized relative quantity of KLK2 transcript in benign, non-involved and prostate cancer cases. KLK2 mRNA (normalized) was significantly reduced in the prostate cancer cases compared to BPH cases.
Figure 1: Quality control amplification check for genomic DNA and KLK2 transcript quantification.
(A)Shows a 150 bp G6PD amplified from gDNA from of archived FFPE. All the samples gave good quality genomic DNA. (B) Shows Sybr safe stains of KLK2 PCR products of genomic DNA from blood samples. Samples 1 to 5 were 578 bp products (exon 1 of five samples), samples 6 to 10 were 410 bp products (exons 2 of same five samples), and samples 11 to 15 were 664 bp products (exon 3+4 of same five samples). The letter L is on the 100bp DNA ladder lane. (C) Normalized relative quantity (NRQ) of KLK2 in FFPE prostate tissue materials. N-I stands for non-involved, PCa stands for prostate cancer and BPH for benign nodular hyperplasia. The NRQ of KLK2 was significantly reduced in PCa cases (P= 0.001). The horizontal lines (bars) represent median values (the distribution was Non-Gaussian). There were 96 cases of prostate cancer, 30 cases of BPH and 12 case of non-involved. The criteria for accepting valid real time PCR results for the KLK2, G6PD and ABL1 transcripts were: i) a slope range of -3.30 to -3.60 for the standard curve involving five dilution points (in triplicate at each point), ii) a Y- intercept of ≤ 40, iii) a quantification cycle (Cq) difference (Δ Cq) of ≤ 1.0 for a triplicate measurement, iv) a maximum relative fluorescence unit plateau value (RFU value) of ≤ 1 and v) a coefficient correlation (R2) of > 0.98 for the standard curve.
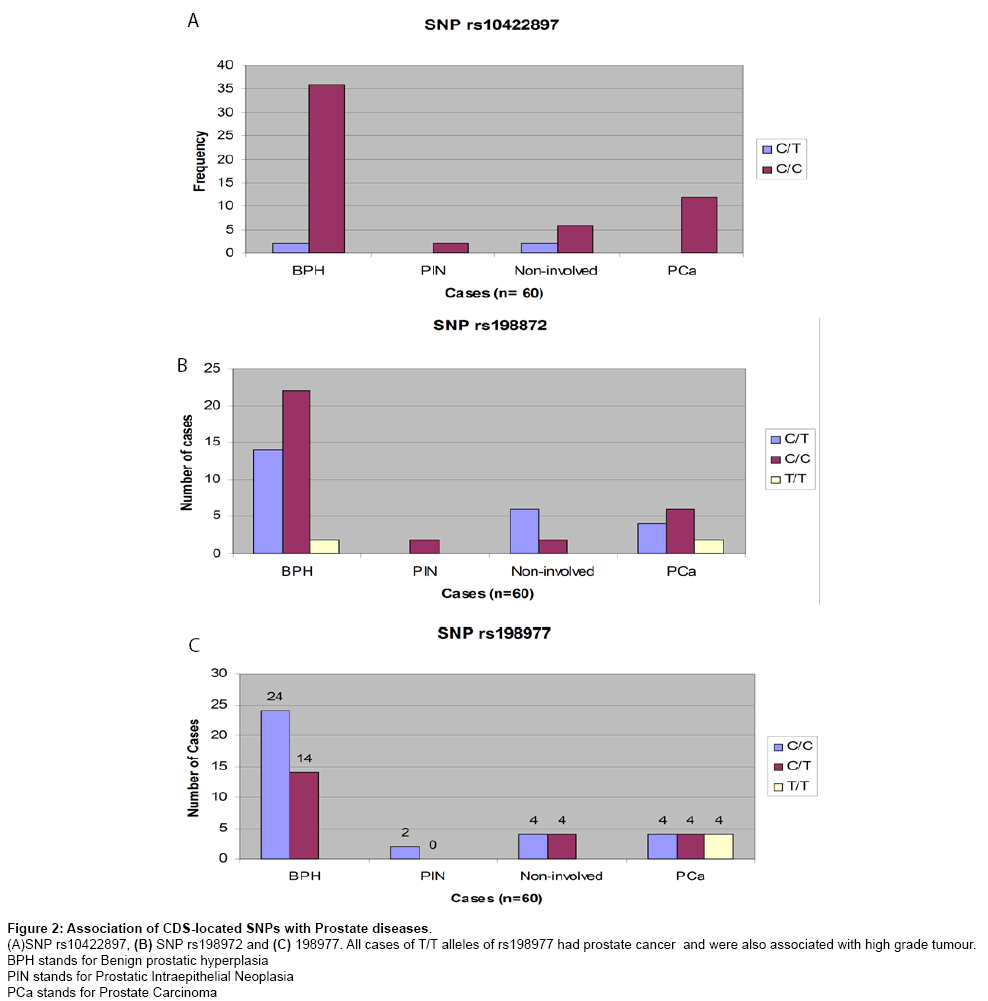
Table 2 contains a summary of all the 18 known SNPs identified in the KLK2 gene. Eleven SNPs (61%) of the 18 SNPs identified in the 60 patients’ cohort through direct DNA sequencing had 100% frequency of the reference (ancestral) allele in the study population, therefore were not investigated further. The remaining 7 SNPs had a heterozygosity of 0.07 to 0.40 in the sample population (n=60). Four of them were intronic SNPs; three were coding sequence (CDS) SNPs. The CDS located SNPs are shown in Figure 2. SNPs rs10422897 and rs198872 did not show any significant association with any prostate disease (P values=0.14 and 0.22 respectively, Chi Square test). The codons in both SNPs are for Arginine (R) and Leucine (L) respectively. However, the SNP rs198977 had a significant