Research Article Open Access
Abnormal Protein Profiles in Hippocampus of Mouse Models of Down Syndrome: Similarities with Alzheimer's Disease
Aaron Block1, A. Ranjitha Dhanasekaran1, Md. Mahiuddin Ahmed1 and Katheleen J Gardiner1,2*
1 Department of Pediatrics, Linda Crnic Institute for Down Syndrome, University of Colorado Denver, Colorado, USA
2 Department of Biochemistry and Molecular Genetics, Human Medical Genetics and Genomics, and Neuroscience Programs, University of Colorado Denver, Colorado, USA
- Corresponding Author:
- Katheleen Gardiner
Department of Pediatrics
Linda Crnic Institute for Down Syndrome
Human Medical Genetics and Genomics
Neuroscience Programs, University of Colorado Denver
Mail Stop 8608, 12700 E 19th Avenue
Aurora, Colorado 80045, USA
Tel: 303-724-0572
Fax: 303-720-5741
E-mail: katheleen.gardiner@ucdenver.edu
Received date: December 20, 2013; Accepted date: January 22, 2014; Published date: February 03, 2014
Citation: Block A, Dhanasekaran AR, Ahmed MM, Gardiner KJ (2014) Abnormal Protein Profiles in Hippocampus of Mouse Models of Down Syndrome: Similarities with Alzheimer’s Disease. J Alzheimers Dis Parkinsonism 4:138. doi:10.4172/2161-0460.1000138
Copyright: © 2014 Block A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Alzheimers Disease & Parkinsonism
Abstract
Down syndrome (DS) is caused by an extra copy of the long arm of human chromosome 21 (HSA21) and the increased expression, due to dosage, of HSA21 encoded genes. In addition to intellectual disability, all individuals with DS develop the neuropathology of Alzheimer’s Disease (AD) by age 30-40. The amyloid precursor protein gene, APP, that is mutated or duplicated in some familial AD (FAD), is encoded by HSA21, over expressed in DS, and a candidate for causing AD in DS. However, only half of those with DS will develop the AD-like dementia by age 50-60, suggesting that additional HSA21 genes may modulate the effects of APP triplication, and/or protect the DS brain from early onset progression to dementia in spite of neuropathology. In sporadic AD and mouse models of FAD, abnormal levels of a diverse set of proteins, including receptors, scaffold proteins, kinases, phosphatases and cytokines, have been documented, but nothing is known about their possible roles in AD in DS. Here, we compare expression of 26 AD-related proteins in hippocampus of four mouse models of DS, the Ts65Dn, Tc1, Dp (10)1Yey and Dp (17)1Yey, that together provided trisomy of partially overlapping subsets of all HSA21 genes or mouse orthologs. In the Dp(10)1Yey, that is trisomic for HSA21 orthologs mapping to mouse chromosome 10, twelve of 26 AD-related proteins were elevated, while in the Tc1, Dp(17)1Yey and the popular Ts65Dn, six, four and two differed from littermate controls. These data suggest that genes mapping to the HSA21 orthologous regions of mouse chromosomes 10 and 17 contribute to protein perturbations in the DS brain, and possibly AD in DS. Considering the different phenotypic features of the four DS mouse models further suggests that some protein abnormalities may be compensatory and protective for brain function and/or that learning and memory deficits may be age-dependent.
Keywords
Alzheimer’s disease; Down syndrome; Trisomy; Agedependent deficits; Mouse chromosome 10
Introduction
Familial Alzheimer’s Disease (FAD) is rare, accounting for fewer than 5% of all cases of AD, and is characterized by early onset, at <60 years of age. Mutations causing FAD have been identified in three genes, the amyloid precursor protein, APP, and the presenilin genes 1 and 2, PSEN1 and PSEN2 [1,2]. Duplications of the genomic region containing the APP gene have also been identified in FAD [3,4]. Sporadic AD (sAD) typically has a later age of onset and is common, estimated to affect as many as 45% of people by the age of 85. The genetic causes of sAD are not known, but are assumed to be complex, involving multiple genes. An allelic variant in the APOE 4 gene is well established as a risk for sAD, but variants affecting susceptibility for AD in many other genes have also been identified [1]. Most recently, incompletely penetrant mutations in the ADAM10 gene have been identified in some late onset families with AD [5].
Down syndrome (DS), trisomy of human chromosome 21 (HSA21), is caused by an extra copy of all or part of the long arm of HSA21 and the increased expression, due to dosage, of some subset of HSA21 encoded genes. In addition to intellectual disability, all individuals with DS develop a neuropathology by the age of 30-40 similar to that seen in AD and approximately 50% will eventually develop an AD-like dementia by the age of 50-60 [6,7]. Because the incidence of DS is approximately one in 750-1000 live births worldwide, with the population in the US alone estimated at approximately 300,000, AD in DS is a significant societal and medical issue [8]. The genetic cause of AD in DS may be a combination of genetic causes similar to those in FAD and sAD. The presence of the APP gene on human chromosome 21 (HSA21), and its consequent triplication and over expression in DS, is a clear candidate, similar to FAD. However, the fact that not all individuals with DS develop an early onset AD-like dementia, in contrast to FAD due to APP duplication [9], suggests that additional genes may modulate the effects of APP triplication, and/or protect the DS brain from early progression to dementia in spite of neuropathology.
FAD, sAD and AD in DS display common features of abnormal processing of the APP protein to neurotoxic Aβ peptides and their accumulation in neuritic plaques; hyperphosphorylation of the microtubule associated protein, Tau, and the formation of neuritic fibrils containing Tau; hyperactivation of glutamate receptors; and neuroinflammation [10-12]. To understand the potential causes of AD in DS at the molecular level, it is helpful to consider functional information regarding the genes of HSA21 and the protein abnormalities observed in brains of patients with AD and mouse models of AD.
HSA21 encodes 161 classical protein coding genes of diverse functions, plus approximately 45 genes encoding keratin associated proteins (some proportion of which may be pseudogenes), five microRNA genes, and more than 300 genes/gene structures of completely unknown functions [13]. Several HSA21 protein coding genes have been shown to affect AD-related cellular and molecular features. S100B, a calcium-binding protein involved in astrocytosis, is over expressed in DS and also AD brains [14]. Over expressing S100B in a mouse model of AD (Tg2576, expressing the Swedish FAD APP double mutation) increases Aβ production and deposition and induces premature expression of pro-inflammatory cytokines [15]. In contrast, over expression in cultured cells of the HSA21 small ubiquitin-like protein modifier, SUMO3, decreases Aβ production; patterns of SUMO2/3 protein modification are altered in brains of AD [16] and of the Tg2576 AD mouse model [17]. For the cysteine protease inhibitor, cystatin B (CSTB), evidence for involvement in AD is indirect. Genetic deletion of CSTB in the TgCRND8 AD mouse model (expressing a triple mutation APP) reduced Aβ levels and prevented development of learning and memory deficits [18]. This suggests that over expression of CSTB in DS could exacerbate Aβ production and cognitive decline. Of potential significance also are the genes encoding the lanosterol synthetase (LSS) and the cholesterol transporter ABCG1 [19] which may contribute to altered levels of cholesterol precursors and transport, seen in AD and AD mice [20,21] and possibly APP processing [22-24].
Multiple HSA21 encoded proteins interact with or contribute to regulation of N-methyl-D-aspartate receptor (NMDAR) activity [25]. Among them is the guanine nucleotide exchange factor, TIAM1 that has specificity for RAC. Through its interaction with NMDAR, TIAM1 mediates signaling downstream of NMDAR activation to alter dendritic spine morphology [26,27]. The multiple domain scaffold protein intersectin1 (ITSN1) interacts with NMDAR through the NUMB protein and also functions in dendritic spine formation [28,29]. ITSN1 is over expressed in DS brain at several ages but, interestingly shows decreased expression in aged DS brains with AD neuropathology [30]. While not yet documented, over expression of TIAM1 and ITSN1 may alter the dynamics of NMDAR activity or downstream signaling.
Two HSA21-encoded proteins, the regulator of calcineurin protein-1, RCAN1, and the protein kinase, DYRK1A exhibit complicated functional interactions. RCAN1 was first described as an inhibitor of the protein phosphatase calcineurin (CaN) [31]. Over expression of RCAN1 has been observed in both DS and AD brains [32], leading to the suggestion that it contributes to the increased levels of phosphorylation observed in AD of CaN substrates, such as Tau and NMDAR [33]. This is, however, inconsistent with observations of increased CaN levels and activity in AD mouse models (Table 1). RCAN1 has targets in addition to CaN. When over expressed in primary neurons, RCAN1 was shown to induce apoptosis through activation of caspase 3 (CASP3) [34]. In the Tg2576 AD mouse model, levels of activated CASP3 increase as the mice age, and one result of this is an increase in the cleavage and activation of one of its targets, CaN. These observations together suggest that over expression of RCAN1 has opposing consequences for CaN activity. This apparent paradox may be related to alternative splicing that, in vivo, produces three protein isoforms for RCAN1. Short term expression of one of these isoforms, RCAN1.1L, has been shown to inhibit CASP3 activation, while longer term expression enhances it, at least in cDNA transfection of cell culture [35]. Detailed information is lacking regarding the dynamics of regulation of RCAN1 isoform expression in vivo during development and in aging, and how this may be perturbed in DS and contribute to AD in DS. As a final complication, RCAN1 is phosphorylated and activated by the HSA21-encoded protein kinase, DYRK1A [36]. Simultaneous over expression of both RCAN1 and DYRK1A likely would alter observations of the effects of RCAN1 alone. DYRK1A also phosphorylates Tau [37], further complicating prediction of the effects of over expression of RCAN1 on Tau dephosphorylation by CaN.
Table 1: Non-HSA21 protein abnormalities in AD and AD mouse models. 3xTg-AD, triple transgenic AD model expressing mutated forms of APP, Tau and PSEN1; Tg2576, transgenic mouse expressing the Swedish APP mutation; APP/PSEN1 AD mice, double transgenic expressing APP plus PSEN1 mutations. L/M, learning/memory; LTD, long term depression.
Nothing is known about how the increased expression of the multiple functions of these nine HSA21 genes, and their isoforms, might integrate with over expression of APP in the DS brain and in the development and progression of AD in DS. Furthermore, given our very limited knowledge of the functions of HSA21 genes, it is not possible to exclude additional genes from contributions to features of the AD phenotype in DS, including protection from dementia in the presence of neuropathology.
