A Rapid and Accurate Microfluidic Detection Method for Amino Acids in Cell Culture Medium
Received: 22-Mar-2022 / Manuscript No. jabt-22-58644 / Editor assigned: 24-Mar-2022 / PreQC No. jabt-22-58644(PQ) / Reviewed: 07-Apr-2022 / QC No. jabt-22-58644 / Revised: 11-Apr-2022 / Manuscript No. jabt-22-58644(R) / Published Date: 18-Apr-2022 DOI: 10.4172/2155-9872.1000448
Abstract
In current biopharmaceutical industry, the development of bioanalytical technology has received more and more attention. The development and optimization of cell culture medium, as one of the key components in biopharmaceutical research and development, largely determines the quality and efficiency of subsequent protein material production. Amino acids are crucial for the cultivation of mammalian cells and the understanding of the concentration of amino acids is important for increasing cell growth and protein productivity in recombinant cell lines. In the process of new drug development, drug proteins are usually expressed by cells or microorganisms. While cells or microorganisms grow and express proteins, nutrients such as amino acids will be continuously consumed. Therefore, different cell culture processes need to monitor the amino acid concentration in the medium to meet the needs of cells for amino acids in time.
Keywords
Amino Acids Analysis; ZipChip CE-MS; Cell Culture Medium
Case Report
The development of medium is an important part of the upstream cell culture process, and the detection of amino acid content can assist in the optimization of the medium process and promote the improvement of the expression of the target protein in cells. In commercial medium, in addition to its high cost, some components are unknown, and in some cases their impact on the quality of protein drugs cannot be accurately assessed. So, biopharmaceutical companies sometimes need to develop their own culture medium. Quick and timely access to the amino acid content of the culture can greatly speed up the medium development process.
At present, the most commonly applied method for determination of amino acid content in cell culture is the chromatography-based method (ion chromatography, reversed-phase liquid chromatography, hydrophilic interaction liquid chromatography). The drawbacks of these chromatography-based methods are low sensitivity, complicated column chemistries, low efficiency, low throughput and not easy to automate. Some methods even only can detect some limited amnio acids [1].
More recently, practical applications involving amino acid analysis are performed using the combination of ESI and/or MALDI-MS/ MS [2]. Among them, HPLC-ESI-MS is a powerful and attractive analytical tool and was recognized to have excellent performance for amino acid quantification. However, the user often faces the problem of ion suppression and blocking of spray needle caused by the complex composition. GC-MS is also proven particularly useful for amino acid analysis because of its excellent performance in separation. However, some challenges still exist in the failure analysis of nonvolatile amino acids (such as Methionine, Tryptophan, Histidine and Tyrosine) [3].
Capillary electrophoresis coupled to mass spectrometry with electrospray ionization (CE-ESI-MS) is recently arisen method used for amino acid analysis, which has been demonstrated to have a great mass selectivity and sensitivity. The method requires no sample derivatization prior to injection, and it simplifies sample preparation [4]. Rodrigues et al [5] reported a CE-MS method used for amino acid analysis of urine. In this method, a Beckman P/ACE MDQ CE coupled to a Bruker HCT Esquire 3000 plus ion trap mass spectrometer was used and samples were centrifuged and diluted with 0.1% formic acid solution before analysis. The total running time is 30mins and the LOQ of amino acids ranged from 1.9 μmol/L (Histidine) to 86 μmol/ L(cysteine).