association with prostate cancer (P=0.0037, Chi Square test). The T/T allele of the rs198977 occurred only in prostate cancer cases. There were 12 prostate cancer cases in the 60 patients’ cohort (representing 20% cancer detection rate by biopsy in this cohort). Four (33%) of the 12 prostate cancer cases had T/T allele of rs198977. At the time of sampling their histology status was not known. The benign hyperplasia cases were 38 (about 63%) of the cohort. Prostatic intraepithelial neoplasia (PIN) was present in 2 cases (about 3% of the cohort) and there were 8 non-involved cases (cases of chronic inflammation, no dysplasia, no carcinoma; about 13% of the cohort).
| SNP ID | Molecular type | Genotypes and their frequency (n = 60) | Reference allele | ||
|---|---|---|---|---|---|
| rs62113073 | intronic | C/G (17%) | C/C (83%) | G/G (0%) | C |
| rs2664155 | intronic | A/G (37%) | G/G (60%) | A/A (3%) | G |
| rs2664156 | intronic | C/T (13%) | C/C (77%) | T/T (10%) | T |
| rs34652810 | 5'UTR | Del/G (0%) | Del/del (100%) | G/G (0%) | del |
| rs2070854 | intronic | A/G (0%) | A/A (100%) | G/G (0%) | A |
| rs198970 | intronic | A/T (0%) | A/A (100%) | T/T (0%) | A |
| rs6072 | CDS-misense | A/C/G (0%) | G/G (100%) | A/C/G (0%) | G |
| rs61750342 | exon | C/T (0%) | C/C (100%) | T/T (0%) | C |
| rs10422897 | CDS-misense | C/T (7%) | C/C (93%) | T/T (0%) | C |
| rs198972 | CDS | C/T (40%) | C/C (53%) | T/T (7%) | C |
| rs1064676 | CDS | C/T (0%) | C/C (100%) | T/T (0%) | C |
| rs6070 | intronic | A/T (40%) | T/T (60%) | A/A (0%) | T |
| rs6071 | intronic | A/G (0%) | A/A (100%) | G/G (0%) | A |
| rs1064703 | CDS | C/T (0%) | T/T (100%) | C/C (0%) | T |
| rs198977 | CDS | C/T (37%) | C/C (56%) | T/T (7%) | C |
| rs60268688 | CDS | A/C (0%) | A/A (100%) | C/C (0%) | A |
| rs1059712 | CDS | A/G (0%) | G/G (100%) | A/A (0%) | G |
| rs11549921 | 3'UTR | C/T (0%) | T/T(100%) | C/C (0%) | T |
Table 2: Frequency distribution of 18 identified SNPs in KLK2 gene.
Note: The SNPs highlighted in yellow showed 100% frequency of the ancestral allele and therefore were not investigated further. UTR stands for untranslated region; CDS stands for amino acid coding sequence. The reference allele is the same as ancestral allele.
Figure 2: Association of CDS-located SNPs with Prostate diseases.
(A)SNP rs10422897, (B) SNP rs198972 and (C) 198977. All cases of T/T alleles of rs198977 had prostate cancer and were also associated with high grade tumour.