Because of the complexity of functional interactions, prediction of the consequences of trisomy of HSA21, and unraveling contributions to AD in DS, cannot be reliably inferred simply from data on genes studied one at a time. An approach to solving the problem of multiple and complex functional interactions is to circumvent HSA21 genes and look downstream at non-HSA21 proteins. Abnormalities in DS of non-HSA21 proteins reflect the integration of contributions from all trisomic genes, over expression of all their isoforms, with endogenous regulatory processes, and the influences of their direct and indirect interactions.
Abnormalities in a number of non-HSA21 proteins have been documented in brains of sAD or in mouse models carrying mutations in APP and/or PSEN1/2 seen in FAD. These proteins include kinases, phosphatases, receptors, scaffold proteins and cytokines, and typically were analyzed because they were known either to interact with or modulate the processing of APP, the activity of NMDAR, phosphorylation of Tau or neuroinflammation. The subsets of proteins investigated here are listed in Table 1 [38-66].
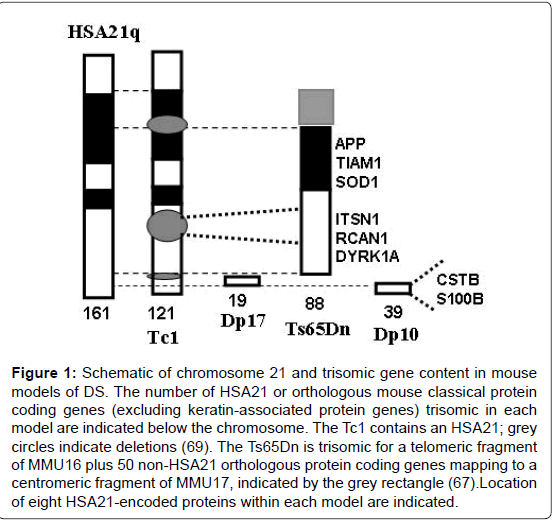
Tissue collections suitable for protein expression analysis are not available from DS with and without AD but can be obtained from the multiple mouse models of DS that have been developed. Multiple models have been required because orthologs of HSA21 genes map to three mouse chromosomal regions: the telomeric segment of mouse chromosome 16 (MMU16), an internal region of mouse chromosome 17 (MMU17) and an internal segment of mouse chromosome 10 (MMU10). The first viable and most popular mouse model of DS, the Ts65Dn, is trisomic for 88 of the 102 orthologs of HSA21 protein coding genes (excluding keratin-associated protein genes) that map to MMU16 [13], plus 50 protein coding genes mapping to the centromeric region of MMU17 that are not orthologous to any HSA21 genes [67]. The Tc1 is the most complete single model of DS available [68]. It carries a human chromosome 21, but due to several internal rearrangements, it is trisomic for only 121 HSA21 protein genes [69,70]. It is trisomic, however, for essentially all the genes that map to MMU17 and MMU10, absent in the Ts65Dn. Two newer mouse models of DS, the Dp(10)1Yey and the Dp (17)1Yey (here abbreviated as Dp10 and Dp17) respectively carry duplications of the 38 and 19 HSA21 orthologous protein coding genes that map to MMU10 and MMU17 [71]. Thus, the Dp10 and Dp17 are trisomic for subsets of genes trisomic in the Tc1, and the Tc1 and Ts65Dn are trisomic for partially overlapping sets of MMU16 genes. Trisomic segments contained in these four mouse models are illustrated in Figure 1. Consistent with their differing trisomic gene content, these DS models have quite different sets of phenotypic features [72]. The Ts65Dn displays many DS or AD-relevant features, including impaired function in learning and memory (L/M) tasks and degeneration of basal forebrain cholinergic neurons after six months of age. The Tc1 mice present with a milder phenotype overall, but still include L/M deficits, specifically short term working memory [73]. The Dp10 mice are not impaired in the standard hippocampal assessments of the Morris Water Maze (MWM) or context fear conditioning (CFC) at 2-4 months of age; they also show normal long term potentiation (LTP) [71]. The Dp17 mice also are not impaired in the MWM or CFC at 2-4 months of age, but in contrast to other models, they show abnormal, but enhanced, LTP [71]. Here, we report analysis of these four mouse modelsat 7-9 months of age for hippocampal expression of the non-HSA21 proteins listed in Table 1. The motivation for these experiments is to describe the influence of trisomy of differing subsets of HSA21 genes on ADrelevant protein expression. The resulting information will help in understanding, or at least more accurately modeling, the molecular mechanisms that lead to the frequent, but not universal, early onset development and progression of AD dementia in DS.
Figure 1: Schematic of chromosome 21 and trisomic gene content in mouse models of DS. The number of HSA21 or orthologous mouse classical protein coding genes (excluding keratin-associated protein genes) trisomic in each model are indicated below the chromosome. The Tc1 contains an HSA21; grey circles indicate deletions (69). The Ts65Dn is trisomic for a telomeric fragment of MMU16 plus 50 non-HSA21 orthologous protein coding genes mapping to a centromeric fragment of MMU17, indicated by the grey rectangle (67).Location of eight HSA21-encoded proteins within each model are indicated.
Materials and Methods
Mice
Breeding pairs of Dp(10)1Yey and Dp(17)1Yey mice (71) were a generous gift of Y. Yu (Roswell Park, New York). Mice for these experiments were bred at the University of Colorado, maintained in a room with HEPA-filtered air and a 14:10 light:dark cycle, fed a 6% fat diet and acidified (pH 2.5-3.0) water ad libitum. All procedures were approved by the University of Colorado Institutional Animal Care and Use Committee and performed in accordance with National Institutes of Health guidelines for the care and use of animals in research. Littermates were housed in the same cage. Male mice, nine controls and ten trisomic littermates from the Dp10, and six controls and five trisomic littermates from the Dp17, aged 7-9 months, were sacrificed by cervical dislocation without anesthetic and the brain quickly removed, frozen in liquid nitrogen, and stored at -80°C until use. All mice were naïve and were sacrificed between noon and 2 PM to maintain a consistent circadian time frame. Optimal preservation of protein profiles precludes use of anesthetic and perfusion.
Tissue processing and preparation of protein lysates
To optimally preserve protein profiles, including phosphorylation, prior to lysate preparation, tissues were heat stabilized in the Stabilizor T1 (Denator, AB) as described previously [74]. Briefly, brains were removed from 80°C and, without thawing, immediately placed in the sample cassette and inserted into the Stabilizor T1 where they were exposed to rapid heating to 95°C under vacuum. After stabilization, hippocampus was dissected out, weighed (Table S1), placed in 10 volumes of IEF buffer (8 M urea, 4% CHAPS, 50 mM Tris) and homogenized by sonication (three bursts of 5 s duration in Branson Sonic Power Co, USA), followed by centrifugation for 30 minutesat 14,000 rpm (Eppendorf 5415C) to remove debris. Protein concentrations, determined using the 660 nM Protein Assay kit (Pierce), were within the range of 9-11 mg/ml for all samples. Information for each mouse on age, littermates, and weights of individual hippocampi is provided in Table S1.
Antibodies and validation for RPPA
Antibodies used in this study are listed in Supplementary Table S2 with supplier and dilution factor. Reverse Phase Protein Arrays (RPPA) require highly specific antibodies. Prior to use in RPPA, each lot of each antibody was tested on Western blots of mouse brain lysates and verified to produce clean band(s) of explainable size in the absence of significant background and non-specific bands.
Array assembly and printing
For each sample, lysates, neat plus four serial dilutions (dilution factor 0.8) and one buffer control were pipetted into V-shaped AB gene 384-well plates (Thermo Fisher Scientific, IL, USA). Samples were printed in triplicate onto nitrocellulose–coated glass slides (Grace Bio-Labs, Inc. OR, USA) using an Aushon BioSystems 2470 Arrayer (Aushon BioSystems, MA, USA) with 185 μm pins and a single touch. Arrays were produced in two major print runs. Slides were stored at 4°C until use. For the small number of proteins measured in the Ts65Dn and Tc1 hippocampus, slides from previous prints [70,75] were used.
Antibody detection and array staining
Procedures for array screening have been described previously [75]. Briefly, slides were incubated in blocking solution (3% BSA (Sigma, USA) in TBST (Tris-buffered saline, 0.1% Tween 20) for 4 hours, followed by overnight incubation at 4°C with primary antibody (antibody dilutions are provided in Table S1). Detection of bound primary antibodies was performed by incubation for 90 min at room temperature with Fluorescence Alexa Fluor 555 goat antimouse or anti-rabbit secondary antibodies (1:2000) (Invitrogen, CA, USA). Slides were washed, dried and scanned on a GenePix 4000B array slide scanner (Axon Instruments, USA) using GenePix Pro 4.0 software or on a Perkin Elmer Scan Array Express HT Microarray Scanner (PerkinElmer Inc. MA, USA). For normalization purposes, total protein in each spot was determined by staining two to three non-sequential slides from each print run with Sypro Ruby reagent (Invitrogen, CA, USA) following the manufacturer’s protocol.
Image analysis, quantification, normalization and statistical analysis
For each slide, the intensity of each spot was quantified using the Scan Array Express software (PerkinElmer Inc. MA, USA). Antibody signal for each spot was normalized to the corresponding SyproRuby signal. Details of quantification and review of data quality and reproducibility were as described previously [75]. After exclusion of technical outliers, all SyproRuby-normalized protein values were included in the statistical analyses; data were transformed to a natural log scale. Mean differences in protein levels between genotypes (trisomy vs. control, reported as a ratio and percent of control) were assessed using a hierarchical three-level mixed effects model to account for the possible correlations and variabilities among replicates and dilution levels within each mouse. A two-tailed p-value <0.05 was considered for overall statistical significance across the entirety of the hypotheses tests; Bonferroni correction and False Discovery Rate (FDR) were applied to correct p-values from pair wise multiple tests. All data analyses were carried out using SAS® version 9.3 (SAS Institute Inc., Cary, NC). Supplemental Table S3 shows results of statistical analyses for both lines for all proteins.
