Mini Review Open Access
A New Perspective on Microbiome and Resource Management in Wastewater Systems
Veera Gnaneswar Gude*Department of Civil and Environmental Engineering, Mississippi State University, USA
- Corresponding Author:
- Veera Gnaneswar Gude
Department of Civil and Environmental Engineering
Mississippi State University, Mississippi State, MS 39762, USA
Tel: 6623250345
E-mail: gude@cee.msstate.edu, gudevg@gmail.com
Received date:: March 03, 2015; Accepted date:: June 17, 2015; Published date:: June 23, 2015
Citation: Gude VG (2015) A New Perspective on Microbiome and Resource Management in Wastewater Systems. J Biotechnol Biomater 5:184. doi:10.4172/2155-952X.1000184
Copyright: © 2015 Gude VG. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
Wastewater treatment systems are predominantly based on biological processes that in principle derive energy from biochemical reactions involving substrate degradation available in the raw waste. The most commonly used technologies are suspended medium (activated sludge process), and attached growth or biofilm based technologies (tricking filters). In these processes, the key players responsible for environmental remediation are the prokaryotic microorganisms that live in communities. The key for optimizing these biological processes is by understanding the microbial ecology of the diverse populations in these systems and formulating appropriate tools of environmental biotechnology to predict their functional roles and their responses to the environmental stress. Environmental biotechnology plays an important role in developing sustainable wastewater treatment systems. This field demands knowledge on microbial ecology to identify the key players and maximize their populations by providing the optimum conditions while mitigating the competitors and other predator species and maintaining the robustness in microbial community structure to promote resilience during environmental stress.
With wastewater treatment systems being increasingly recognized as resource (energy, nutrients and water) recovery facilities, the role of microbiome and resource management is crucial for sustainable process development. Recent developments in molecular biology tools such as meta-omics have enhanced our ability to better understand these biological processes. Much greater discoveries or discovery-driven experimental planning and analytical approaches are needed to develop robust and resilient environmental systems. This mini-review focuses on the use of meta-omic tools to understand wastewater microbiology and potential integrated approaches for simultaneous and simple yet reliable analysis of the microbial systems in anaerobic digestion and microbial fuel cell systems which share several common features including the ability to produce energy and other valuable chemical and energy products under anaerobic conditions with complex microbial ecology.
Keywords
Microbiome; Wastewater; Metagenomics; Metatranscriptomics; Molecular biology; Metaproteomics; Metabolomics
Microbiome and Resource Management (MiRM)
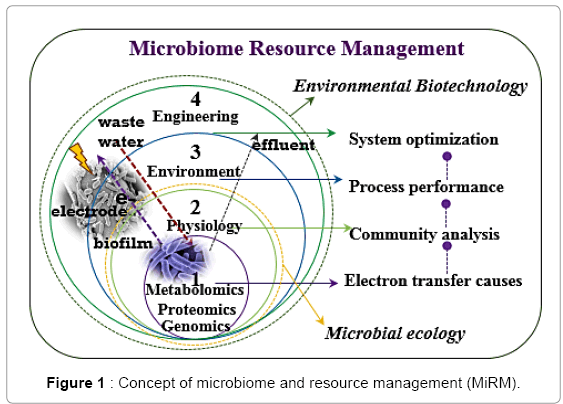
Indigenous microbial communities are the key players that fulfill essential functions of the biological processes designed for wastewater treatment. It is of paramount importance to manage these microbial communities for reliable and sustainable operations. Microorganisms in the wastewater treatment systems live under either suspended medium or attached growth conditions. In the suspended medium, the active biomass along with other inactive cell biomass is kept in suspension facilitating ready access to the available substrates in the wastewater. In attached growth systems, the microorganisms form assemblages that comprise one or more species. Biofilms also provide for highly structured matrix-enclosed communities separated by a network of open water channels which provides an excellent environment for cellcell interactions [1]. These interactions could result in intracellular exchange of genetic material, communication signals and metabolites that enable diffusion of necessary nutrients to the biofilm community. Study of biofilm communities would facilitate the development of better techniques for the bioremediation of contaminated sites and wastewaters. Microbiome and resource management (MiRM) essentially involves two major steps – (1) understanding the microbial ecology (populations, functions and responses to the environment) and (2) formulating appropriate environmental biotechnology that enables high performance of these biological systems designed for environmental remediation or energy and nutrient recovery [2,3]. A MiRM concept for electricity production from wastewaters is shown in Fig. 1. It shows that the fundamental understanding about the molecular biology techniques to characterize the wastewater microbiological communities for their significance, response to the environmental stressors