Research Article Open Access
A Submersible Holographic Microscope for 4-D In-Situ Studies of Micro-Organisms in the Ocean with Intensity and Quantitative Phase Imaging
Lina M. Rotermund1, John Samson2 and Hans Jürgen Kreuzer3*
1Department of Physics & Atmospheric Science, Dalhousie University, Halifax, NS B3H 4R2, Canada
24Deep Inwater Imaging, 6589 Chebucto Rd, Halifax, NS B3L 1L9, Canada
3Department of Physics & Atmospheric Science, Dalhousie University, Halifax, NS B3H 4R2, Canada
- *Corresponding Author:
- Hans Jürgen Kreuzer
Department of Physics & Atmospheric Science
Dalhousie University, Halifax
NS B3H 4R2, Canada
Tel: +19024946594
E-mail: h.j.kreuzer@dal.ca
Received date: January 03, 2016 Accepted date: January 21, 2016 Published date: January 27, 2016
Citation: Rotermund LM, Samson J, Kreuzer HJ (2016) A Submersible Holographic Microscope for 4-D In-Situ Studies of Micro-Organisms in the Ocean with Intensity and Quantitative Phase Imaging. J Marine Sci Res Dev 6:181. doi:10.4172/2155-9910.1000181
Copyright: © 2016 Rotermund LM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Marine Science: Research & Development
Abstract
Digital holographic reconstructions yield the complex wave amplitude throughout a 3-D volume from which intensity and quantitative phase images can be obtained. A submersible digital holographic microscope with a point source has been used to demonstrate the feasibility of continuous monitoring of micro-organisms such as algae at any depth in oceans, lakes and rivers. The local refractive index can be obtained throughout the organisms. A tracking algorithm is used to study their mobility.
Keywords
4-D imaging; In water; Intensity images; Quantitative phase imaging
Introduction
With the steady increase of aquaculture sites in coastal areas monitoring water quality becomes mandatory to avoid pollution and resulting economic setbacks. Standard procedure to monitor water quality is to take samples which are then analyzed in the lab for chemical pollutants and for micro-organisms. This procedure is not only time consuming and costly but also leads to delays for any response to correct difficulties arising. This could be avoided by continuous in-situ remote monitoring. An instrument to do this for micro-organisms has been developed and is successfully used at the research level, namely the submersible holographic microscope [1] that not only is capable of registering and identifying micro-organisms and microscopic solids but also record variations as a function of time. It can be used at shallow depths but also at great depth such as in an exploration down to 6000 m in the South Atlantic [2], and even under extreme conditions such as the high Arctic [3]. In this feasibility study we have used such a microscope over the summer of 2015 to record micro-organisms in the North West Arm of Halifax Harbour in Nova Scotia, Canada. The aim is to show that this technique can be used for continuous in-situ monitoring of oceans, rivers and lakes.
Materials and Methods
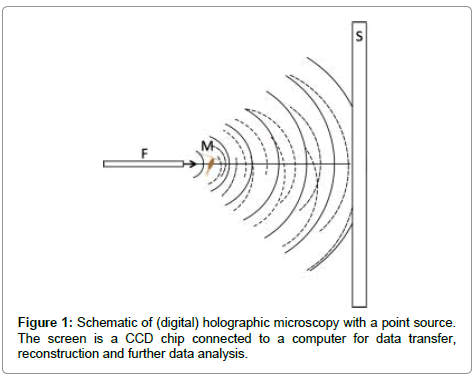
A brief introduction to digital in-line holographic microscopy (DIHM) with a point source is in order. Illumination is done with a point source which is either a pinhole illuminated by a laser or the end of a fibre optic cable. Both must be of the order of the wavelength of the laser so that the emanating waves are spherical as sketched in Figure 1. These waves travel through the volume to be imaged and scatter off any objects therein such as algae or oil drops, etc. The interference of the scattered waves with the primary wave from the point source creates an interference pattern, called a hologram that is recorded by a CCD chip. This hologram has no resemblance to the objects imaged. Therefore, in a second step the hologram is used in numerical reconstruction to produce a 3D image of all the objects in the volume illuminated. The reconstruction is based on the Helmholtz-Kirchhoff transform
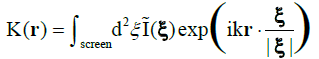 (1)
(1)
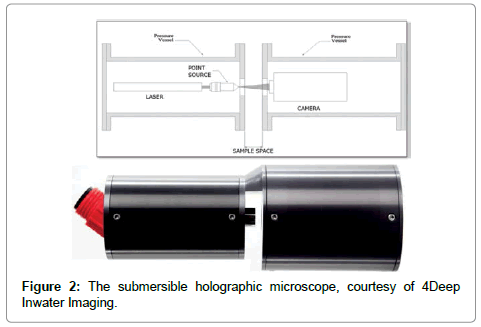
The screen is located at coordinates ξ = (X,Y,L), where L is the distance from the source to the screen centre, k = 2π/λ is the wave number of the radiation and I(ξ) is the (contrast) hologram. The algorithm is explained in detail in recent reviews, covering both the technical details and wide range of applications [4-6] and, in a second review [7], applications in the marine environment are discussed. What is important is the fact that K is a wave amplitude so that we obtain both intensity images, I(r) = KK*, and phase images, φ(r) = tan-1(im(K)/real(K)) where real(K) and im(K) are real and imaginary parts of K. As shown elsewhere [8] DIHM can determine the local phase throughout the object quantitatively. We will rely heavily on this duality of intensity and phase images. The schematics of the submersible microscope1 are shown in Figure 2 together with an instrument.
