Research Article Open Access
Advances in Understanding Cold Sensing and the Cold-Responsive Network in Rice
| Qi Zhang1, Nan Jiang1, Guo-Liang Wang1,2, Yahui Hong1 and Zhilong Wang1* | ||
| 1Hunan Provincial Key Laboratory of Crop Germplasm Innovation and Utilization, College of Biological Science and Technology, College of Agronomy, Hunan Agricultural University, Changsha, Hunan 410128, China | ||
| 2Department of Plant Pathology, Ohio State University, Columbus, Ohio 43210, USA | ||
| Corresponding Author : | Zhilong Wang Hunan Provincial Key Laboratory of Crop Germplasm Innovation and Utilization College of Biological Science and Technology College of Agronomy, Hunan Agricultural University Changsha, Hunan 410128, China E-mail: zhilongwang@126.com |
|
| Received January 31, 2013; Accepted March 06, 2013; Published March 16, 2013 | ||
| Citation: Zhang Q, Jiang N, Wang GL, Hong Y, Wang Z (2013) Advances in Understanding Cold Sensing and the Cold-Responsive Network in Rice. Adv Crop Sci Tech 1:104. doi:10.4172/2329-8863.1000104 | ||
| Copyright: © 2013 Qi, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | ||
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
||
Visit for more related articles at Advances in Crop Science and Technology
Abstract
Cold stress reduces the growth and production of many crops including rice (Oryza sativa L.), which is a staple food crop and model monocot plant. During the past two decades, significant progresses have been made in understanding the cold-responsive network in rice. Various genes and gene products functioning in cold sensing and transcriptional regulation and post-transcriptional processing of the response to cold have been identified and these include OsDREBs, OsNACs, OsMAPs, OsCDPKs, miR-171, and miR-444a. The important roles of calcium, Reactive Oxygen Species (ROS) and Abscisic Acid (ABA) in cold sensing and signaling during both vegetative and reproductive stages have also been revealed. The new findings summarized in this review will facilitate the breeding of cold tolerance in rice and will also be useful for studying and understanding cold sensing and signaling in other crops.
| Keywords | |
| Cold stress; Sensing; Responsive network; Rice | |
| Introduction | |
| Temperature severely affects plant growth and basically determines the geographical distribution of plants. The changes in mean annual temperatures predicted by climate change models will affect both soil organic matter turnover and cropping patterns in agriculture [1,2]. Rice is one of the most important stable crops globally and also a monocot model plant for molecular research. Like other plant species originating in tropical or sub-tropical areas, rice is sensitive to low temperatures with prolonged exposure resulting in chilling injury [3]. In tropical areas, especially in Indonesia and Malaysia, cold stress rarely occurs and is not the major problem for rice breeding in those areas. Nevertheless, rice is cultivated well beyond tropical environments to temperate and high altitude areas. In these areas, temperatures fluctuate greatly across seasons and rice is often exposed to temperatures below 20°C during the most sensitive stages of development (i.e., germination, seedling, and reproduction) [4]. Low temperature impairs seed germination, reduces seedling vigor, weakens photosynthetic ability by inducing leaf discoloration, reduces plant height, produces degenerated spikes, delays days to heading, reduces spikelet fertility, causes irregular grain maturity, and poor grain quality [5]. About 30.7 million ha of rice in China are grown over a wide area from 53°27’N to 18°90’N, and almost the entire area can be harmed by cold injury caused by low temperatures. Annual losses are 3-5 million tonnes. Low temperature during the reproductive stage of rice in the Republic of Korea caused 17, 78, and 20% damage to the total rice area in 1971, 1980, and 1993, respectively, with maximum yield loss of milled rice of 3.9 t/ha in 1980. Yield loss of 1-2 t/ha due to low temperature during the reproductive stage in 1995-96 was also reported in Australia and an annual yield loss of 3-5 million tons was recorded in China [5,6]. The mechanisms of cold stress in rice have been extensively investigated in the past two decades, and cold-responsive strategies that differ from those in other plant species such as Arabidopsis and tomato have been detected in rice. The identification of QTLs for cold stress shows that