and functions in the waste removal is essential for designing suitable techniques for successful operation of these facilities. For example, as shown in Fig. 1, the organic substrates in the wastewater can be directly converted into clean bioelectricity by employing the indigenous exoelectrogenic (electron releasing) microbial communities on the electrodes [4]. But this requires an understanding of the microbial community (genomics tools to identify “who is there”), and their physiology (metabolomics and proteomic tools to determine “what they are capable of doing”) to predict their responses to the environmental stressors such as the wastewater characteristics [3]. All this information will embedded together to provide suitable environment and biotechnological parameters that help enhance the survival of important key microbial communities and their performance for the desired environmental remediation application. Since wastewater systems are complex and diverse microbiological systems and different communities respond to the environmental stressors in relation to other communities, it is crucial to develop knowledge on how these communities are interacting with each other and whether that phenomenon is beneficial to the desired environmental remediation. This can be called “microbiome management”. in addition, wastewater is a source for various valuable products (water itself is a major resource), the whole system management can then be termed as “Microbiome and Resource Management” (MiRM) [5].
This paper discusses the importance of microbiome and resource management in wastewater systems in terms of functional key players and environmental stressors and responses that address the physiology of a microbiome. Activated sludge process was described to elaborate these two items followed by discussion on the molecular biological tools such as next generation technologies for microbiome management. The use of meta-omic tools to understand wastewater microbiology and development of integrated “omics” approaches for simultaneous and simple yet reliable analysis of the microbial systems in anaerobic digestion and microbial fuel cell systems which share several common features such as the ability to produce energy and other valuable chemical/fuel products under anaerobic conditions with complex microbial ecology are discussed.
MiRM in Wastewater Treatment Systems
Wastewater treatment systems are predominantly based on the biological processes that in principle derive energy from biochemical reactions and thrive on the substrates available in the raw waste. The most commonly used technologies are activated sludge process, aerated lagoons, oxidation ditch, biofilm and attached growth based technologies such as tricking filters, rotating biological contactors and moving bed or anaerobic fluidized bed reactors. Wastewater treatment systems have long been viewed as feasible means to provide suitable effluents that remove waste and provide environmental protection. The role of these facilities has been limited to the environmental sanitation until recently [6]. It was also acknowledged that the vast portion of the microbial communities that played major roles in the biological treatment of wastewater remained unknown [7]. With latest developments in microbiology assays and molecular biology tools, our knowledge of these “microsystems” has improved to a great extent. This enabled us to realize that the wastewater treatment systems can be designed with appropriate biological formulae to produce high quality effluents, to generate various forms of energy carriers and to recover valuable nutrients.
Activated sludge process
Various configurations/modifications of the activated sludge process are widely used for wastewater treatment throughout the world. Aerobic-anoxic-anaerobic units operations and processes are designed with the goal to remove organic (carbon) and nutrient (nitrogen and phosphorous) compounds from wastewaters. The prokaryotic microorganisms are reportedly predominant in these processes and are accounted for the biochemical reactions responsible for successful degradation of environmental pollutants [7,8]. Often, the performance of these systems is impeded by dominance of other microorganisms that manifest in activated sludge bulking, foaming and poor settling characteristics. All of this suggests that the resilience, robustness, environmental response and removal efficiencies depend on the composition, activities and cell to cell interactions of these communities. Nearly a hundred years of operating experience can be found in this technological area. However, the knowledge related to microbiology and its composition was not understood until too long ago. The microbial resource in the AS or any other biological process was conventionally managed by two essential steps: 1) identifying functional groups (key players); and 2) providing suitable environmental conditions.