The left pressure chamber contains the laser and fibre optic cable from which light shines through a window into the gap between the two cylinders which together with the light scattered by the objects enters through a window into the right chamber and is recorded by a CCD chip. The data are either recorded on a hard drive or sent via an Ethernet cable ashore for further processing. Software packages are available not only for the reconstruction of the images but also for data analysis such as obtaining the size of the objects and their number both of which are recorded automatically and continuously on histograms as a function of time. The overriding advantage of this instrument is that all objects in the volume illuminated by the light source are recorded whereas a conventional microscope can only image objects in a narrow focal plane of typically less than 10 μm for a high resolution instrument. The DIHM instrument has a spatial resolution of better than 1 μm if a blue laser is used, and a depth of field of several millimeters. This is achieved by two tricks: (1) because the holograms are formed in water the wavelength of the laser is reduced [9] from λ = 0.408μm to λ/nw = 0.3 μm. (2) As shown elsewhere [10] two deconvolutions are used, one for the finite size of the source and a second for the finite size of the CCD chip. The temporal resolution is controlled by the frame rate of the CCD chip. The software also generates movies to study the motion of the micro-organisms in 3-D. This method of quantitative phase measurements has been used successfully to study cell death [11]. The instrument used here has several advantages: (1) it is a submersible microscope that allows in-situ, real time experiments. (2) the simplicity of the optical components (fibre optic cable, laser, CCD chip) with no lenses or mirrors as in off-line holography guarantee robustness of the instrument, and (3) the software allows real time reconstructions in a sub-second time frame.
Results and Discussion
This paper presents a brief survey of the microorganisms found in the North West Arm of Halifax Harbour using digital in-line holographic imaging. Most of the images were obtained with the submersible DIHM but some experiments were also done with the desktop version using water samples from the same location.
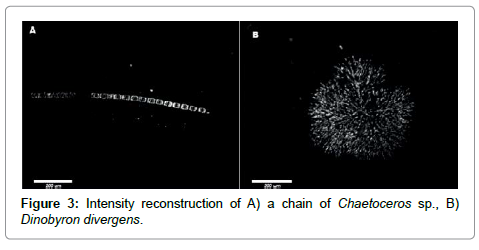
To demonstrate the quality of intensity images we show in Figure 3 a short chain of the diatom Chaetoceros socialis in panel A and a fresh water organism, Dynobyrn duvergens, in panel B. The latter were very abundant after heavy rains and were most likely washed into the ocean by run-off.