Network display and databases
Components of the Alzheimer’s disease (AD) pathway and their relationships were obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG http://www.genome.jp/kegg/) database. Protein interaction partners of each AD pathway protein component were obtained from the IntACT (http://www.ebi.ac.uk/intact/), HPRD (Human Protein Reference Database, http://www.hprd.org/) and BioGRID (Biological General Repository for Interaction Datasets, http://thebiogrid.org/) databases; all interacting proteins that were analyzed by RPPA were retained, filtered for Gene Ontology (GO) terms and those sharing Cellular Component GO terms were added to the AD pathway network. The expanded network was constructed using Cytoscape 3.0.2.
Results
We measured a total of 34 proteins/phospho-proteins in hippocampus of trisomic Dp10 and Dp17 mice and corresponding littermate controls at 7-9 months of age. Eight proteins were encoded by HSA21. The 26 non-HSA21 proteins were chosen based on their functional interactions with APP, Tau or subunits of the NMDAR and reports of abnormal levels in brains of AD and/or mouse models of AD (Table 1). They included six proteins for which levels of whole protein plus one or two phosphorylated forms were measured, and 13 for which either whole or phosphorylated levels were measured. This is the first time that this many proteins with functional links to AD have been measured simultaneously. We also compared these results to those in Tc1 and Ts65Dn mice of similar ages, data largely from previous reports [70,75], with three additional proteins screened here.
Altered expression of HSA21 encoded proteins
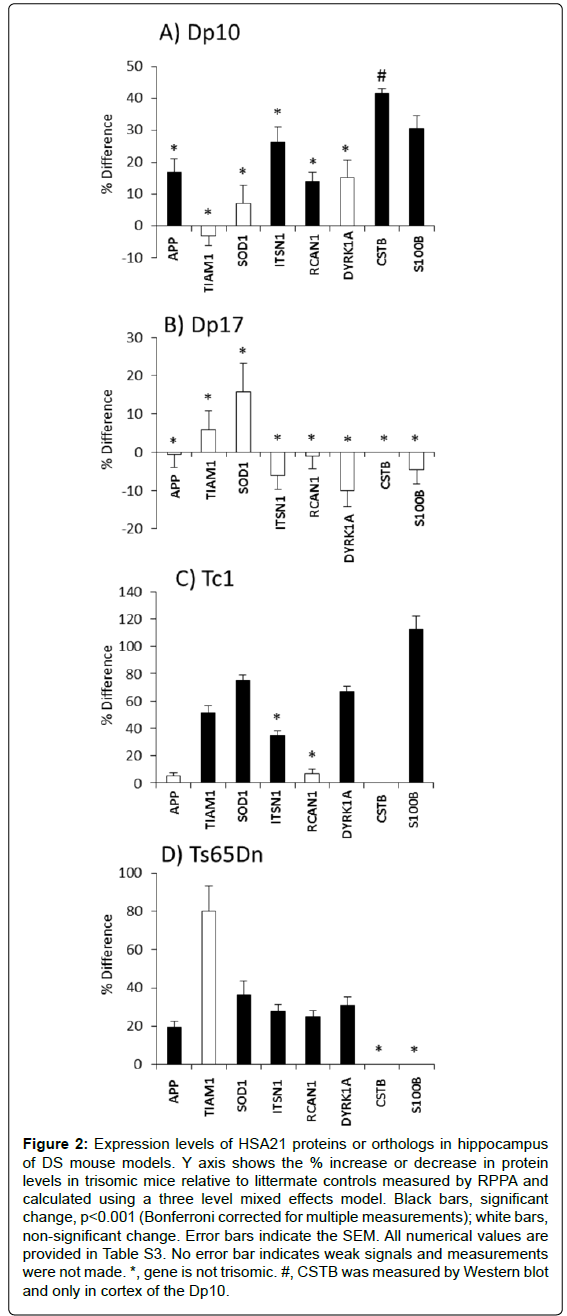
Figure 2 shows the level of each HSA21 protein in trisomy mice vs. littermate controls. It is clear that trisomy does not predict expression level. In the Dp10 (Figure 2A), levels of S100B and CSTB were elevated, consistent with their triplication. However, also elevated, by 17%, 26% and 14%, respectively, were levels of APP, ITSN1 and RCAN1. Genes encoding these proteins map to MMU16 and are not trisomic in the Dp10. Their increased expression is an indirect result of trisomy of the Dp10 segment.
All genes trisomic in the Dp10 are also trisomic in the Tc1 mice but the effects on expression of non-trisomic HSA21 orthologs differ between the two lines (Figure 2C). Similar to the Dp10, levels of S100B, which is trisomic in the Tc1, and ITSN1, which is not trisomic, are both elevated. Unlike the Dp10, non-trisomic HSA21 proteins, APP and RCAN1, do not show elevated expression levels in the Tc1.
In the Ts65Dn, proteins encoded by the trisomic genes, APP, SOD1, ITSN1, RCAN1 and DYRK1A, all show elevated expression, but TIAM1, that is also trisomic, is not elevated. TIAM1 however is over expressed in the Tc1, as are the trisomic proteins SOD1 and DYRK1A. Lastly, no genes trisomic in the Dp17 were evaluated because of the lack of adequate antibodies; none of the HSA21 proteins measured showed altered expression in the Dp17.
Figure 2: Expression levels of HSA21 proteins or orthologs in hippocampus of DS mouse models. Y axis shows the % increase or decrease in protein levels in trisomic mice relative to littermate controls measured by RPPA and calculated using a three level mixed effects model. Black bars, significant change, p<0.001 (Bonferroni corrected for multiple measurements); white bars, non-significant change. Error bars indicate the SEM. All numerical values are provided in Table S3. No error bar indicates weak signals and measurements were not made. *, gene is not trisomic. #, CSTB was measured by Western blot and only in cortex of the Dp10.
Altered expression of AD-relevant non-HSA21 encoded proteins
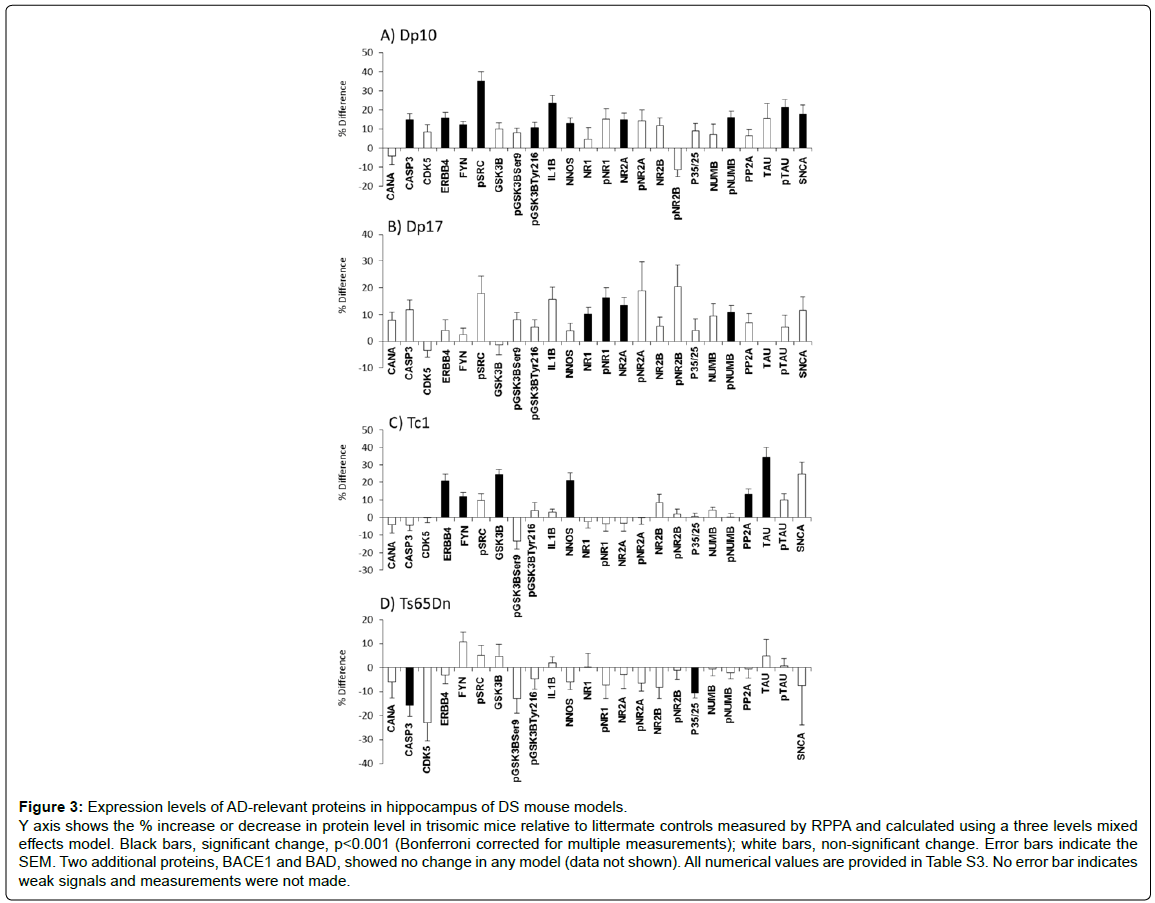
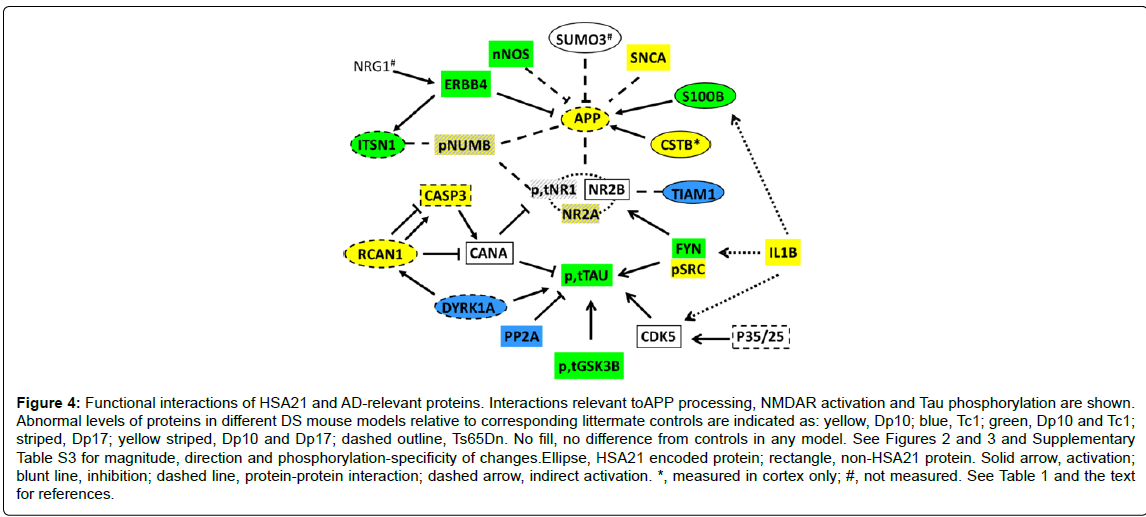
The non-HSA21 proteins measured have a broad range of functions. They include Tau and subunits of the NMDAR because abnormalities in the phosphorylation and activation of these proteins are key components of AD pathology and cognitive impairment. Other proteins analyzed included the neuregulin 1(NRG1) receptor ERBB4, the scaffold protein NUMB, kinases CDK5, GSK3B and FYN (phosphorylated and non-phosphorylated forms), subunits of the phosphatases PP2A and calcineurin (CaN) and the cytokine IL1B. Results are shown in Figure 3 and Supplementary Table S3. The first general observation is that the Dp10 mice show the greatest number of abnormalities, with 11 of 26 proteins elevated compared with littermate controls. In contrast, Dp17, Tc1 and Ts65Dn mice showed abnormalities in only 4, 6 and 2 proteins, respectively. In Figure 4, the abnormalities in protein levels are superimposed on a network showing the functional interactions of all proteins measured, anchored to APP, NMDAR and Tau.