Functional groups (key players)
Whether in natural systems or engineered (artificial) systems microbial communities are complex ranging from a simple microbial structure consisting of a few members such as in acid mine drainage to entirely diverse such as in soils, oceans and human guts [9-11]. The networks of biochemical functions within a microbial community are synchronized to perform critical functions to sustain an ecosystem, whether it be geochemical cycling, maintaining the health of an individual or treating waste for environmental remediation. A microbial community is also highly dynamic and is constantly responding to external stimuli [12]. Identifying the key players (functional microbial groups) responsible for desired environmental remediation needs is the first step to microbiome resource management. These functional groups often do not represent a single and consistent phylogenetic group [13]. A few examples include nitrifiers in activated sludge, and methanogens in anaerobic digesters. Wastewater engineers and microbiologists are in search of the “superbug” that survives process discrepancies and functions in extreme conditions while being effective, reliable and predictive. The rationale behind this is to optimize the process conditions suitable for these superbugs so that the processes can be designed to be most effective and economical. However, the implications from these approaches could be very diverse.
Physiological conditions (Environmental response)
Physiology refers to the functioning of a microorganism within the cell boundary and beyond in response to the environmental conditions (stressors). The microorganisms in the wastewater systems possess the capability to respond and acclimate to the changing influent concentrations such as organic and inorganic substances (potential substrates) and other inhibitory effects posed by toxic chemical elements, aerobic and anaerobic conditions and other pH and temperature influences. Understanding microbial physiology is a critical step in designing appropriate biotechnological tools for environmental remediation in wastewater systems and other systems in general.
Wastewater sources are very inconsistent in flow and composition and the characteristics. Prediction of microbiome performances is very complex since it consists of many unrealized communities that could play a significant role in environmental remediation. The functional performance of cells as well as sub-communities, including, for example, their metabolic pathways and replication, their interrelation and response to environmental parameters have to be considered and should be properly represented [14]. Every wastewater treatment plant is a complex ecosystem because wastewater destined for treatment includes fats, proteins, carbohydrates and many other substances that serve as nutrients for the resident bacteria. Countless bacterial species adapt to the living conditions in the water, compete for resources and each find a niche in which they can best survive.
The performance of wastewater treatment systems can depend on creating the conditions that induce the presence of specific types of bacteria, for example a balance of filamentous and zoogleal bacteria in activated sludge flocs, and certain types of denitrifiers in systems operated for nutrient control [7]. Few recent studies analyzed the core microbial communities in various wastewater treatment plants. One study reported that 60 genera of bacterial populations commonly shared by 14 different wastewater treatment plant samples, including Ferruginibacter, Prosthecobacter, Zoogloea, Subdivision 3 genera incertae sedis, Gp4, Gp6, etc., indicating a core microbial community in the microbial populations of wastewater treatment palnts at different geographic locations. The microbial community varies strongly in relation with wastewater temperature, conductivity, pH, and dissolved oxygen content [15]. The variance in these microbial communities could be independent of the geographic location of wastewater treatment plants and operating parameters.