different loci are involved in cold tolerance at different growth stages in rice [7,8]. And these findings are useful to facilitate the selection and development of improved cold-tolerant genotypes with high percent seed set for their cultivation in temperate environments and high-altitude areas. According to common cold-responsive network in various plants, previous reviews provide useful information, but the review which specifically focuses on the studies in crops, especially in rice, is scarce. In this review, we summarize recent advances on cold sensing and transcriptional regulation in rice. In addition, we discuss intriguing findings on the role of post-transcriptional regulation in chilling and/ or freezing tolerance in rice. | |
| Cold sensing | |
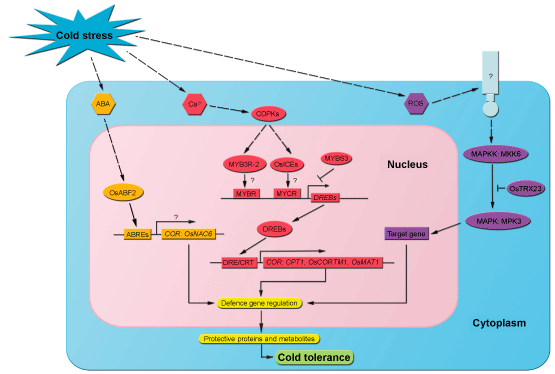
| Plant cells can sense cold stress based on changes in membrane rigidity, changes in the physical state of membrane proteins, and changes in the concentrations of metabolites. At low temperatures, an increase in electrolyte leakage (EL) commonly occurs in rice cultivars because of increased membrane rigidity [9-11]. Ca2+ influx into the cystol is an important and early event caused by cold stress, an influx which may be mediated by membrane rigidification-activated mechano-sensitive or ligand-activated Ca2+ channels [12]. Subsequently, the Ca2+ signatures are interpreted and amplified by calcium sensors, such as Calmodulin (CaM) and Calmodulin-like Proteins (CMLs), Calcineurin B-like Proteins (CBLs), and Calcium-Regulated Protein Kinases (CDPKs) (Figure 1). The CDPK gene OsCDPK7 is induced by chilling in rice, and overexpression of this gene enhances cold tolerance in transgenic rice [13]. Other calcium-regulated protein kinase genes such as OsCDPK13, OsCBF1, and OsNAC5 are also cold-induced and involved in the calcium signaling pathway [14-16]. | |
| As a cold-responsive signaling in cold-stressed plants, abscisic acid (ABA) accumulates and then initiates the cold-sensing period (Figure 1) [17]. An ABRE (ABA-responsive elements)-binding bZIP transcription factor OsABF2 plays a role in the cold response, and significantly increases at the transcriptional level [18]. In a rice mutant deficient in the ABA-responsive pathway ABI5-Like1 (abl1), over one-third of the genes are down-regulated in response to cold and other abiotic stresses, indicating that ABA may play a positive and complex role in the sensing of cold stress in rice [19]. | |
| Reactive Oxygen Species (ROS) accumulate in cells when plants are challenged with biotic or abiotic stresses. ROS, which include superoxide (O2-), hydrogen peroxide (H2O2), and the hydroxyl radical, play two roles under cold stress: they induce ROS scavengers and initiate protective mechanisms [20]. When plants are exposed to biotic and abiotic stimuli, an increase in ROS level can damage membranes, induce alterations of cellular ion conductance, and trigger mitogen-activated protein kinase (MAPK) cascades (Figure 1) [19]. Transgenic rice overexpressing OsAPXa (ascorbate peroxidase) exhibits an elevated cold tolerance that is negatively correlative with the levels of H2O2 and lipid peroxidation, which have been scavenged by increased ascorbate peroxidase activity during cold treatment [21]. Moreover, ROS regulate the OsMKK6 (MAPK kinase)-OsMPK3 (MAPK) pathway through redox control of OsTRX23 [22]. Interestingly, an analysis of the transcriptional regulatory network in japonica rice subjected to cold treatment revealed that oxidative-mediated clusters (several H2O2-induced gene such as bZIsP, ERF, and MYB genes) are activated earlier than ABA-mediated clusters (factors acting on ABRE-like enriched clusters), and that the former may play a more important role in early cold sensing [8,23-25]. | |
| CDPKs pathway | |
| Calcium-Dependent Protein Kinases (CDPKs) belong to a family of Ser/Thr protein kinases that were first discovered in plants. By playing a central role in calcium-dependent pathways and by controlling the calcium content in the cell, CDPKs have been thought to coordinate the sensing of and responses to abiotic stress [26,27]. Expression by all 29 CDPK (CPK) genes in rice has been evaluated under various abiotic stresses. Nine CDPK genes (OsCPK7, OsCPK13, OsCPK15, OsCPK17, OsCPK20, OsCPK21, OsCPK23, OsCPK24, and OsCPK29) are induced by chilling temperatures in rice seedlings [28]. Moreover, OsCDPK7 and OsCDPK13 overexpression transgenic lines exhibit higher recovery rates after