Five non-HSA21 proteins, NUMB, pNUMB, nNOS, ERBB4 and SNCA, influence APP processing or cytotoxicity of APP peptides. All but NUMB were increased in the Dp10. nNOS and ERBB4 were also increased in the Tc1. ERBB4 levels are of particular interest. ERBB4 is proteolytically processed by γ-secretase and the C-terminal intracellular fragment translocates to the nucleus where it functions in transcriptional activation [76,77]. While the molecular mechanism causing the increased levels of ERBB4 here is not known, it is known that among the targets of ERBB4 transcriptional activation is ITSN1 [77], suggesting a cause for the elevated levels of ITSN1 in both Dp10 and Tc1. Increased levels of ITSN1 may indirectly influence APP through interaction with NUMB. In the Dp17, the only APP influence is an elevated level of pNUMB. None of the proteins influencing APP was altered in the Ts65Dn.
Levels of pTau-Thr212, but not total Tau, were increased in the Dp10, while the reverse was observed in the Tc1. Tau can be phosphorylated at multiple sites and eight of the proteins measured, subunits of the phosphatases CaN and PP2A, and kinases/phosphokinases, GSK3B, pGSK3B-Ser9, pGSK3B-Tyr216, CDK5, FYN and pSRC, can affect phosphorylation of Tau at sites other than Thr212 [38,78]. Levels of the CaN and PP2A subunits were unchanged in the Dp10. Levels of kinases/phospho-kinases, GSK3B, pGSK3B-Tyr216, FYN and pSRC were increased, which could contribute to increased phosphorylation of Tau at Thr212 and additional sites. Levels of IL1B are uniquely elevated in the Dp10 and may be indirectly influencing the increased levels of FYN/pSRC [79].
The picture in the Tc1 is also complex; levels of the PP2A phosphatase subunit and kinases DYRK1A, GSK3B and FYN were increased. Tau-Tyr18 is a target of both PP2A and FYN, and potential competition between the two makes the outcome of their increases difficult to predict. Changes in levels of PP2A subunits and decreased PP2A activity have been documented in AD, potentially contributing to the observations of hyperphosphorylated Tau [50-52]. In the Ts65Dn, the only perturbation directly influencing Tau is the elevated level of DYRK1A; however, although DYRK1A can increase both levels of Tau and phosphorylation of Tau at Thr212 [37,80], increases in neither total Tau nor Tau phosphorylation were detected in the Ts65Dn. An indirect influence on Tau in the Ts65Dn is the decreased level CDK5 regulatory proteins p35/25. p35 and its calpain-cleaved p25 fragment mediate, respectively, transient and extended activation of CDK5 [81]. Because the antibody used here detects both p35 and p25, it is not known if only one or both forms are decreased, and therefore, the consequences for the dynamics of CDK5 activity are not known. No Tau-relevant abnormalities were detected in the Dp17. No perturbations in the CaN subunit level were found in any of the four models. This is in spite of the direct influences on CaN activity and phosphorylation potentially caused by elevated levels of CASP3, RCAN1 and DYRK1A in the Dp10 and/or the Ts65Dn.
Figure 3: Expression levels of AD-relevant proteins in hippocampus of DS mouse models. Y axis shows the % increase or decrease in protein level in trisomic mice relative to littermate controls measured by RPPA and calculated using a three levels mixed effects model. Black bars, significant change, p<0.001 (Bonferroni corrected for multiple measurements); white bars, non-significant change. Error bars indicate the SEM. Two additional proteins, BACE1 and BAD, showed no change in any model (data not shown). All numerical values are provided in Table S3. No error bar indicates weak signals and measurements were not made.
Figure 4: Functional interactions of HSA21 and AD-relevant proteins. Interactions relevant toAPP processing, NMDAR activation and Tau phosphorylation are shown. Abnormal levels of proteins in different DS mouse models relative to corresponding littermate controls are indicated as: yellow, Dp10; blue, Tc1; green, Dp10 and Tc1; striped, Dp17; yellow striped, Dp10 and Dp17; dashed outline, Ts65Dn. No fill, no difference from controls in any model. See Figures 2 and 3 and Supplementary Table S3 for magnitude, direction and phosphorylation-specificity of changes.Ellipse, HSA21 encoded protein; rectangle, non-HSA21 protein. Solid arrow, activation; blunt line, inhibition; dashed line, protein-protein interaction; dashed arrow, indirect activation. *, measured in cortex only; #, not measured. See Table 1 and the text for references.
Among the non-HSA21 proteins measured, phosphorylation of NMDAR subunits can be affected by CaN and FYN/pSRC [44], and downstream signaling from NMDAR is impacted by interactions with NUMB and the HSA21 protein TIAM1. The Dp17 showed unique elevations in both pNR1 and total NR1, while levels of NR2A and pNUMB were increased in both the Dp17 and the Dp10. In spite of altered levels of interacting proteins, no changes in levels of NMDAR subunits or their phosphorylation were detected in either the Tc1 or the Ts65Dn.
Discussion
All individuals with DS develop the neuropathology of AD and approximately half will also develop an associated early onset dementia [6]. Triplication of the APP gene encoded by HSA21 may be a primary cause of AD in DS, similar to cases of FAD where affected individuals carry a duplication of the APP locus on one HSA21. However, several HSA21 genes have been shown to influence processing of APP to Aβ, the phosphorylation of Tau and activation of NMDAR. The effects of their combined over expression as in DS are not understood and currently not predictable. That only half the individuals with DS will develop an AD-like dementia by age 50-60, as compared to almost all that carry a duplication of APP in FAD, suggests that there are also genes on HSA21 that provide protection from the effects of over expression of APP.
The majority of people with DS are trisomic for a complete HSA21. In contrast, the popular and best studied mouse model of DS, the Ts65Dn, is trisomic for only 88 of 161 HSA21 protein coding genes [13]. Studies with the Ts65Dn therefore leave unexplored the contributions to DS phenotypic features of the HSA21 orthologs that map to the MMU10 and 17 regions. While the Dp10 and Dp17 mice have been analyzed for L/M in the MWM and CFC and for LTP at 2-4 months of age, nothing has been reported regarding abnormalities in protein expression [71]. Together with the Tc1, the Ts65Dn, Dp10 and Dp17 provide trisomy of partially overlapping subsets of all HSA21 genes conserved in mouse (Figure 1).
In this work, levels of eight HSA21 proteins and 26 non-HSA21 proteins were measured in whole hippocampal lysates of these DS mouse models. Measurements at the protein and protein phosphorylation level provide a view of gene expression that more closely reflects functional consequences than measurements at the mRNA level can. Furthermore, use of RPPA allows simultaneous measurement of protein levels in a large number of samples (here, 5-10 individuals, trisomic and controls, from two lines of mice, for a total of 31 samples) with a sufficient number of replicates to detect significant differences as small as 10% [70,75]. Because RPPA requires small amounts of lysate, even small tissues such as hippocampus yield sufficient material for measurement of tens of proteins. However, because RPPA requires highly specific antibodies, some proteins of interest, e.g. additional phosphorylation sites of Tau and NMDAR subunits, could not be measured.
Protein profiles and trisomic gene content
From analysis of 34 proteins in the four mouse models, the most striking general observation is the extraordinarily large proportion of protein abnormalities displayed by the Dp10. Of the 32 proteins not trisomic in the Dp10, 15 showed elevated expression levels, including three, APP, ITSN1 and RCAN1, of the six non-trisomic HSA21 proteins. The molecular mechanisms leading to increases in APP and RCAN1 are not known, but elevated ITSN1 could be due to transcriptional activation produced by elevated levels of ERBB4 [77]. In the Dp17, Tc1 and Ts65Dn, 34, 30 and 27 of the proteins measured were not trisomic, and only four, seven and two, respectively, were altered. The results for Dp10 are particularly noteworthy because the non-HSA21 proteins measured were not chosen for their functional interactions with genes encoded by the MMU10 region, but only for their abnormal levels seen in brains from AD or AD mouse models. These data therefore suggest that trisomy of the HSA21 MMU10 orthologous region, which occurs in the large majority of DS, may contribute to AD-relevant protein perturbations. Interpreting the extent of such contributions, however, must also consider observations in the Tc1. With the exception of rearrangements/deletions affecting a few genes, Tc1 mice are trisomic for all the genes in the Dp10 region [69]. However, they are also trisomic for genes in the MMU17 region and a large proportion of those mapping to MMU16. Consequently, the Tc1 show both similarities and differences with the Dp10 mice. Similar to the Dp10, Tc1 mice showed elevated levels of the non-trisomic HSA21 protein ITSN1, and the non-HSA21 proteins nNOS, ERBB4 and FYN. Unlike the Dp10, the TC1 showed normal levels of non-trisomic HSA21 proteins APP and RCAN1 and the non-HSA21 proteins pNUMB, NR2A, CASP3, SNCA, pSRC and IL1B. In addition, Tc1 showed elevated levels of total Tau and GSK3B, not of the phosphorylated forms seen in the Dp10. Unique to the Tc1 were increased levels of the PP2A regulatory subunit. Similarly, although the Dp17 and Tc1 share trisomy of the MMU17 region genes, elevated levels of pNUMB, pNR1, total NR1 and total NR2A are seen in the Dp17 but not in the Tc1.It is noteworthy too that, with no trisomic genes in common, both the Dp10 and the Dp17 have elevated levels of NR2A and pNUMB. Together, these differences and similarities suggest that, in DS, trisomy of MMU10 and MMU17 orthologs may cause unique non-HSA21 protein perturbations that will be further modulated by influences of additional trisomic HSA21 genes. That the Tc1 is trisomic for human genes in a mouse background could contribute to some differences from the Dp10 and Dp17, if transcriptional, translational and/or post-translational regulatory mechanisms differ between human and mouse genes. These observations do emphasize however the potential pitfalls of predicting perturbations in full trisomy HSA21 from observations only in the Ts65Dn.