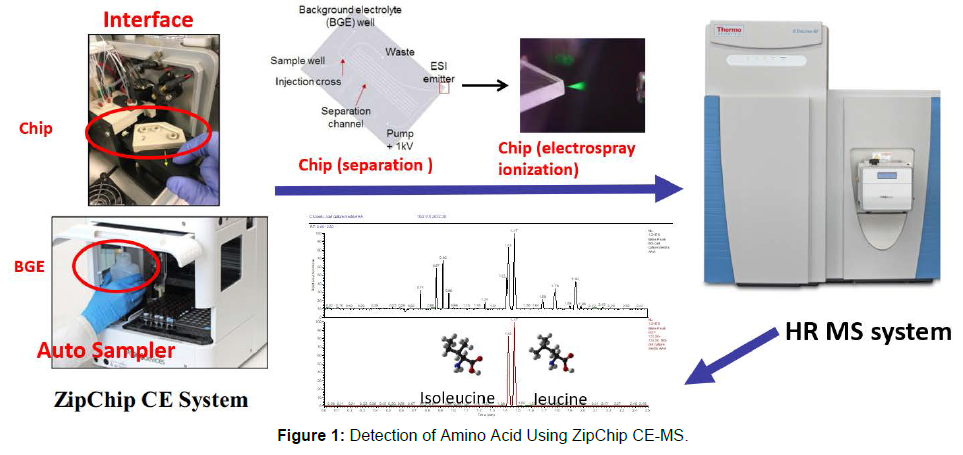
In current study, a faster, simpler and more sensitive amino acid detection method using 908 Devices ZipChip CE coupled to ThermoFisher QE Orbitrap Mass Spectrum (ZipChip CE-MS) was proposed. ZipChip microfluidic capillary electrophoresis developed by 908 devices is a novel capillary electrophoresis technology. To enable high throughput analysis, the workflow efficiently couples the microfluidic CE device with a robotic autosampler. The integration of multiple functional elements in a simple microfluidic platform to provide an important path to perform the sample handling, separation, and direct coupling of the CE separation to MS, leading to the rapid, sensitive and efficient separations for analytes [6].
The cell culture medium sample is thawed and centrifuged at 13000rpm for 15min, then the samples were diluted with 200 times of volume of diluent buffer (100mM NH4Ac/50%MeOH in H2O). The amino acid standard solution with a concentration of 1mM per amino acid was also diluted to the 0.01μM~60μM with the diluent buffer. A High-Speed (HS) Chip was loaded into the interface which was used to separate and ion spray with volatile background electrolyte (50%MeOH in H2O). The ZipChip parameters include the Field Strength of 1000V/cm, Injection Volume of 2.0nL, Replicate Delay of 30sec and Analysis time of 2.0min. The MS parameters comprise a scan range of 70~500m/z, resolution of 17500, scan type of Full MS, polarity of positive, Sheath gas flow rate of 20psi, and Capillary temp of 200℃. After acquisition, the MS data was analyzed and reported by the Xcalibur (Quan). From sample loading to completion of data collection, to output of quantitative analysis data, to detection report of 21 amino acids (20 common amino acids plus cystine) in a sample can be completed in as little as 3 minutes.
Compared with traditional CE-ESI-MS technology, it has high ionization efficiency and low sample load (up to nanoscale level), which improves the sensitivity and detection limit of mass spectrometry, extremely fast, sample detection within minutes, and enabling highthroughput detection. Based on the above considerations, we applied ZipChip CE-MS technology to the detection of amino acids in cell cultures to give full play to its advantages and obtain a product with strong specificity, high sensitivity, high accuracy, and analytical throughput (Figures 1 and 2).
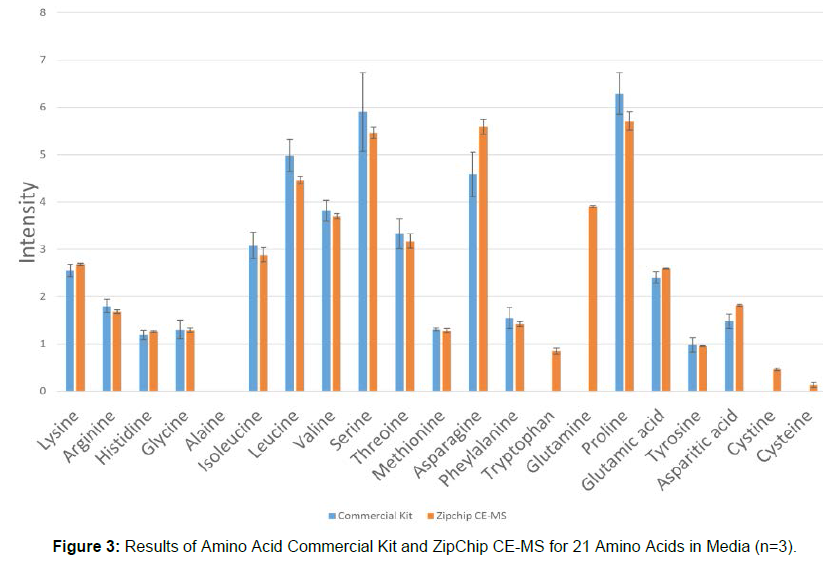
ZipChip CE-MS can detect amino acids in such a brief time, with very good accuracy. The analysis results of ZipChip CE-MS with a commercial amino acid analysis kit (HPLC method) were compare and shown as below (Figure 3).