cold stress than the vector control. An in situ hybridization localized the expression of RAB16A, one of the proposed target genes regulated by the OsCDPK7 signaling pathway. CDPKs have also been identified as downstream components in Gibberellin (GA) and Brassino Steroids (BRs) signaling pathways [29-31]. With transgenic analysis in rice, the expression of OsCDPK13 is up-regulated in response to both GA3 and cold treatment, suggesting that OsCDPK13 may mediate the GA-induced signaling and the Ca2+ influx [32]. A reduction in GA content of cold-treated Arabidopsis, modulated through the stimulation of expression of GA-inactivating GA 2-oxidase (GA2ox) genes is associated with the accumulation of a green fluorescent protein-tagged DELLA protein [33]. Because CBF1 acts downstream in the CDPK pathway, a question arises about whether there is a feedback regulation between CBFs, GA, and CDPKs that explains their changes in response to cold stress. Accordingly, further research in the function of the rice CDPK genes would be useful for increasing our understanding of signal transduction pathways in cold stress responses. | |
| OsMAPKs | |
| The MAPK pathway is a well-characterized signal transduction cascade system in animals, yeast, and plants. The MAPK cascade consists of three subsequently interactive phosphorylation kinases: MAP kinase kinase kinase (MAPKKK), MAP kinase kinase (MAPKK), and MAPK [34,35]. MAPK, the last component of the phosphorylation cascade, is activated by MAPKK, which is in turn activated by MAPKKK. The MAPK cascade regulates cell division, development, and differentiation in response to stress stimuli in animals and yeast [36,37]. In recent years, a series of genes encoding MAPKs, MAPKKs, and MAPKKKs have been identified from different plant species. An increasing body of evidence has shown that MAPKs play important roles in signal transduction in response to drought, ROS, pathogen attack, wounding, and low temperature in plants [38-42]. | |
| Of the 20 MAPK genes in Arabidopsis, AtMPK3 and AtMPK6, have been implicated in the tolerance to multiple abiotic and biotic stresses [39,43]. Nine MAPK genes have been identified from rice, and each MAPK encodes a distinct protein kinase that plays a role in mediating abiotic tolerance [44]. BWMK1, which was the first MAPK gene found in rice and which was detected in the indica-type cultivar IR36, is induced by infection by the blast pathogen (Magnaporthe oryzae) and by mechanical wounding [45]. Recently, a time-based transcriptional regulation mechanism controlled by OsTRX23 has been reported for OsMPK3/OsMAP1 and OsMPK6/OsSIPK which are strongly induced by low temperature treatment (12°C) at the transcriptional level [22]. In the first 24 h of cold treatment, transcripts of OsMPK3 and OsMPK6 rapidly increase, and then, with an increasing transcription of OsTRX23, they sequentially decrease, suggesting that the MAPK pathway could be a rapid stress-responsive element associated with the transient accumulation of H2O2 during chilling stress. These results also indicate that some unknown late-responsive element mediated by the inhibition of OsMPK3 and OsMPK6 by OsTRX23 leads to cold tolerance in rice. Recently, a yeast two-hybrid screen designed to identify partner MAPKs for OsMKK6 revealed specific interactions of OsMKK6 with OsMPK3 and OsMPK6. A constitutively active form of OsMKK6 and OsMKK6DD, showed elevated phosphorylation activity against OsMPK3 and OsMPK6 in vitro. OsMPK3, but not OsMPK6, was constitutively activated in transgenic plants overexpressing OsMKK6DD, indicating that OsMPK3 is an in vivo target of OsMKK6. Chilling tolerance was enhanced in transgenic plants overexpressing OsMKK6DD [46]. Taken together, these data suggest that the MAPK signaling cascade is one of the low-temperature signaling pathways in rice and positively regulates cold stress tolerance. | |
| The transcriptional-regulatory network associated with cold response | |