Protein profiles and phenotypic features
With the exception of the elevated levels of APP observed here in the Dp10, there are currently no reports of analysis of the Dp10 and Dp17 for AD-related features of aberrant APP expression, enlarged early endosomes, loss of basal forebrain cholinergic neuronal markers, or impaired nerve growth factor transport, features that in the Ts65Dn have been best documented after 12 months of age [82-84]. Instead, the Dp10 and Dp17 have been analyzed only for behavior and LTP and only in mice 2-4 months of age [71] (for a detailed review of behavioral features of all four models, [72]). Nevertheless, comparisons of phenotypic features among the four models are of interest, not least because such features would not predict the patterns and extent of abnormal protein expression, either generally or specifically, observed here. The greatest contrast between behavioral and molecular phenotype is seen in comparing the Dp10 with the Ts65Dn. The Dp10 is not impaired in the MWM or CFC [71], while the Ts65Dn is impaired in both [72]. Similarly, the Dp10 has shown normal LTP while the Ts65Dn has reduced LTP. There are several possible explanations for the relative protein expression patterns. One possibility is that the protein perturbations observed in the Dp10 are irrelevant to L/M and synaptic plasticity. While this could be true for some of them, it seems unlikely to be true for all. Indeed, with a few exceptions, the non-HSA21 proteins measured here have been shown to cause L/M or synaptic plasticity abnormalities when mutated or knocked out in mouse (Table S2) [85]. Additionally, in human populations, mutations in three proteins, the NMDAR subunits, NR1, NR2A and NR2B are associated with intellectual disability. Certainly an elevated level of a protein would be expected to have different consequences from its mutation or deletion; nevertheless, it is difficult to conclude that an abnormal level of every protein observed in the Dp10 could be individually, or in combination, benign.
A second possibility is that the Dp10 mice may show age-dependent L/M deficits more reflective of protein abnormalities. Mice here were aged 7-9 months, while mice showing no learning deficits were tested at 2-4 months [71]. It is possible therefore that assessing learning in older Dp10 mice and generating protein profiles from younger mice would reveal deficits that are associated with age-dependent protein expression abnormalities. Such a scenario is not without precedent. In the Ts65Dn, protein levels of APP and SOD1 were not elevated in brain at 4 months, but were elevated at 12 months [84]. In the Tc1 mice, elevated levels of pTau-Thr212 and pGSK3B-Ser9 were found at 12 but not at two months [86].
Protective effects of protein perturbations?
An interesting alternative, or addition, to postulating irrelevance and age-dependence is the possibility that some of the perturbations measured in the Dp10 may be compensatory responses, serving to ameliorate more direct negative consequences of increased expression of Dp10 genes. This concept has been suggested by others [87] although no experimental evidence was provided and candidate causes were confined to genes trisomic in the Ts65Dn. Several examples of protective responses relevant to Dp10 abnormalities come from analysis of individual proteins in other AD model systems. (i) Knockdown of FYN in the 3 X Tg triple transgenic mouse model of AD (carrying mutations in APP, Tau and PSEN1 genes) resulted in both increased Aβ levels and exacerbation of L/M deficits [43]. The abnormally high levels of FYN in Dp10, therefore, may be a protective response to some other perturbation. (ii) NRG1 can prevent the cytotoxic effects in cultured neurons of exposure to APP C-terminal fragments; however, if ERBB4, a receptor for NRG1, is inhibited, the benefits of NRG1 treatment are eliminated [58]. Elevated levels of ERBB4 in the Dp10 may therefore facilitate the signaling, and protective effects, downstream of endogenous NRG1. (iii) The J20 AD mouse model (expressing APP carrying the Swedish plus Indiana mutations) shows cerebral amyloid angiopathy, characteristic of AD. Treatment with rapamycin induced, among other effects, phosphorylation and activation of nitric oxide synthase (NOS) and rescued the cerebral angiopathy; when NOS was inhibited, rescue was reversed [64]. (iv) Cytokine activation, including increased levels of IL1B, has been associated with negative consequences of neuroinflammation in response to injury and neurodegeneration [88]. However, earlier data suggests neuroprotective roles of elevated IL1B in its ability to increase nerve growth factor levels and signaling [89]. (v) A neuroprotective role may also be played in the Dp10 by the induced increase in the level of the calcineurin inhibitor, RCAN1. Inhibition of CaN activity in the Tg2576 mouse model of AD reverses Aβ-induced abnormalities in synaptic plasticity, dendritic spines and learning/memory [54,55]. Elevated activity of RCAN1, which could occur in the Dp10 due both to elevated RCAN1 protein levels and to increased RCAN1 phosphorylation by increased levels of pGSK3BTyr216, could facilitate inhibition of CaN activity. As discussed in the Introduction, the functions and regulation of RCAN1 activity are complex and intertwined with that of DYRK1A. However, increased expression of RCAN1, from its endogenous genomic context, without increases in DRYK1A, as occurs in the Dp10, may perturb CaN activity in a protective manner. (vi) S100B has both neurotoxic and neuroprotective roles, with functions governed by concentration, cellular localization and oxidative state [90]. S100B is neurotrophic at lower concentrations, and intracellular S100B is neuroprotective and anti-apoptotic. Measurement here of elevated levels of S100B in whole hippocampal lysates lacks information on local concentrations and intracellular proportions. More detailed analysis, in the context of the Dp10 segmental trisomy, at young ages, where no deficits have so far been described, through older ages, is required to reveal if and when S100B exerts positive effects in trisomy.
Perturbations in the Dp10 are modest, with most proteins altered by 15%-20% relative to controls. Only FYN and nNOS are <15%, while pSRC and IL1B are both ~25%-30%. Such modest alterations could be key to neuroprotective effects, with larger perturbations leading to neurotoxicity.
Additional insight may be obtained from abnormalities observed in the Dp17. Dp17 are also not impaired in L/M and show enhanced LTP at 2-4 months. Common to the Dp17 and the Dp10, and not seen in either the Tc1 or the Ts65Dn at 7-9 months of age, are increased levels of NR2A and pNUMB. Increases in NR2A-containing NMDAR have been shown to be protective of various neurological insults [91,92]. The functional consequences of increases in pNUMB are currently not known, however, NUMB is expressed in multiple protein isoforms that differentially participate in regulation of APP processing and transport in an isoform-specific fashion [60]. Levels of NUMB isoforms are altered in brains of both AD and the triple transgenic AD mouse model [62]. While these isoforms could not be distinguished here, it would be of interest to compare isoform levels between the Dp10 and Dp17, that are not impaired in L/M at 2-4 months, with those in the Tc1 and Ts65Dn that are impaired.
Altered protein activity cannot be inferred solely from observation of an abnormal protein level. For example, levels of only single subunits of PP2A and calcineurin were measured. Changes in levels of other components of these phosphatase complexes, or the lack of such changes, would have consequences for changes in phosphatase activities. Patterns of post-translational modification (PTM) are also critical. Phosphorylation could be measured for only six proteins and at only one or two sites each. Phosphorylation at multiple sites, plus additional PTMs contributes to regulation of protein activities, and the extents, varieties and dynamics of PTMs are poorly documented. Lastly, the analysis here focused on how altered levels of AD-related proteins might impact APP, Tau and NMDAR, however, each of the proteins measured has additional targets, substrates and interactions, the majority of which are probably unknown. Given these gaps in knowledge, protein abnormalities observed in the Dp10 and Dp17 in addition to those discussed above, and indeed abnormalities observed in the other mouse models, similarly could be serving compensatory or protective roles.
Consequences for complete trisomy
It is clear that the molecular consequences of segmental trisomies are not simply additive and there is ample evidence that the behavioral phenotypic consequences are also not additive. For example, a single gene transgenic over expressing the DYRK1A kinase is impaired in the MWM while the Ts1Rhr model, trisomic for 33 genes that include DYRK1A, is not [93] (although it does display other DS-relevant phenotypic features). The Ts1Yah is trisomic for nine genes from the Mmu17 segment and shows enhanced learning in the MWM [94] while the Dp17, which is trisomic for the same 9 genes plus 10 others, does not [71]. In addition, while trisomy of the MMU17 region alone, as in the Dp17, shows no impairment in L/M, it does contribute to L/M impairments in the complete trisomy DS model composed of trisomy of the MMU16, MMU17 and MMU10 regions [95]. In contrast, deleting the MMU10 region from the Tc1 did not change the Tc1 phenotype [67]. It is premature, however, to conclude that genes encoded by the MMU10 syntenic region make no contribution to any significant DS phenotypic feature. Differences in genetic backgrounds are known to alter observed phenotypes [67], and different phenotypic features, more or less severe, might be observed if the Dp10 were bred to genetic backgrounds more prone to L/M deficits or other behavioral abnormalities. Alternatively, increasing the complexity and the diversity of the phenotypic assessments of DS mice, either the Dp10 alone, the Tc1 deleted for the MMU10 segment, or the Dp16/Dp17/ Dp10 compared with the Dp16/Dp17, might reveal contributions of the Dp10 region. Regardless, the possibility that MMU10 region orthologs contribute to neuroprotection in DS is worth consideration. The differences in electrophysiological and behavioral phenotypes identified so far among the Dp10, Dp17, Tc1 and Ts65Dn models at young ages and in molecular profiles at this intermediate age of 7-9 months makes clear that the identity of the trisomic genes modulates the phenotypic features at all levels.