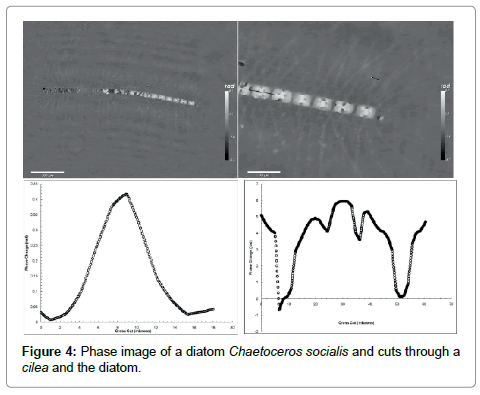
To examine the diatom further we show in Figure 4 its phase image. Two things are worth noticing: (1) the part that was black, i.e. not visible in the intensity plot can be seen in the phase image but only their outline indicating that they are empty of any material except for water. A suggestion would be that they are dead; yet they hold the whole structure together. In the enlargement of panel B we can also see the network of setae or cileae. To obtain further information we make a cross cut through one cilea, the short black line, depicted in panel C. The bell-like shape suggests that the cross section of the cilea is circular which results in a maximum phase shift relative through the center to the surrounding seawater
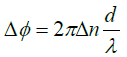 (2)
(2)
Here Δn is the difference in the refractive index of the cilea relative to water and a is its diameter. This shows clearly that our measurement of the index of refraction is a microscopic implementation of the Jamin-Lebedeff interferometer where the reference “beam” is simply that portion of the illuminating light that does not cross the sample [12]. The result is a local measurement of the index of refraction throughout the sample at the micron scale. A fit to the cross section gives d approximately 7 μm and Δn approximately 0.004 i.e. a refractive index of the cilea not much larger than water.
The cut through the cell in panel D is quite revealing. The cell is roughly 40 μm long. The dips to zero phase shift (relative to water) is the gap between two cells which is about 4 μm as one would expect for a van der Waals attraction through a water layer of that thickness. The big peak in the middle is most likely the nucleus of the cell and the two smaller peaks are perhaps oil drops. The cell wall or frustule made of SiO2 , shows up as the break in the flanks of the profile.
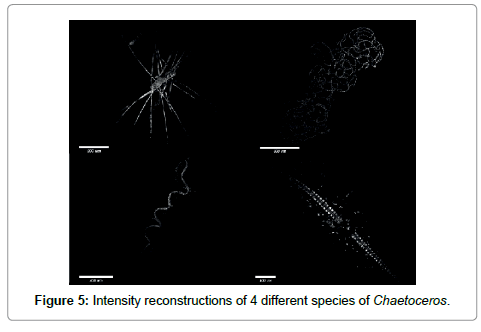
Going to the macroscopic scale we show in Figure 5 large aggregates of cells abundant in early spring. The large setae or cilea of the diatoms are capable of collecting debris (the bright spots surrounding the organisms). Their purpose seems to be defensive i.e. to irritate the gills of larger marine organisms.
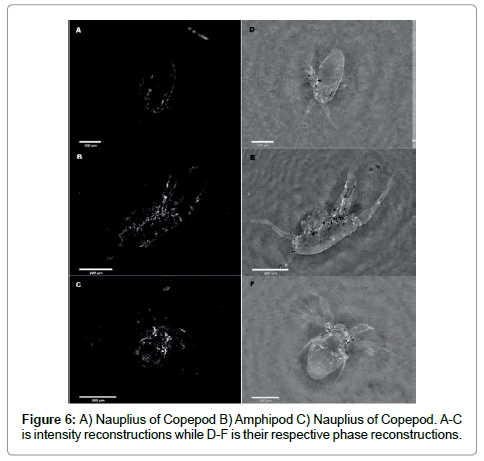
To demonstrate the use of both intensity and phase images we show examples for three organisms in Figure 6.
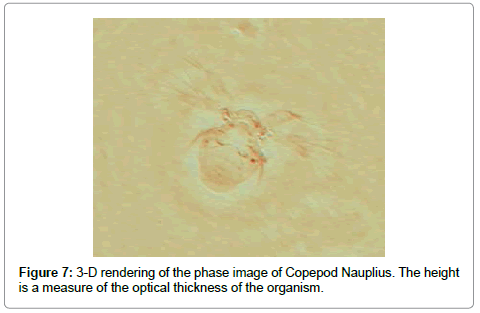
To dramatize phase imaging further we show in Figure 7 the phase image of Copepod Nauplius in a 3-D rendering which is a true representation of the optical thickness of the species. Of interest is the fact that the back of the organism is of the same (optical) thickness as the cilea.