| The transcriptional regulatory network involved in low temperature response prolongs the expression of cold acclimation genes in rice. Of the 2,604 genes that are upregulated in response to chilling in japonica rice, about 6% (148) have been estimated to be Transcription Factors (TFs) based on the classification of the Database of Rice Transcription Factors. These putative TFs include members of AP2/ERF, bZIP, MYB, WRKY, bHLH, and NAC families, and are characterized by waves of induction at different time periods. For example, the ‘early rapid response’ group (phase-1) is induced during the initial 6 h of cold treatment, and the ‘early slow response’ group (phase-2) is induced between 6 and 24 h. A few TFs exhibited ‘late response’ profiles (phase-3) with no significant induction until after 24 h. Most TFs that are upregulated by chilling are activated at phase-1 and phase-2, indicating that early responses are critical for cold stress tolerance and that late responses may be involved in unknown mechanisms that protect rice growth after 24 h of exposure to low temperatures [10]. In the last two decades, researchers have made significant progress in clarifying the function of key TFs responding to cold stress, as discussed in the next section. | |
| DREB-CRT/DRE pathway | |
| DREBs (dehydration-responsive element-binding proteins, also known as CBFs for C-repeat binding factors) have been the most intensively studied of the TFs involved in plant responses to cold. In rice, seven members in the DREB family have been studied in depth: OsDREB1A/CBF3, OsDREB1B/CBF1, OsDREB1C/CBF2, OsDREB1D, OsDREB1F, OsDREB2A, and OsDREB2B [47]. Overexpression of DREBs in transgenic rice significantly increases cold tolerance; relative to wild-type plants, the transgenic plants exhibit increased survival and growth, and changes in ROS scavenging, membrane transport, hormone metabolism, proline concentration, and accumulation of sugars such as raffinose, sucrose, glucose, and fructose [14,48]. | |
| DREBs belong to the APETALA2/Ethylene response factor (AP2/EREBP) family of TFs. These TFs can bind to GCC-box and C-repeat/dehydration-responsive elements (CRT/DRE) to promote the Cold responsive (COR) gene. To analyze the DNA-binding specificity of OsDREBs, researchers have selected OsDREB1A as a typical DREB1-type gene and OsDREB2A as a typical DREB2-type gene. The ability of the OsDREB1A and OsDREB2A fusion proteins to bind the wild-type or mutated DRE sequences in the rd29A promoter has been identified by the gel mobility shift assay. Furthermore, the transactivating DRE-dependent transcription capability of OsDREB1A and OsDREB2A in rice cells has been determined by transactivation experiments. Protoplasts are co-transfected with a β-glucuronidase (GUS) reporter gene fused to the dimeric 75-bp fragments containing the DRE motif and the effector plasmid. The GUS activity is distinctly upregulated in the presence of OsDREB1A and OsDREB2A relative to the vector control, indicating that the DREB1A and DREB2A proteins act as transcription activators in rice protoplasts [49]. | |
| In Arabidopsis, ICE1 (Inducer of CBF expression1), a MYC-type basic helix-loop-helix transcription factor, is a common TF that promotes the expression of DREB1A. ICE1 binds to MYC recognition (MYCR) elements in the DREB1A promoter region and is important for the expression of DREB1A during cold acclimation [12]. So far, there is no evidence that any ICE can function as a transcriptional factor for DREBs in rice. The Basic Helix-loop-Helix (bHLH) protein gene OrbHLH001, encoding an ICE1-like protein containing multiple homopeptide repeats, has been characterized and isolated from Dongxiang Wild Rice (Oryza rufipogon). Overexpression of OrbHLH001 enhances the tolerance to freezing stress in transgenic Arabidopsis. Examination of the expression of cold-responsive genes in transgenic Arabidopsis shows that the effect of OrbHLH001 in cold response differs from that of ICE1 and is independent of the CBF/ DREB1 cold-response pathway [50]. Recently, two ICE homologs in rice, OsICE1 and OsICE2, have been identified by BLAST searches with the nucleotide sequence of Arabidopsis ICE1 in the Rice Annotation Project Data Base [51]. Unlike the increased expression of Arabidopsis ICE1 under cold stress, the expression of OsICE1 and OsICE2 remains constant under cold stress as indicated by semi-quantitative RT-PCR; immunoblot with the anti-ICE specific antibody showed, however, that the levels of OsICE1 and OsICE2 proteins are upregulated by both cold and salt stresses. Although the expression of OsDREB1B increases under cold treatment, no evidence of a direct interaction between OsDREB1B and OsICE1/OsICE2 has been provided, There is little information available concerning the role of the MYC-type transcription factor in the DREB pathway but many genes have been found that upregulate the expression of DREB in rice [10,52,53]. OsMYB3R-2, a nuclear-localized R1R2R3 MYB TF, plays a role