BPH stands for Benign prostatic hyperplasia
PIN stands for Prostatic Intraepithelial Neoplasia
PCa stands for Prostate Carcinoma
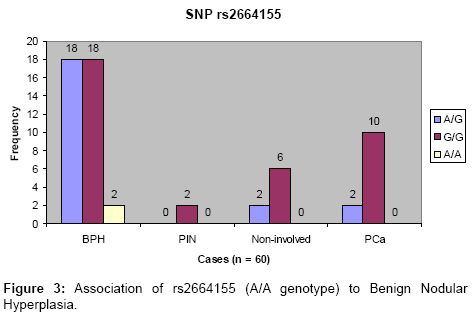
None of the four intronic SNPs had any significant association with prostate cancer (P>0.05, Chi Square test). However, the A/A allele of the SNP rs2664155 occurred only in benignhyperplasia patients (5.26% of the BPH cases) as shown in Figure 3. For SNP genotyping in the 138 archived FFPE prostate tissue sections, only two SNPs were genotyped by the TaqMan allelic discrimination assay (real time PCR system). However, there were 100% valid results when the TaqMan SNP genotyping was used on the 60 samples that had been previously evaluated by direct DNA sequencing. The SNP rs198972 determined by TaqMan assay did not show any significant association with any prostate disease (P=0.76, Chi Square test). The result was similar to previous result on the 60 samples evaluated by resequencing. For the rs198977 the T/T allele was also significantly associated with prostate cancer, all the T/T genotypes were found only in the prostate cancer tissues (4.2% of 96 prostate cancer tissues contained in the 138 archived samples). The average heterozygosity of the rs198977 for the sample population (British whites) in this study was 0.38.
Overall, there were 7 cases of T/T allele of rs198977 identified in this study using direct DNA resequencing and TaqMan SNP genotyping assay of 198 samples in total. Two of the seven cases had Gleason score 8-10; four cases had Gleason Score 6 and one case had Gleason score 7. There were no low grade cancers (Gleason score 4-5) identified with the T/T allele, indicating a possible association with high tumour grade.
Discussion
Of the 18 known SNPs identified in the KLK2 gene through direct DNA re-sequencing of 60 patients, only two SNPs showed a significant association with prostate diseases. The T/T allele of rs198977 was significantly associated with presence of prostate cancer at biopsy. The sample size in this study was relatively small. However, in many SNP association studies, categories of prostate diseases such BPH, PIN and chronic inflammation are usually all grouped into ‘no tumour category’ and are compared with ‘tumour group’. This practice could skew results because the detection rate of prostate cancer by histology in a pool of patients attending TRUS biopsy is at best 30% [9], and in this cohort the cancer detection rate was even lower (20%). There was a higher frequency of BPH in prostate lesions. Therefore, larger population studies incorporating these sub-groups are necessary to confirm the association of SNPs with prostate diseases. Contrary to the report by Nam et al., our study showed that the rs2664155 was associated with risk of benign hyperplasia rather than prostate cancer. In addition, our study showed that the occurrence of T/T allele of rs198977 was associated with high tumour grade. Larger studies are still required to confirm these associations. This study did not evaluate the association of these SNPs in haplotypes. In addition, genotyping SNPs from archived tissue samples could be problematic since possible somatic mutations were differentiated from germ line variations.
The normalized quantification of KLK2 showed that the transcript level was significantly reduced in prostate cancer cases compared to benign cases (P<0.05). Although the protein product of KLK2 is known to be increased in prostate cancer progression, our study, in agreement with some other studies [10,11], showed that the normalized mRNA level was reduced in prostate cancer. It is therefore unclear how the KLK2 gene is up-regulated during prostate cancer progression.
The TaqMan SNP genotyping is a cheap, flexibly high throughput and was found suitable for genotyping gDNA from archived paraffin tissue materials and blood samples. Both the rs198977 and rs2664155 could be determined in the same plate reaction for each patient sample using the TaqMan SNP assay. However, known allelic variants (calibrators for the three possible variants determined by re-sequencing) must always be included in each run for quality control purposes. Again the amount of gDNA used in the TaqMan real time PCR for genotyping is very important (our optimization assay showed that 50 to 100ng amount of gDNA was required for good quality base calling).
SNPs could provide additional genetic information to help stratify patients into risk groups. It might help identify males who will benefit from prostate cancer annual screening programmes, which are, in many countries, strongly debated [12]. Males who have the T/T allele of rs198977, for example, could be enrolled into annual screening programme before the age of 50 (the recommended age in some countries). SNPs could also inform prognosis, and may help in choice of treatment options. For example, if it is confirmed that the T/T allele of rs198977 was associated with higher tumour grade, this can influence patients with localised prostate cancers to choose a radical treatment option earlier on. Although the rs198977 SNP is located in the CDS exons of KLK2 gene, the resulting codon is a ‘coding synonymous’,which means there is no change in peptide sequence for whichever allele of this particular SNP the patient has. It is therefore unclear how allelic variation affects the function of the KLK2 gene product. The NRQ of the KLK2 gene also showed that it was down regulated in prostate cancer cases compared to benign cases.