From Figure 3 it is obvious that the amino acid analysis results of ZipChip CE-MS are highly consistent with those of the commercial amino acid analysis kit, and can also detect amino acids such as alanine, tryptophan, and cysteine that cannot be detected by the kit. The amino acid detection cycle of the kit is three days. In addition, the detection cost of this kit is several times higher than that of ZipChip CE-MS. It can be seen that the amino acid analysis technology based on ZipChip CE-MS is far superior to the traditional HPLC/UPLC method in terms of detection cycle and detection cost. Since the detection results of the two technologies are highly consistent, so the ZipChip CE-MS detection method can replace commercial kits for medium amino acids detection (Table 1).
| Items | Conventional HPLC Method | ZipChip CE-MS Method |
|---|---|---|
| Sample Pre-treatment | 1-2 day | < 20 min |
| Time of Analyzing | 0.5 hr – 1 hr | < 3 min |
| Types of Amino Acids | Only detect 17 amino acids (cannot detect Alanine, Tryptophan, Cysteine) | detect 20 amino acids (including Cystine) and some cell metabolites |
| Cost of Detection | Expensive | Cheap |
| Detection of Limit | 10-6mol/L-10-3mol/L | 10-9mol/L |
| Range of Linearity | Small | Broad |
| High Throughput | No | Yes |
| Data Analysis | Complicated integration, some amino acids are hard to be separated at baseline | Simple integration, complete separation at baseline, high degree of automation |
| Matrix Interference | Big interference | No or little interference |
| Method Development Difficulty | Hard | Easy |
Table 1: Comparison between HPLC and ZipChip CE-MS.
Due to the extremely low amount sample needed in ZipChip CE-MS, the sample pre-treatment is simple with good linearity. The loading capacity of ZipChip CE-MS is up to one thousandth of that of conventional HPLC method, due to the high analyte ionization efficiency and low background matrix interference, the amino acid signal detected by ZipChip CE-MS is greatly improved.
Using ZipChip CE-MS to analyze the amino acid composition of cell culture medium has the advantages of fast analysis speed, high data quality, and low cost. It is a high-throughput amino acid analysis technology. Compared with the mainstream amino acid analysis technology on the market, it can better meet the needs of rapid detection of amino acids in cell culture medium and plays a key role in promoting the development of cell culture.
References
- Dhillon M, Kumar S, Gujar G (2014) A common HPLC-PDA method for amino acid analysis in insects and plants . Indian J Exp Biol 52(1): 73-79.
- Dally JE, Gorniak J, Bowie R, Bentzley CM (2003) Quantitation of underivatized free amino acids in mammalian cell culture media using matrix assisted laser desorption ionization time-of-flight mass spectrometry. Anal Chem 75(19): 5046-5053.
- Williams BJ, Cameron, CJ, Workman R, Broeckling CD, Sumner LW, et al. (2007) Amino acid profiling in plant cell cultures: an inter-laboratory comparison of CE-MS and GC-MS . Electroohoresis 28(9): 1371-1379.
- Mayboroda OA, Christian N, Pelzing M, Zurek G, Derks R, et al. (2007) Amino acid profiling in urine by capillary zone electrophoresis-mass spectrometry . J Chromatogr A 1159: 149-153.
- Rodrigues KT, Mekahli D, Tavares MFM, Schep dael AV (2018) CS-MS for the analysis of amino acids. 37: 1039-1047.
- Erin A, Redman, Ramos-Payan M, Mellors JS, Ramsey JM (2016) Analysis of hemoglobin glycation using microfluidic CE-MS: a rapid, mass spectrometry compatible method for assessing diabetes management. Anal Chem 88: 5324-5330.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: Zhang Q, Tie L, Shi L (2022) A Rapid and Accurate Microfluidic Detection Method for Amino Acids in Cell Culture Medium. J Anal Bioanal Tech 10: 448. DOI: 10.4172/2155-9872.1000448
Copyright: © 2022 Zhang Q, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Tools
Article Usage
- Total views: 1895
- [From(publication date): 0-2022 - Apr 05, 2025]
- Breakdown by view type
- HTML page views: 1421
- PDF downloads: 474