in cold response in rice [54]. Arabidopsis transgenic plants overexpressing OsMYB3R-2 show increased tolerance to cold, drought, and salt stresses, and the expression of some cold-related genes such as DREB2A and CBF1/2/3 is increased in the OsMYB3R-2-overexpression plants. These results suggest that OsMYB3R-2 acts as a master switch in cold tolerance. A more recent study has shown that overexpression of OsMYB3R-2 in transgenic rice leads to higher transcript levels of several G2/M phase-specific genes and OsCPT1, which are putatively targeted by the DREB genes [53]. The role of R2R3-type MYB TF (OsMYB2), which is localized in the nucleus and which has transactivation activity, in tolerance to cold stress has been functionally characterized by generating overexpression and RNAi transgenic rice plants of OsMYB2. In the OsMYB2-overexpression plants, the expression of OsDREB2A is upregulated compared with the wild type under low temperature treatment, suggesting that OsMYB2 is an upstream regulon for OsDREB2A [55]. | |
| A negative regulator of the CBF regulon, MYBS3, operates via a distinct pathway to help rice plants tolerate cold stress. MYBS3 is a single DNA-binding repeat MYB TF. By using genotypes that overexpress or underexpress MYBS3 to identify genes in the MYBS3-mediated cold signaling pathway, Su et al. [56] determined that MYBS3 responds slowly to cold stress and enables rice seedling to tolerate 4°C for at least 1 week. Surprisingly, MYBS3 represses the well-known DREB1/CBF fast-acting and short-term signaling pathway at the transcriptional level in rice [56]. By repressing the expression of the DREB1 regulon, the slow acting MYBS3 may prevent unnecessary expression of DREB1. | |
| NAC TFs | |
| NAC proteins represent one of the largest families of TFs. There are at least 105 putative NAC TFs in Arabidopsis, 140 in rice, 205 in soybean, and 152 in tobacco (Nicotiana tabacum) [10]. NAC TFs are key regulators of stress sensation and developmental programs, and contain an N-terminal NAC domain. Although the NAC TF family is widely distributed in plants, it has not been found in other eukaryotes [57,58]. | |
| Analysis of the NAC domain has been conducted by database searches with a comprehensive analysis of NAC family genes both in rice and Arabidopsis [59]. Five subdomains, each typically ~50 amino acids long, have been identified in the DNA-binding domain of a typical NAC protein. A comprehensive in silico analysis of the NAC TF family in rice recently identified 36 putative motifs within the NAC family. Based on the pattern of these motifs, the NAC family of rice can be classified into 15 types (types A-O). Most rice NAC proteins (97 of 140) contain a complete NAC DNA-binding domain with five major subdomains, which are classified into types A-E [60,61]. The sequences in the 1.5 kb promoter region of OsNAC6 include ABREs, MYBRS, MYCRS, W-boxes, GCC boxes, and as-1 motifs [24]. ABREs, MYBRS, and MYCRS play important roles in ABA signaling and abiotic stress response. The W-boxes and GCC boxes are recognition sites for WRKY and ERF transcription factors, respectively, and are involved in the regulation of various plant-specific physiological processes such as pathogen defense and senescence. The as-1 motifs are known as oxidative stress-responsive elements. These sequences may function as abiotic and/or biotic stress-responsive cis-acting elements in the OsNAC6 promoter, and they may sense cold stress from the upstream transcription factors induced by ABA, ROS, and other metabolite changes. | |
| Regarding the OsNAC5-dependent tolerance to abiotic stress in rice, research has demonstrated that the overexpression line is more tolerant of cold stress and that the OsNAC5 RNAi line is less tolerant of cold stress than the wild type [16,62]. Moreover, knockdown and overexpression of OsNAC5 enhances and reduces, respectively, the accumulation of malondialdehyde and H2O2, suggesting that knockdown of OsNAC5 renders RNAi plants more sensitive to oxidative damage. In addition, the sensitivity of seed germination to ABA is increased in overexpression plants and decreased in RNAi plants. These results indicate that OsNAC5 is important for cold tolerance in rice and that the effect of OsNAC5 is associated with an ABA-related transcriptional regulation network. | |