Conclusion
Two SNPs in the KLK2 gene: rs198977 and rs2664155 were significantly associated with prostate cancer risk and benign hyperplasia respectively. The T/T allele of rs198977 occurred only in prostate cancer cases and was also associated with high tumour grade. The A/A allele of rs2664155 was, contrary to previous reports, associated with benign nodular hyperplasia. The normalized mRNA level of KLK2 was significantly reduced in prostate cancer tissues compared to benign cases. The TaqMan SNP genotyping assay provided a cheap and flexibly high throughput assay for clinical allelic discrimination in samples including gDNA from archived FFPE prostate tissue blocks and blood samples.
Acknowledgements
We are grateful to Dr Wazil Mohammed, Dr Harbinder Sharma, Mrs Gill Flack and Dr Margaret Wilkins of Bedford hospital, United Kingdom for their assistance in sample collection. We also thank Dr Linmarie Ludeman of the Royal Gloucestershire Hospital, Cheltenham for helping in the sample collection. We are greatly indebted to the Alan and Nesta Fergusson Trust for research grant to Dr E Nna.
References
- Marth GT, Korf I, Yandell MD, Yeh RT, Gu Z, et al. (1999) A general approach to single-nucleotide polymorphism discovery. Nat Genet 23: 452-456.
- Collins FS, Guyer MS, Charkravarti A (1997) Variations on a theme: cataloging human DNA sequence variation. Science 278: 1580-1581.
- International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437: 1299-1320.
- Nam RK, Zhang WW, Klotz LH, Trachtenberg J, Jewett MAS, et al. (2006) Variants of the hK2 protein gene (KLK2) are associated with serum Hk2 levels and predict the presence of prostate cancer at biopsy. Clin Cancer Res 12: 6452-6458.
- Helfand BT, Loeb S, Meeks JJ, Fought AJ, Kan D, et al. (2009) Pathological outcomes associated with the 17q prostate cancer risk variants. J Urol 181: 2502-2507.
- Zheng SL, Stevens VL, Wiklund F, Isaacs SD, Sun J, et al. (2009) Two independent prostate cancer risk-associated Loci at 11q13. Cancer Epidemiol Biomarkers Prev 18: 1815-1820.
- McCormack RT, Rittenhouse HG, Finlay JA, Sokoloff RL, Wang TJ, et al. (1995) Molecular forms of prostate-specific antigen and the human kallikrein gene family: a new era. Urology 45: 729-744.
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45.
- Welch HG, Fisher ES, Gottlieb DJ, Barry MJ (2007) Detection of prostate cancer via biopsy in the Medicare-SEER population during the PSA era. J Natl Cancer Inst 99: 1395-1400.
- Magklara A, Scorilas A, Stephan C, Kristiansen GO, Hauptmann S, et al. (2000) Decreased concentrations of prostate-specific antigen and human glandular kallikrein 2 in Malignant versus non-malignant prostatic tissue. Urology 56: 527-532.
- Pannek J, Rittenhouse HG, Evans CL, Finlay JA, Bruzek DJ, et al. (1997) Molecular forms of Prostate specific antigen and human kallikrein 2 (hk2) in Urine are not clinically useful for early detection and staging of prostate cancer. Urology 50: 715-721.
- World Health Organization (2004) should mass screening for prostate cancer be introduced at national level?
Relevant Topics
- Anal Cancer Diagnosis
- Biomarkers
- Bladder Cancer Diagnosis
- Blood Cancer Diagnosis
- Brain Cancer Diagnosis
- Breast Cancer Diagnosis
- Cancer Diagnosis
- Cervical Cancer Diagnosis
- Colon Cancer Diagnosis
- Diagnostic Algorithms
- Early Detection
- Esophageal Cancer Diagnosis
- Genetic Mutations
- Imaging Techniques
- Kidney Cancer Diagnosis
- Leukemia Diagnosis
- Liver Cancer Diagnosis
- Lung Cancer Diagnosis
- Lymphoma Diagnosis
- Mesothelioma Diagnosis
- Mouth Cancer Diagnosis
- Ovarian Cancer Diagnosis
- Pancreatic Cancer Diagnosis
- Prostate Cancer Diagnosis
- Rectal Cancer Diagnosis
- Skin Cancer Diagnosis
- Testicular Cancer Diagnosis
- Thyroid Cancer Diagnosis
- Uterine Cancer Diagnosis
Recommended Journals
- Journal of Lung Cancer Diagnosis & Treatment
- Cancer Prevention Journal
- Advances in Cancer Prevention
- Breast Cancer: Current Research
- Cancer Surgery Journal
- International Journal of Inflammation, Cancer and Integrative Therapy
- Cervical Cancer: Open Access
- Journal of Oncology Research and Treatment
- Journal of Prostate Cancer
Article Tools
Article Usage
- Total views: 12992
- [From(publication date):
December-2016 - Apr 03, 2025] - Breakdown by view type
- HTML page views : 12063
- PDF downloads : 929