“Omics” as Robust Tools for Mirm in Wastewater Systems
As shown in Figure 1, wastewater treatment systems engineering depends on understanding the metabolic pathways of each identified functional microbial group i.e. microbial ecology and their response to the inconsistent environment. The goal of microbial ecological studies is to enable prediction, modeling and management of the complex and heterogeneous microbes within a community to achieve the optimal process activity whether it be a wastewater treatment process, soil remediation and air pollution treatment and other processes of environmental concern. This analysis should be done accurately, rapidly and in economically efficient manner. Microbial communities in biological wastewater treatment processes have been analyzed by culture-dependent bottom-up reductionist approach which does not provide a comprehensive analysis of the microbial communities in the real wastewaters. The fact is that over 99 percent of the microbial communities are not cultivable. This could lead to a biased representation of certain populations in the samples. With the advent of culture-independent high-resolution technologies, it has now become possible to identify a wide array of these microbial communities with accuracy and precision. Among the “omic” tools or technologies are the genomics, transcriptomics, proteomics, and metabolomics. Genomics provides the analysis on the key players (i.e. who the community is and what it is capable of doing), while transcriptomics provides a functional assay of the environmental interactions and responses of the microbial community. Although the transcriptomics is limited by the technical challenges such as the depletion of the ribosomal RNA, this technique has already been applied on marine, fecal, and many other communities. Microbial functions are mediated by proteins, as such protein analysis could serve as the most convenient and direct estimator of the potential activity of the microbial community [16]. The protein extraction methods have recently advanced along with the development of shotgun proteomics using more sophisticated two-dimensional liquid chromatography separation methods and subsequent tandem mass spectrometry analysis.
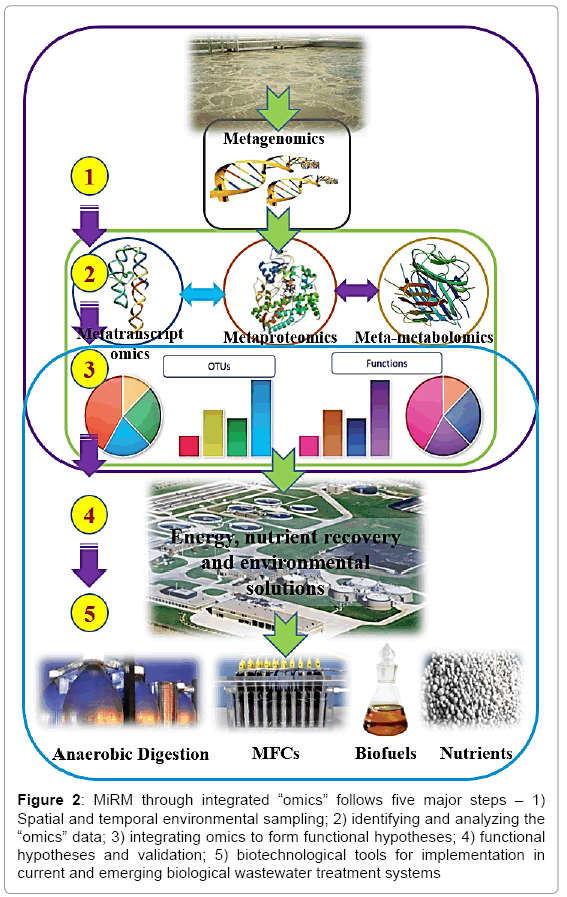
With the advent of next generation shot-gun technologies more breadth and depth of these fields is being analyzed known as “metagenomics”, “meta-transcriptomics”, “meta-proteomics”, and “metametabolomics” respectively (Figure 2). High throughput via nextgeneration sequencing platforms (e.g. Illumina HiSeq or Roche 454 FLX Titanium) are the state-of-the-art sequencing platforms which can sequence the genomes of single isolates as well as those belonging to a complex mixture of organisms via shotgun metagenomic sequencing [17,18]. This massive parallel sequencing has also enhanced the depth of 16S ribosomal RNA (rRNA) gene surveys much greater than Sanger sequencing [19-21] and the ability to increase the sequencing depth of phylogenetic markers broadened our knowledge of the diversity of a microbial community in various environments [22,23]. Further, the expression and regulation of transcripts and proteins provide a deeper insight into the metabolism and cell-to-cell interaction and the dynamic roles between members of a community. Transcriptomic (meta-) analysis of complex microbial communities [24,25], isolates and enriches messenger RNA (mRNA) from total RNA. Sequenced mRNA reads can be mapped back to the sequenced genomic data to quantitatively examine gene transcription [21,26-28]. Meta-proteomics provide important information on the proteins which catalyze the biochemical reactions within cells. This analysis is carried through liquid chromatography to separate the peptides, mass spectrometers and in silico algorithms to match the experimental mass fragments against those generated from genomic sequencing [29]. Mass-spectrometry results not only provide a profile of the protein expressed, but they can also be used to annotate genomes by more accurately identifying gene-encoding products [30]. In some cases, differences in amino acid sequences can be further used to differentiate strains within a microbial community [31].