| However, the target genes of NAC TFs in rice remain largely unknown. The rice NAC gene SNAC1 (stress-responsive NAC 1) is predominantly induced in guard cells by drought and encodes NAC TF with transactivation activity [63]. SNAC1-overexpression plants show a greater sensitivity to ABA and an increased rate of stomatal closure that reduces water loss. In the field, the drought tolerance of transgenic plants is significantly greater than that of wild-type plants at anthesis, and the increased drought tolerance is not associated with other phenotypic changes or with yield reduction. The distinctive binding character of SNAC1 suggests that a special NAC binding site exists in rice. | |
| Other TFs | |
| In addition to the well-characterized rice TF genes in the cold responsive pathway that were described in the previous sections, many other genes also display changes in transcriptional expression upon low temperature treatment and are involved in the cold-sensing cascade. | |
| Proteins with the A20/AN1 zinc-finger domain are present in all eukaryotes and are well characterized as common elements in the stress response of animals and plants [64]. The stress-associated proteins (SAPs) contain the AN1 domain, which has a dimetal (zinc)-bound alpha/beta fold and often combines with A20 zinc finger domains (SAP8) or C2H2 domains (SAP16) including ascidian posterior end mark 6 (PEM-6) protein and human AWP1 protein (associated with PRK1) [65,66]. The human AWP1 protein is expressed during early embryogenesis, and mutations in SMUBP-2 (human immunoglobulin mu binding protein 2) cause muscular atrophy with respiratory distress type 1 [67]. OsiSAP8 is an early cold-responsive gene in rice, and the OsiSAP8 protein fused to GFP is specifically localized in the cytoplasm, indicating that, unlike many zinc-fingers containing proteins, OsiSAP8 is a cytoplasmic protein. Furthermore, the A20 and AN1 type zinc-finger domains of OsiSAP8 interact with each other in yeast two-hybrid assays, and OsiSAP8 overexpression transgenic rice plants grow better than wild-type plants under cold stress [68]. According to a microarray analysis of the rice A20/AN1-type zinc finger genes, four genes (ZFP177, ZFP181, ZFP176, ZFP173), two genes (ZFP181 and ZFP176), and one gene (ZFP157) are significantly induced by cold, drought, and H2O2 treatments, respectively [69]. The promoter region of ZFP177, which is a specific cold-induced A20/AN1-type zinc finger gene, contains no stress-associated cis-acting elements other than HSE, suggesting that the heat response of ZFP177 might be regulated by heat shock factors but that the cold response is not directly mediated by CBF/DREB transcription factors. | |
| The TFIIIA-type zinc finger protein was first detected in Xenopus oocytes [70] and contains at least one TFIIIA-type zinc finger motif with the consensus of CX2–4CX3FX5LX2HX3-5H. A number of TFIIIA-type zinc finger proteins are related to stress response [71]; for example, Arabidopsis STZ/ZAT10 and ZAT7 are involved in salt tolerance [72-74], soybean SCOF-1 is involved in cold tolerance [52], and Arabidopsis ZAT12 is involved in cold and oxidative stress [75]. The first TFIIIA-type zinc finger protein identified in rice, ZFP245, is involved in cold and drought tolerance [9]. Overexpression of ZFP245 in rice leads to increased cold tolerance and increased sensitivity to exogenous ABA treatment, suggesting that ZFP245 may play a role in the ABA signal transduction pathway during stress responses. Another TFIIIA-type zinc finger protein, ZFP182, helps protect cell membrane integrity under cold stress (as indicated by reduced electrolyte leakage) and is associated with the DREB pathway [76]. | |
| Recent research has also demonstrated the involvement of many other genes in the cold signal pathway, including bHLH1, OsP5CS2, LIP19, OSPGYR, ZFP245, OVP1, bZIP52/RISBZ5, TEF1 and SlCZFP1. The CBF-independent gene OrbHLH001 from Dongxiang wild rice can enhance tolerance to low temperature, suggesting that wild rice may have a distinct stress responsive pathway that enables a perennial life history [9,23,50,77-82]. | |
| Post-transcriptional Regulation | |
| mRNA processing | |