Perturbation of the AD network
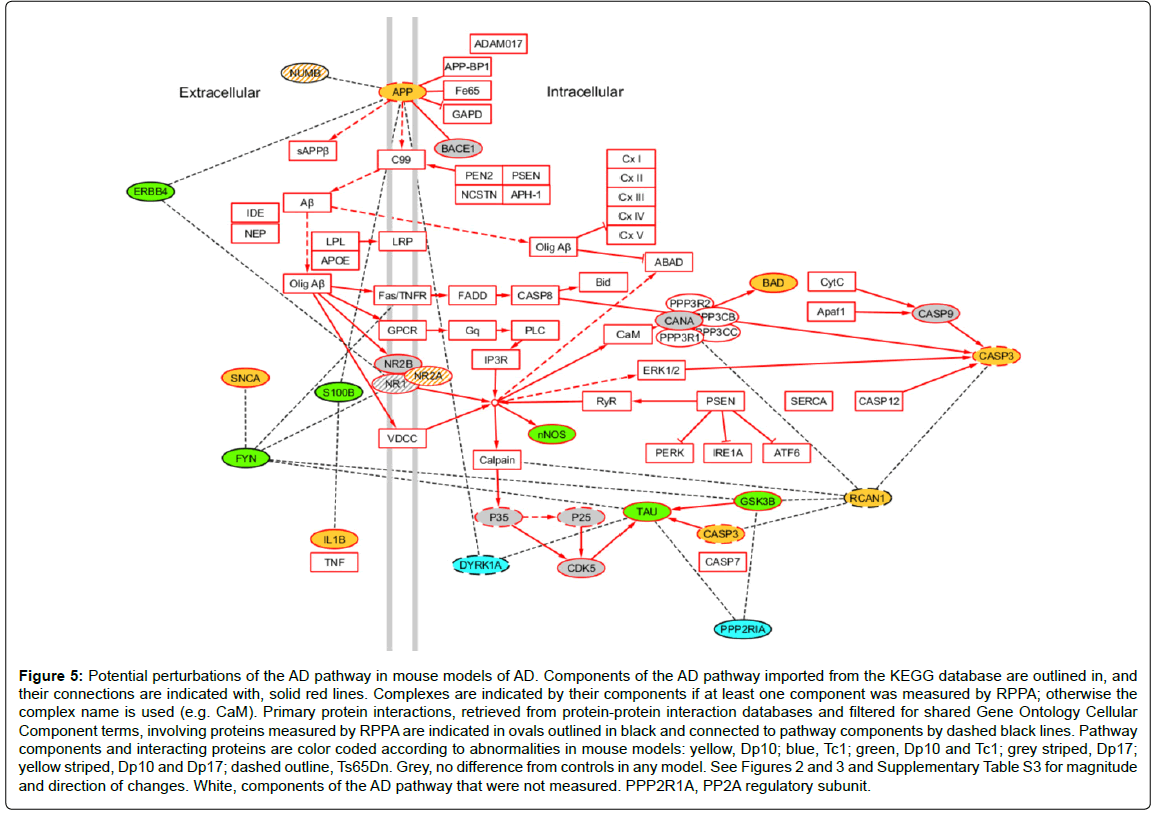
As a last illustration of the consequences of protein interactions, and the challenges in interpreting their contributions to full trisomy HSA21 in people with DS, we superimposed the observed protein abnormalities from these mouse models on the AD pathway obtained from KEGG, where all protein components (or complexes) and their relationships are manually curated from the primary literature [96]. To this pathway, we added proteins that have been shown to interact with any component of the pathway (filtered for common cellular component GO terms). Color coding for all proteins measured here provides a graphical representation of how trisomies for different sets of genes impact the pathway (Figure 5). Fifteen protein components (plus phosphorylated forms for six) of the AD pathway were measured here. Twelve of these, including one HSA21 protein, APP, were altered in one or more of the four DS models. This suggests that the pathway could be perturbed in each model, but at different points and in different ways, possibly leading to different outcomes. A further seven proteins measured, including the HSA21 proteins RCAN1 and DYRK1A, interact with a total of 11 different components; abnormal levels of each of these interacting proteins could potentially further perturb the pathway in model-specific patterns. The model-specific features, and the associated trisomic genes that underlie them, emphasize our lack of adequate understanding of the molecular mechanisms that may lead to AD in DS. It is reasonable to propose, however, that APP is unlikely to be the sole contributing gene. The complexity of these data also makes clear that conventional attempts [95] proposed to identify contributions of individual HSA21 genes to the DS phenotype, including AD in DS, by deleting single HSA21 genes from a complete trisomy, will remain confounded by the complexities of the same interactions.
Figure 5: Potential perturbations of the AD pathway in mouse models of AD. Components of the AD pathway imported from the KEGG database are outlined in, and their connections are indicated with, solid red lines. Complexes are indicated by their components if at least one component was measured by RPPA; otherwise the complex name is used (e.g. CaM). Primary protein interactions, retrieved from protein-protein interaction databases and filtered for shared Gene Ontology Cellular Component terms, involving proteins measured by RPPA are indicated in ovals outlined in black and connected to pathway components by dashed black lines. Pathway components and interacting proteins are color coded according to abnormalities in mouse models: yellow, Dp10; blue, Tc1; green, Dp10 and Tc1; grey striped, Dp17; yellow striped, Dp10 and Dp17; dashed outline, Ts65Dn. Grey, no difference from controls in any model. See Figures 2 and 3 and Supplementary Table S3 for magnitude and direction of changes. White, components of the AD pathway that were not measured. PPP2R1A, PP2A regulatory subunit.
The data in Figure 5 can also be used to consider the potential consequences of proposed pharmacoptherapeutics. For example, memantine, an uncompetitive antagonist of the NMDAR, approved for use in moderate to severe AD, has been used in both acute and chronic treatment of Ts65Dn mice of different ages. In each case, partial or complete rescue of L/M deficits was observed [97-99]. In clinical trials, however, neither younger nor older individuals with DS showed significant improvement with memantine treatment [100,101]. To begin to dissect the molecular correlates of such disparate results, we have generated protein profiles from mice treated with memantine. Memantine treatment alone, in control mice, produced changes in the levels of twelve of the 34 proteins measured here, among them, NMDAR subunits and phosphorylation, Tau, pNUMB, PP2A, CDK5 and P35/25, and interestingly, both RCAN1 and TIAM1 [102]. Because abnormal levels of each of these proteins are present in one or more of the DS mouse models studied here and in particular in the Dp10, it will be informative to determine the molecular responses to memantine in both partial and complete trisomy DS models.
Mouse models of DS have provided valuable insight into potential molecular mechanisms subserving the phenotypic features of DS. Used appropriately, and with appreciation of the complexity and diversity of molecular interactions, application of mouse models to the study of AD in DS may inform, not only DS, but also important aspects of FAD and sAD, including underlying molecular mechanisms, associated molecular perturbations, and potential targets for therapeutic interventions.
Funding
This work was supported by the National Institute of Child Health and Human Development (HD071585) the Linda Crnic Institute for Down Syndrome and the Fondation Jerome Lejeune.
References
- Schellenberg GD, Montine TJ (2012) The genetics and neuropathology of Alzheimer's disease. Acta Neuropathol 124: 305-323.
- Tanzi RE (2012) The genetics of Alzheimer disease. Cold Spring Harb Perspect Med 2.
- Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerrière A, et al. (2006) APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet 38: 24-26.
- Sleegers K, Brouwers N, Gijselinck I, Theuns J, Goossens D, et al. (2006) APP duplication is sufficient to cause early onset Alzheimer's dementia with cerebral amyloid angiopathy. Brain 129: 2977-2983.
- Kim M, Suh J, Romano D, Truong MH, Mullin K, et al. (2009) Potential late-onset Alzheimer's disease-associated mutations in the ADAM10 gene attenuate {alpha}-secretase activity. Hum Mol Genet 18: 3987-3996.
- Lott IT (2012) Neurological phenotypes for Down syndrome across the life span. Prog Brain Res 197: 101-121.
- Teipel SJ, Hampel H (2006) Neuroanatomy of Down syndrome in vivo: a model of preclinical Alzheimer's disease. Behav Genet 36: 405-415.
- Irving C, Basu A, Richmond S, Burn J, Wren C (2008) Twenty-year trends in prevalence and survival of Down syndrome. Eur J Hum Genet 16: 1336-1340.
- Hooli BV, Mohapatra G, Mattheisen M, Parrado AR, Roehr JT, et al. (2012) Role of common and rare APP DNA sequence variants in Alzheimer disease. Neurology 78: 1250-1257.
- Paula-Lima AC, Brito-Moreira J, Ferreira ST (2013) Deregulation of excitatory neurotransmission underlying synapse failure in Alzheimer's disease. J Neurochem 126: 191-202.
- Danysz W, Parsons CG (2012) Alzheimer's disease, β-amyloid, glutamate, NMDA receptors and memantine--searching for the connections. Br J Pharmacol 167: 324-352.
- Heneka MT, O'Banion MK (2007) Inflammatory processes in Alzheimer's disease. J Neuroimmunol 184: 69-91.
- Sturgeon X, Gardiner KJ (2011) Transcript catalogs of human chromosome 21 and orthologous chimpanzee and mouse regions. Mamm Genome 22: 261-271.
- Griffin WS, Stanley LC, Ling C, White L, MacLeod V, et al. (1989) Brain interleukin 1 and S-100 immunoreactivity are elevated in Down syndrome and Alzheimer disease. Proc Natl Acad Sci U S A 86: 7611-7615.
- Mori T, Koyama N, Arendash GW, Horikoshi-Sakuraba Y, Tan J, et al. (2010) Overexpression of human S100B exacerbates cerebral amyloidosis and gliosis in the Tg2576 mouse model of Alzheimer's disease. Glia 58: 300-314.
- Li Y, Wang H, Wang S, Quon D, Liu YW, et al. (2003) Positive and negative regulation of APP amyloidogenesis by sumoylation. Proc Natl Acad Sci U S A 100: 259-264.