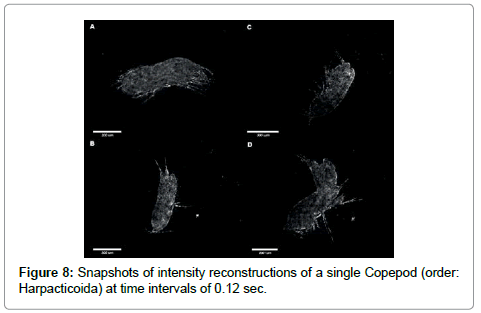
Our last demonstration is concerned with the 4-D capability of DIHM. By taking a sequence of holograms we can follow the swimming motion using three approaches. We can simply show the reconstructed intensity or phase images at successive times as fast as the frame rate of the CCD chip which can be shown as a movie or as a collection of still pictures. An example is given in Figure 8 for a Copepod.
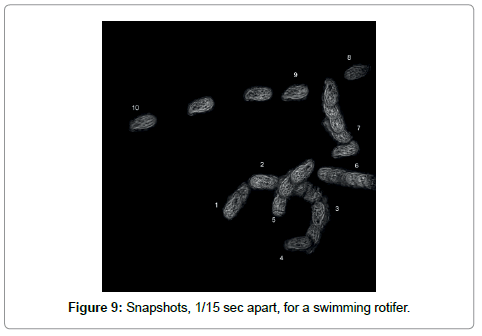
A second approach is to construct a composite hologram in which many holograms taken at different times are “added”. Technically this is possible because holograms are linear in the scattering amplitude and thus the superposition principle holds. Note that classical scattering is quadratic in the amplitude and superposition of exposures leads to a mess. The “addition” works as follows: Take holograms h1, h2, h3, h4,…. at times t1, t2, t3, t4,…. and then combine these holograms as (h1- h2)+(h3-h4) +....This packages n holograms into one of the same size. In addition to reducing the size this procedure also has the advantage of eliminating any static features that are not involved in the motion. Standard reconstruction will then produce the 3-D trajectories of many objects simultaneously. The trajectory of a rotifer is shown in Figure 9.
The actual measurement can be done in the Octopus software as shown in Figure 10.
From the length scale known in the reconstruction and the frame rate of the CCD camera we can estimate the swimming speed to be of the order of ν ~ 300 μm/s. Because we are in the low Reynolds number regime where frictional forces dominate we can go further and equate the c to the frictional energy mv2/2 = Ffrd where Ffr = 3πηdvλ is Stokes’ drag force. If we approximate the cell by a sphere of equal volume we have λ = 1 and if we approximate it by two touching spheres it is λ = 1.3. η is the viscosity of water. Likewise, the power needed to maintain a constant swim speed is this energy divided by the time it takes to swim the distance d. More on such analyses can be found elsewhere for the swimming behaviour of dinoflagellates [13].
Conclusions
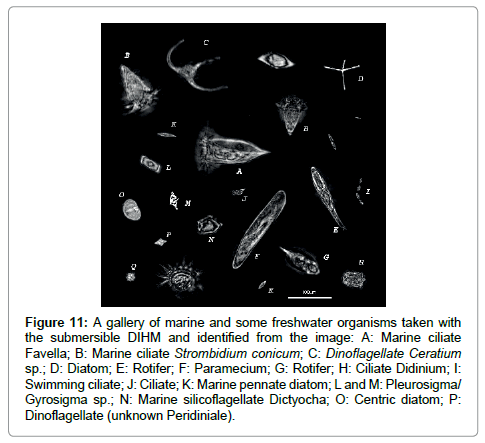
The purpose of this paper is to demonstrate that the submersible holographic microscope is the ideal tool to not only image microorganisms in-situ at any depth of water in oceans, rivers and lakes but to collect in-situ microscopic structural data on the organisms by also generating, from the same set of holographic data, phase images that by applying cross sectional cuts (in the data and NOT of the organism!) from which internal structure is revealed that previously could only be obtained by sophisticated, i.e. expensive and time-consuming methods like x-ray or electron microscopy. As we hope to have demonstrated the resolution is of the order of microns. To do better than that a high resolution desktop holographic microscope can be used to image organisms still alive and swimming in a water sample. Finally we show a gallery of micro-organisms in Figure 11.
Figure 11: A gallery of marine and some freshwater organisms taken with the submersible DIHM and identified from the image: A: Marine ciliate Favella; B: Marine ciliate Strombidium conicum; C: Dinoflagellate Ceratium sp.; D: Diatom; E: Rotifer; F: Paramecium; G: Rotifer; H: Ciliate Didinium; I: Swimming ciliate; J: Ciliate; K: Marine pennate diatom; L and M: Pleurosigma/ Gyrosigma sp.; N: Marine silicoflagellate Dictyocha; O: Centric diatom; P: Dinoflagellate (unknown Peridiniale).