| Pre-mRNA processing is an important mechanism of post-transcriptional regulation of gene expression in eukaryotes. For conversion into mature mRNA, pre-mRNA requires various modifications such as the addition of a 5’ methyl cap and poly (A) tail and intron splicing. Precursor mRNA with more than one intron can undergo alternative splicing to produce functionally different proteins from a single gene. In plants, about 20% of genes require alternative splicing [83]. Alternative splicing affects many important processes and characteristics in plants including photosynthesis, flowering, grain quality in cereals, and stress responses. For example, the Arabidopsis COR15A gene, which encodes a chloroplast stromal protein with cryoprotective activity, plays an important role in conferring freezing tolerance to chloroplasts; the Arabidopsis stabilized1 (sta1) mutant is defective in the splicing of the cold-induced COR15A pre-mRNA and is hypersensitive to chilling, ABA, and salt stress [84,85]. STA1 encodes a nuclear pre-mRNA splicing factor and is upregulated by cold stress. STA1 catalyzes the splicing of COR15A, which is necessary for cold tolerance. Furthermore, pre-mRNAs of serine/arginine-rich (SR) proteins, which are involved in the regulation or execution of mRNA splicing, also undergo alternative splicing under cold and heat stresses in Arabidopsis. | |
| In rice, recent studies on the mRNA processing of OsDREB2B, a member of the DREB-CRT/DRE pathway, have revealed an efficient mRNA processing strategy for cold acclimation [86,87]. To recognize the two types of OsDREB2B transcripts, the authors of these papers designed two primers that are specific for each transcript. In the non-stress plants, OsDREB2B1 is approximately five times more abundant than OsDREB2B2. However, in the cold stress plants, OsDREB2B2 accumulates to a level equal to or greater than that of OsDREB2B1. To determine whether OsDREB2B2 is modified from OsDREB2B1 or is spliced alternatively from the OsDREB2B gene, the authors introduced OsDREB2B1 or OsDREB2B2 fused to the synthetic green fluorescent protein (sGFP) genes into two kinds of transgenic rice plants. RT-PCR analysis did not detect sGFP-OsDREB2B2 in the Ubi:sGFP:OsDREB2B1 plants but did detect the accumulated endogenous OsDREB2B2 transcript. This result indicated that OsDREB2B2 is produced directly from the OsDREB2B2 gene and is not modified from OsDREB2B1. In addition, a transcriptional analysis showed that transcription is greater for OsDREB2B2 than for OsDREB2A under stress conditions. These data suggest that mRNA processing is an important regulatory strategy for OsDREB2B expression as part of the abiotic stress response in rice. | |
| Small RNAs | |
| Small RNAs (sRNAs) are sequence-specific regulatory elements that mediate endogenous gene silencing in eukaryotes. Plant sRNAs have been divided into four classes based on their origins and structures: microRNAs (miRNAs) and three types of small interfering RNAs (siRNAs), including trans-acting siRNAs (ta-siRNAs), natural cis-antisense transcripts-derived siRNAs (nat-siRNAs), and repeat-associated siRNAs (ra-siRNAs) [88-90]. One major difference between miRNAs and siRNAs is that miRNAs result from the processing of a single-stranded hairpin precursor while siRNAs are generated from long double-stranded RNAs (dsRNAs) [91]. Plant miRNAs, a class of short non-coding RNAs (~22 nucleotides), are processed from primary miRNA transcripts through two sequential cleavages by Dicer-like1 (DCL1), and mature miRNAs are loaded into argonaute proteins to guide cleavage of target mRNAs or translational repression [92]. The biogenesis of ta-siRNAs is initiated by miRNA-mediated cleavage of non-coding transcripts. The cleaved RNAs are copied into dsRNAs by RNA-dependent RNA polymerase 6 (RDR6) and are processed by DCL4 into phased siRNAs from the end defined by miRNA-mediated cleavage. The production of ra-siRNAs requires the activity of DCL3, RDR2, and polymerase (Pol) IV, a plant-specific DNA-dependent RNA polymerase [93,94]. | |
| The rice genome contains approximately 250,000 transposable elements (TEs), constituting approximately 35% of the genome sequence [95,96]. TEs can lead to the folding of RNA sequences into hairpin structures (reminiscent of the pre-miRNAs), and many miRNAs derived from TEs have been found in rice [97]. Global expression profiling of rice miRNAs showed that 18 miRNAs respond rapidly to cold stress. Interestingly, most of these miRNAs are down-regulated, indicating that the expression of target genes controlled by these miRNAs is induced in an adaptive response to cold stress [96]. | |