- McMillan LE, Brown JT, Henley JM, Cimarosti H (2011) Profiles of SUMO and ubiquitin conjugation in an Alzheimer's disease model. Neurosci Lett 502: 201-208.
- Yang DS, Stavrides P, Mohan PS, Kaushik S, Kumar A, et al. (2011) Therapeutic effects of remediating autophagy failure in a mouse model of Alzheimer disease by enhancing lysosomal proteolysis. Autophagy 7: 788-789.
- Javitt NB (2013) Alzheimer's disease: neuroprogesterone, epoxycholesterol, and ABC transporters as determinants of neurodesmosterol tissue levels and its role in amyloid protein processing. J Alzheimers Dis 35: 441-450.
- Kölsch H, Heun R, Jessen F, Popp J, Hentschel F, et al. (2010) Alterations of cholesterol precursor levels in Alzheimer's disease. Biochim Biophys Acta 1801: 945-950.
- Vanmierlo T, Rutten K, Dederen J, Bloks VW, van Vark-van der Zee LC, et al. (2011) Liver X receptor activation restores memory in aged AD mice without reducing amyloid. Neurobiol Aging 32: 1262-1272.
- Tansley GH, Burgess BL, Bryan MT, Su Y, Hirsch-Reinshagen V, et al. (2007) The cholesterol transporter ABCG1 modulates the subcellular distribution and proteolytic processing of beta-amyloid precursor protein. J Lipid Res 48: 1022-1034.
- Burgess BL, Parkinson PF, Racke MM, Hirsch-Reinshagen V, Fan J, et al. (2008) ABCG1 influences the brain cholesterol biosynthetic pathway but does not affect amyloid precursor protein or apolipoprotein E metabolism in vivo. J Lipid Res 49: 1254-1267.
- Maulik M, Westaway D, Jhamandas JH, Kar S (2013) Role of cholesterol in APP metabolism and its significance in Alzheimer's disease pathogenesis. Mol Neurobiol 47: 37-63.
- Siddiqui A, Lacroix T, Stasko MR, Scott-McKean JJ, Costa AC, et al. (2008) Molecular responses of the Ts65Dn and Ts1Cje mouse models of Down syndrome to MK-801. Genes Brain Behav 7: 810-820.
- Tolias KF, Bikoff JB, Burette A, Paradis S, Harrar D, et al. (2005) The Rac1-GEF Tiam1 couples the NMDA receptor to the activity-dependent development of dendritic arbors and spines. Neuron 45: 525-538.
- Tolias KF, Bikoff JB, Kane CG, Tolias CS, Hu L, et al. (2007) The Rac1 guanine nucleotide exchange factor Tiam1 mediates EphB receptor-dependent dendritic spine development. Proc Natl Acad Sci U S A 104: 7265-7270.
- Thomas S, Ritter B, Verbich D, Sanson C, Bourbonnière L, et al. (2009) Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J Biol Chem 284: 12410-12419.
- Nishimura T, Yamaguchi T, Tokunaga A, Hara A, Hamaguchi T, et al. (2006) Role of numb in dendritic spine development with a Cdc42 GEF intersectin and EphB2. Mol Biol Cell 17: 1273-1285.
- Hunter MP, Nelson M, Kurzer M, Wang X, Kryscio RJ, et al. (2011) Intersectin 1 contributes to phenotypes in vivo: implications for Down's syndrome. Neuroreport 22: 767-772.
- Rothermel B, Vega RB, Yang J, Wu H, Bassel-Duby R, et al. (2000) A protein encoded within the Down syndrome critical region is enriched in striated muscles and inhibits calcineurin signaling. J Biol Chem 275: 8719-8725.
- Ermak G, Morgan TE, Davies KJ (2001) Chronic overexpression of the calcineurin inhibitory gene DSCR1 (Adapt78) is associated with Alzheimer's disease. J Biol Chem 276: 38787-38794.
- Lloret A, Badia MC, Giraldo E, Ermak G, Alonso MD, et al. (2011) Amyloid-β toxicity and tau hyperphosphorylation are linked via RCAN1 in Alzheimer's disease. J Alzheimers Dis 27: 701-709.
- Sun X, Wu Y, Chen B, Zhang Z, Zhou W, et al. (2011) Regulator of calcineurin 1 (RCAN1) facilitates neuronal apoptosis through caspase-3 activation. J Biol Chem 286: 9049-9062.
- Wu Y, Song W (2013) Regulation of RCAN1 translation and its role in oxidative stress-induced apoptosis. FASEB J 27: 208-221.
- Jung MS, Park JH, Ryu YS, Choi SH, Yoon SH, et al. (2011) Regulation of RCAN1 protein activity by Dyrk1A protein-mediated phosphorylation. J Biol Chem 286: 40401-40412.
- Woods YL, Cohen P, Becker W, Jakes R, Goedert M, et al. (2001) The kinase DYRK phosphorylates protein-synthesis initiation factor eIF2Bepsilon at Ser539 and the microtubule-associated protein tau at Thr212: potential role for DYRK as a glycogen synthase kinase 3-priming kinase. Biochem J 355: 609-615.
- Wang JZ, Xia YY, Grundke-Iqbal I, Iqbal K (2013) Abnormal hyperphosphorylation of tau: sites, regulation, and molecular mechanism of neurofibrillary degeneration. J Alzheimers Dis 33 Suppl 1: S123-139.
- D'Amelio M, Cavallucci V, Middei S, Marchetti C, Pacioni S, et al. (2011) Caspase-3 triggers early synaptic dysfunction in a mouse model of Alzheimer's disease. Nat Neurosci 14: 69-76.
- Hooper C, Killick R, Lovestone S (2008) The GSK3 hypothesis of Alzheimer's disease. J Neurochem 104: 1433-1439.
- Shirazi SK, Wood JG (1993) The protein tyrosine kinase, fyn, in Alzheimer's disease pathology. Neuroreport 4: 435-437.
- Lee G, Thangavel R, Sharma VM, Litersky JM, Bhaskar K, et al. (2004) Phosphorylation of tau by fyn: implications for Alzheimer's disease. J Neurosci 24: 2304-2312.
- Minami SS, Clifford TG, Hoe HS, Matsuoka Y, Rebeck GW (2012) Fyn knock-down increases Aβ, decreases phospho-tau, and worsens spatial learning in 3×Tg-AD mice. Neurobiol Aging 33: 825.
- Boehm J (2013) A 'danse macabre': tau and Fyn in STEP with amyloid beta to facilitate induction of synaptic depression and excitotoxicity. Eur J Neurosci 37: 1925-1930.
- Zhang J, Johnson GV (2000) Tau protein is hyperphosphorylated in a site-specific manner in apoptotic neuronal PC12 cells. J Neurochem 75: 2346-2357.
- Zhang J, Cicero SA, Wang L, Romito-Digiacomo RR, Yang Y, et al. (2008) Nuclear localization of Cdk5 is a key determinant in the postmitotic state of neurons. Proc Natl Acad Sci U S A 105: 8772-8777.
- Zhang J, Herrup K (2008) Cdk5 and the non-catalytic arrest of the neuronal cell cycle. Cell Cycle 7: 3487-3490.
- Cheung ZH, Ip NY (2012) Cdk5: a multifaceted kinase in neurodegenerative diseases. Trends Cell Biol 22: 169-175.
- Cruz JC, Kim D, Moy LY, Dobbin MM, Sun X, et al. (2006) p25/cyclin-dependent kinase 5 induces production and intraneuronal accumulation of amyloid beta in vivo. J Neurosci 26: 10536-10541.
- Sontag E, Luangpirom A, Hladik C, Mudrak I, Ogris E, et al. (2004) Altered expression levels of the protein phosphatase 2A ABalphaC enzyme are associated with Alzheimer disease pathology. J Neuropathol Exp Neurol 63: 287-301.
- Sontag JM, Nunbhakdi-Craig V, White CL 3rd, Halpain S, Sontag E (2012) The protein phosphatase PP2A/Bα binds to the microtubule-associated proteins Tau and MAP2 at a motif also recognized by the kinase Fyn: implications for tauopathies. J Biol Chem 287: 14984-14993.
- Martin L, Latypova X, Wilson CM, Magnaudeix A, Perrin ML, et al. (2013) Tau protein phosphatases in Alzheimer's disease: the leading role of PP2A. Ageing Res Rev 12: 39-49.
- Abdul HM, Sama MA, Furman JL, Mathis DM, Beckett TL, et al. (2009) Cognitive decline in Alzheimer's disease is associated with selective changes in calcineurin/NFAT signaling. J Neurosci 29: 12957-12969.
- Dineley KT, Hogan D, Zhang WR, Taglialatela G (2007) Acute inhibition of calcineurin restores associative learning and memory in Tg2576 APP transgenic mice. Neurobiol Learn Mem 88: 217-224.
- Cavallucci V, Berretta N, Nobili A, Nisticò R, Mercuri NB, et al. (2013) Calcineurin inhibition rescues early synaptic plasticity deficits in a mouse model of Alzheimer's disease. Neuromolecular Med 15: 541-548.
- Kitazawa M, Cheng D, Tsukamoto MR, Koike MA, Wes PD, et al. (2011) Blocking IL-1 signaling rescues cognition, attenuates tau pathology, and restores neuronal β-catenin pathway function in an Alzheimer's disease model. J Immunol 187: 6539-6549.
- Woo RS, Lee JH, Yu HN, Song DY, Baik TK (2011) Expression of ErbB4 in the neurons of Alzheimer's disease brain and APP/PS1 mice, a model of Alzheimer's disease. Anat Cell Biol 44: 116-127.
- Ryu J, Yu HN, Cho H, Kim HS, Baik TK, et al. (2012) Neuregulin-1 exerts protective effects against neurotoxicities induced by C-terminal fragments of APP via ErbB4 receptor. J Pharmacol Sci 119: 73-81.
- Roncarati R, Sestan N, Scheinfeld MH, Berechid BE, Lopez PA, et al. (2002) The gamma-secretase-generated intracellular domain of beta-amyloid precursor protein binds Numb and inhibits Notch signaling. Proc Natl Acad Sci U S A 99: 7102-7107.
- Kyriazis GA, Wei Z, Vandermey M, Jo DG, Xin O, et al. (2008) Numb endocytic adapter proteins regulate the transport and processing of the amyloid precursor protein in an isoform-dependent manner: implications for Alzheimer disease pathogenesis. J Biol Chem 283: 25492-25502.