Seawater samples were looked at in the lab and in situ measurements were taken. Up to 15 types of microorganisms were tentatively identified, however some were left unknown. Many more organisms (up to a few hundred) within the raw data were left out due to space and time limitations. While individual identification may be in error, it is impressive that each image has superb resolution showing minute features such as antennae, setae, ciliae and tails. Furthermore, there are 3 videos in the supplementary information showing time sequences of A) scallop larvae swimming, B) a Copepod and C) a Ceratium in the tidal current. In the Appendix we show a few more examples of micro-organisms found in our survey. Supplementary information accompanies the manuscript on the Light: Science & Applications website (http://www.nature.com/lsa/).
Acknowledgements
This work was supported by grants (HJK) from the Natural Sciences Research Council of Canada and from the Office of Naval Research, Washington DC. LMR would like to thank the Larouche lab at Dalhousie University for allowing John and I to use their submersible DIHM.
References
- 4Deep In water Imaging.
- Bochdansky AB, Jericho MH, Herndl GJ (2013) Development and deployment of a point-source digital inline holographic microscope for the study of plankton and particles to a depth of 6000m. LimnolOceanogr Methods 11:28-40.
- Jericho SK, Klages P, Nadeau J, Dumas EM, Jericho MH, et al. (2010) In-line digital holographic microscopy for terrestrial and exobiological research. Planet Space Sci 58: 701-705.
- Xu W, Jericho MH, Meinertzhagen IA, Kreuzer HJ (2001) Digital in-line holography for biological applications. Proc Natl Acad Sci 98: 11301-11305.
- Garcia-Sucerquia J, Xu W, Jericho SK, Klages P, Jericho MH, et al. (2006) Digital in-line holographic microscopy. Appl Optics 45: 836-850.
- Jericho MH, Kreuzer HJ (2011) In: Ferraro P, Wax A, Zalevsky Z (eds.) Point Source Digital In-line Holographic Microscopy. Coherent Light Microscopy. Berlin Heidelberg: Springer-Verlag 46: 3-30.
- Jericho SK, Jericho MH, Kreuzer HJ (2012) In: Emmanuel Reynaud (ed.) Holographic Microscopy of Marine Organisms. Imaging Marine Life: Modern Imaging Techniques in Marine Biology. Weinheim: Wiley-VCH 48-66.
- Jericho M, Kreuzer HJ, Kanka K, Riesenberg R (2012) Quantitative Phase and Refractive Index Measurements with Point Source Digital In-line Holographic Microscopy. Appl Optics 51: 1503-1515.
- Garcia-Sucerquia J, Xu W, Jericho MH, Kreuzer HJ (2006) Immersion digital in-line holographic microscopy. Opt Lett 31: 1211-1213.
- Nickerson BS, Kreuzer HJ (2013) Deconvolution for digital in-line holographic microscopy. Adv Opt Techn 2: 337-344.
- Missan S, Hrytsenko O (2015) Quantitative phase contrast imaging with digital inline holographic microscopy for viability assessment of mammalian cells in culture. Proc SPIE 9336: 93361-933615.
- Leertouwer HL, Wilts BD, Stavenga DG (2011) Refractive index and dispersion of butterfly chitin and bird keratin measured by polarizing interference microscopy. Opt Express 19:24061-24066.
- Lewis NI, Cembella AD, Xu W, Jericho MH, Kreuzer HJ (2006) Swimming Speed of three species of the marine dinoflagellate Alexandrium as determined by Digital In-line Holography. Phycologia 45: 61-70.
Relevant Topics
- Algal Blooms
- Blue Carbon Sequestration
- Brackish Water
- Catfish
- Coral Bleaching
- Coral Reefs
- Deep Sea Fish
- Deep Sea Mining
- Ichthyoplankton
- Mangrove Ecosystem
- Marine Engineering
- Marine Fisheries
- Marine Mammal Research
- Marine Microbiome Analysis
- Marine Pollution
- Marine Reptiles
- Marine Science
- Ocean Currents
- Photoendosymbiosis
- Reef Biology
- Sea Food
- Sea Grass
- Sea Transportation
- Seaweed
Recommended Journals
Article Tools
Article Usage
- Total views: 12752
- [From(publication date):
specialissue-2016 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 11722
- PDF downloads : 1030