| miRNAs also affect plant hormones, which regulate many important aspects of growth and development as well as responses to environmental stresses. miR-167 helps regulate the auxin signal by cleaving two auxin-response factors and might play a role in cold tolerance by affecting auxin-signaling pathways [96]. miR-171 is a large and conserved miRNA family. A microarray-based analysis has revealed that 6 h of cold stress upregulated miR-171a in Arabidopsis but down-regulated miR-171a in rice [98]. These results demonstrate that miRNAs belonging to the same family can display opposite patterns in response to cold stress, suggesting that they may perform different functions. The results also suggest that differences in their expression in rice vs. Arabidopsis may represent species-specific differences in the response to cold stress. Generally, miRNA accumulation negatively correlates with the level of target transcripts. Two MADS-box genes, MADS 57 and MADS 27, are targets of miR-444a [96,99]. How miR-444a differentially regulates the two MADS genes under cold stress warrants further investigation. | |
| Conclusion | |
| Intensive research has greatly increased our understanding of cold sensing, transcriptional networks, post-transcriptional regulation, and other responses of rice to cold stress. Rice plants use cold-induced calcium influx and changes in levels of ABA and ROS as signals to activate a response to cold stress. The DREB-CRT/DRE pathway has an important effect on the rice response to cold, and all DREB1 and DREB2 family members in the pathway have been identified and characterized by transgenic analysis in rice. The MAP kinase pathway and ABF/AREB pathway, whose effects have been well characterized in responses to other biotic and abiotic stress such as pathogen attack, heat, hyperosmotic stress, and oxidative stress, are involved in the cascade of cold stress responses in rice. Recent microarray analysis has revealed that small RNAs are important for cold responses. The recent research on cold acclimation also indicates that these cold-relevant genes can be used for molecular-assisted selection and transgenic research [100]. | |
| However, development of breeding techniques for rice shows that breeding of cold tolerant rice is still a challenge. Although many QTLs in cold tolerant rice varieties are identified, those QTLs are mostly stage specific and only effect germination stage, vegetative stage, or reproductive stage. Concerning current breeding techniques, varieties which tolerate to cold stress in all stages are hard to breed for the reason that the convergence of those QTLs into one rice variety is time and labor consuming. It is important, therefore, to develop alternative strategies for the breeding of crops and to discover more cold tolerant varieties and put them into breeding practices, including those wild rice varieties which live in high altitude areas and have an inborn tolerance under cold stress [101]. As the cold-responsive network is complex, characterizations of key factors and components among different cold signal transduction pathways are also useful for the breeding of cold tolerance in rice. | |
| Although the number of cold-responsive genes found continues to increase, the biological complexity underlying cold acclimation has not decreased. An increased understanding of the molecular basis of cold tolerance in rice should facilitate the development of varieties of rice and of other crops that are tolerant to cold stress. | |
| Acknowledgements | |
| This work was supported, in part, by grants from the National Transgenic Project (2012ZX08009001), the National 973 Project (2012CB723000), and the National Natural Science Foundation of China (31071674). Z. W. and G. L. W. were also supported by Program for Innovative Research Team in University (IRT1239), the Aid Program for Science and Technology Innovative Research Team in Higher Educational Institutions of Hunan Province, the Hunan Provincial Key Laboratory of Crop Germplasm Innovation and Utilization (11KFXM01) and Hunan Agricultural University (11YJ13). | |
| References | |
References
|
|
Figures at a glance
 |
| Figure 1 |
Relevant Topics
- Agricultural science
- Agronomy
- Climate impact on crops
- Crop Productivity
- Crop Sciences
- Crop Technology
- Field Crops Research
- Hybrid Seed Technology
- Irrigation Technology
- Organic Cover Crops
- Organic Crops
- Pest Management
- Plant Genetics
- Plant Breeding
- Plant Nutrition
- Seed Production
- Seed Science and Technology
- Soil Fertility
- Weed Control
Recommended Journals
Article Tools
Article Usage
- Total views: 15146
- [From(publication date):
May-2013 - Apr 02, 2025] - Breakdown by view type
- HTML page views : 10471
- PDF downloads : 4675