- Chigurupati S, Madan M, Okun E, Wei Z, Pattisapu JV, et al. (2011) Evidence for altered Numb isoform levels in Alzheimer's disease patients and a triple transgenic mouse model. J Alzheimers Dis 24: 349-361.
- Ntelios D, Berninger B, Tzimagiorgis G (2012) Numb and Alzheimer's disease: the current picture. Front Neurosci 6: 145.
- Tokumitsu H, Hatano N, Inuzuka H, Sueyoshi Y, Yokokura S, et al. (2005) Phosphorylation of Numb family proteins. Possible involvement of Ca2+/calmodulin-dependent protein kinases. J Biol Chem 280: 35108-35118.
- Lin AL, Zheng W, Halloran JJ, Burbank RR, Hussong SA, et al. (2013) Chronic rapamycin restores brain vascular integrity and function through NO synthase activation and improves memory in symptomatic mice modeling Alzheimer's disease. J Cereb Blood Flow Metab 33: 1412-1421.
- Zhihui Q (2013) Modulating nitric oxide signaling in the CNS for Alzheimer's disease therapy. Future Med Chem 5: 1451-1468.
- Ramakrishna N, Meeker HC, Patel S, Brown TW, El Idrissi A (2012) Regulation of α-synuclein expression in Down syndrome. J Neurosci Res 90: 1589-1596.
- Duchon A, Pothion S, Brault V, Sharp AJ, Tybulewicz VL, et al. (2011) The telomeric part of the human chromosome 21 from Cstb to Prmt2 is not necessary for the locomotor and short-term memory deficits observed in the Tc1 mouse model of Down syndrome. Behav Brain Res 217: 271-281.
- O'Doherty A, Ruf S, Mulligan C, Hildreth V, Errington ML, et al. (2005) An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science 309: 2033-2037.
- Gribble SM, Wiseman FK, Clayton S, Prigmore E, Langley E, et al. (2013) Massively parallel sequencing reveals the complex structure of an irradiated human chromosome on a mouse background in the Tc1 model of Down syndrome. PLoS One 8: e60482.
- Ahmed MM, Dhanasekaran AR, Tong S, Wiseman FK, Fisher EM, et al. (2013) Protein profiles in Tc1 mice implicate novel pathway perturbations in the Down syndrome brain. Hum Mol Genet 22: 1709-1724.
- Yu T, Liu C, Belichenko P, Clapcote SJ, Li S, et al. (2010) Effects of individual segmental trisomies of human chromosome 21 syntenic regions on hippocampal long-term potentiation and cognitive behaviors in mice. Brain Res 1366: 162-171.
- Rueda N, Flórez J, Martínez-Cué C (2012) Mouse models of Down syndrome as a tool to unravel the causes of mental disabilities. Neural Plast 2012: 584071.
- Morice E, Andreae LC, Cooke SF, Vanes L, Fisher EM, et al. (2008) Preservation of long-term memory and synaptic plasticity despite short-term impairments in the Tc1 mouse model of Down syndrome. Learn Mem 15: 492-500.
- Ahmed MM, Gardiner KJ (2011) Preserving protein profiles in tissue samples: differing outcomes with and without heat stabilization. J Neurosci Methods 196: 99-106.
- Ahmed MM, Sturgeon X, Ellison M, Davisson MT, Gardiner KJ (2012) Loss of correlations among proteins in brains of the Ts65Dn mouse model of down syndrome. J Proteome Res 11: 1251-1263.
- Lee HJ, Jung KM, Huang YZ, Bennett LB, Lee JS, et al. (2002) Presenilin-dependent gamma-secretase-like intramembrane cleavage of ErbB4. J Biol Chem 277: 6318-6323.
- Allison JG, Das PM, Ma J, Inglis FM, Jones FE (2011) The ERBB4 intracellular domain (4ICD) regulates NRG1-induced gene expression in hippocampal neurons. Neurosci Res 70: 155-163.
- Drewes G, Mandelkow EM, Baumann K, Goris J, Merlevede W, et al. (1993) Dephosphorylation of tau protein and Alzheimer paired helical filaments by calcineurin and phosphatase-2A. FEBS Lett 336: 425-432.
- Viviani B, Bartesaghi S, Gardoni F, Vezzani A, Behrens MM, et al. (2003) Interleukin-1beta enhances NMDA receptor-mediated intracellular calcium increase through activation of the Src family of kinases. J Neurosci 23: 8692-8700.
- Qian W, Jin N, Shi J, Yin X, Jin X, et al. (2013) Dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A) enhances tau expression. J Alzheimers Dis 37: 529-538.
- Shukla V, Skuntz S, Pant HC (2012) Deregulated Cdk5 activity is involved in inducing Alzheimer's disease. Arch Med Res 43: 655-662.
- Cataldo AM, Petanceska S, Peterhoff CM, Terio NB, Epstein CJ, et al. (2003) App gene dosage modulates endosomal abnormalities of Alzheimer's disease in a segmental trisomy 16 mouse model of down syndrome. J Neurosci 23: 6788-6792.
- Salehi A, Delcroix JD, Belichenko PV, Zhan K, Wu C, et al. (2006) Increased App expression in a mouse model of Down's syndrome disrupts NGF transport and causes cholinergic neuron degeneration. Neuron 51: 29-42.
- Choi JH, Berger JD, Mazzella MJ, Morales-Corraliza J, Cataldo AM, et al. (2009) Age-dependent dysregulation of brain amyloid precursor protein in the Ts65Dn Down syndrome mouse model. J Neurochem 110: 1818-1827.
- Sturgeon X, Le T, Ahmed MM, Gardiner KJ (2012) Pathways to cognitive deficits in Down syndrome. Prog Brain Res 197: 73-100.
- Sheppard O, Plattner F, Rubin A, Slender A, Linehan JM, et al. (2012) Altered regulation of tau phosphorylation in a mouse model of down syndrome aging. Neurobiol Aging 33: 828.
- Head E, Lott IT, Patterson D, Doran E, Haier RJ (2007) Possible compensatory events in adult Down syndrome brain prior to the development of Alzheimer disease neuropathology: targets for nonpharmacological intervention. J Alzheimers Dis 11: 61-76.
- Shaftel SS, Griffin WS, O'Banion MK (2008) The role of interleukin-1 in neuroinflammation and Alzheimer disease: an evolving perspective. J Neuroinflammation 5: 7.
- Strijbos PJ, Rothwell NJ (1995) Interleukin-1 beta attenuates excitatory amino acid-induced neurodegeneration in vitro: involvement of nerve growth factor. J Neurosci 15: 3468-3474.
- Donato R, Sorci G, Riuzzi F, Arcuri C, Bianchi R, et al. (2009) S100B's double life: intracellular regulator and extracellular signal. Biochim Biophys Acta 1793: 1008-1022.
- Choo AM, Geddes-Klein DM, Hockenberry A, Scarsella D, Mesfin MN, et al. (2012) NR2A and NR2B subunits differentially mediate MAP kinase signaling and mitochondrial morphology following excitotoxic insult. Neurochem Int 60: 506-516.
- Liu Y, Wong TP, Aarts M, Rooyakkers A, Liu L, et al. (2007) NMDA receptor subunits have differential roles in mediating excitotoxic neuronal death both in vitro and in vivo. J Neurosci 27: 2846-2857.
- Olson LE, Roper RJ, Sengstaken CL, Peterson EA, Aquino V, et al. (2007) Trisomy for the Down syndrome 'critical region' is necessary but not sufficient for brain phenotypes of trisomic mice. Hum Mol Genet 16: 774-782.
- Pereira PL, Magnol L, Sahún I, Brault V, Duchon A, et al. (2009) A new mouse model for the trisomy of the Abcg1-U2af1 region reveals the complexity of the combinatorial genetic code of down syndrome. Hum Mol Genet 18: 4756-4769.
- Zhang L, Meng K, Jiang X, Liu C, Pao A, et al. (2014) Human chromosome 21 orthologous region on mouse chromosome 17 is a major determinant of Down syndrome-related developmental cognitive deficits. Hum Mol Genet 23: 578-589.
- Kanehisa M, Goto S, Kawashima S, Nakaya A (2002) The KEGG databases at GenomeNet. Nucleic Acids Res 30: 42-46.
- Rueda N, Llorens-Martín M, Flórez J, Valdizán E, Banerjee P, et al. (2010) Memantine normalizes several phenotypic features in the Ts65Dn mouse model of Down syndrome. J Alzheimers Dis 21: 277-290.
- Lockrow J, Boger H, Bimonte-Nelson H, Granholm AC (2011) Effects of long-term memantine on memory and neuropathology in Ts65Dn mice, a model for Down syndrome. Behav Brain Res 221: 610-622.
- Costa AC, Scott-McKean JJ, Stasko MR (2008) Acute injections of the NMDA receptor antagonist memantine rescue performance deficits of the Ts65Dn mouse model of Down syndrome on a fear conditioning test. Neuropsychopharmacology 33: 1624-1632.
- Boada R, Hutaff-Lee C, Schrader A, Weitzenkamp D, Benke TA, et al. (2012) Antagonism of NMDA receptors as a potential treatment for Down syndrome: a pilot randomized controlled trial. Transl Psychiatry 2: e141.
- Hanney M, Prasher V, Williams N, Jones EL, Aarsland D, et al. (2012) Memantine for dementia in adults older than 40 years with Down's syndrome (MEADOWS): a randomised, double-blind, placebo-controlled trial. Lancet 379: 528-536.
- Ahmed MM, Dhanasekaran AR, Block A, Costa AC, Gardiner KJ (2013) Protein profiles associated with context fear conditioning and their modulation by memantine. Molec Cell Proteomics, in press.
Relevant Topics
- Advanced Parkinson Treatment
- Advances in Alzheimers Therapy
- Alzheimers Medicine
- Alzheimers Products & Market Analysis
- Alzheimers Symptoms
- Degenerative Disorders
- Diagnostic Alzheimer
- Parkinson
- Parkinsonism Diagnosis
- Parkinsonism Gene Therapy
- Parkinsonism Stages and Treatment
- Stem cell Treatment Parkinson
Recommended Journals
Article Tools
Article Usage
- Total views: 15491
- [From(publication date):
April-2014 - Apr 02, 2025] - Breakdown by view type
- HTML page views : 10863
- PDF downloads : 4628