Figure 2: MiRM through integrated “omics” follows five major steps – 1) Spatial and temporal environmental sampling; 2) identifying and analyzing the “omics” data; 3) integrating omics to form functional hypotheses; 4) functional hypotheses and validation; 5) biotechnological tools for implementation in current and emerging biological wastewater treatment systems
Integrated “omics” technologies – the way forward
Integrating genomics, transcriptomics, proteomics and metabolomics tools over spatially and temporally might give exciting results and more detailed and critical information on the microbiome and resource management especially for the intended energy and valuable resource recovery options. The immense computational challenge of assembling these vast and diverse meta-omic data to describe entire ecosystems, identifying how the genes interact with one another within and between bacteria to produce metabolites, and quantifying and understanding how these processes change over time and in response to the environment, is a complex task but can be achieved by developing appropriate molecular biology tools [32]. Integrating these new technologies with model organisms and culturebased approaches and then application to the natural environmental samples will help us to unravel key processes for sustainable wastewater treatment [33]. For example, environmental (meta-) genomics provides previously unobtainable insight into the ecophysiology and evolution of wastewater microorganisms, including uncultured functional key players. Comparative genomics can yield clues about the genomic variability among functionally similar organisms, thus increasing our understanding of structure–function relationships in microbial communities and potentially leading to better knowledge about the stability of key processes in wastewater systems [34]. Other metagenomics studies have revealed critical microbial species that have the capability for ladderane lipid synthesis and hydrazine metabolism in anammox bacteria [35]. This played a key role in developing energy and chemical efficient nitrogen removal process called one-stage oxygen limited autotrophic nitrification/denitrification (OLAND) process [36-38]. This process eliminates the need for carbon addition, reduces 90% sludge production and 60% oxygen requirements compared with other conventional nitrification-denitrification process. Also, about 40% overall costs can be reduced by treating the wastewaters at low BOD (biochemical oxygen demand) to N (nitrogen) ratio. While metagenomics studies have greatly contributed to the clarification of the functional capabilities of specific populations of interest, metaproteomics [39], meta-transcriptomics [40], and the combination of these two approaches have furthered our knowledge about the actual expression of relevant genes involved in key processes under different environmental conditions such as alternating anaerobic/aerobic phases. Thus, integrated “omics” over space and time [41] in combination with recorded physico-chemical parameters will allow reconstruction of the ecological networks and detailed definition of organismal niches. Such knowledge may then be used to identify key determinants of overall microbial community structure and function, which in turn may be harnessed for comprehensive reclamation of energy and other resource management in wastewater systems [42].
Integrated tools to optimize energy production and nutrient recovery
Since wastewater treatment plants are increasingly being recognized as “water and resource recovery facilities” (WRRF) due their potential to help derive various forms of valuable energy products and recover nutrients in the form of biosolids and fertilizers, the paradigm shift is to design these systems with pragmatic considerations that result in maximized recovery of energy, water and nutrient sources [6]. The pragmatic approach will need to focus on the theoretical possibility of the fundamental bioelectrochemical reactions and then considering practical solutions to develop a feasible technology. In this context, the molecular biology tools described earlier will advance our ability to connect the fundamental microbial ecology functions with the practical environmental biotechnology solutions to develop energy-positive wastewater treatment options with recovery of valuable resources such as water and nutrients. For example, Microthrix parvicella is considered a nuisance microorganism due to foam forming issues on the surface of aeration tanks. However, this bacterium has a capability to accumulate oils, lipids and fats available in wastewater systems. Understanding the microbial ecology and metabolism of these microorganisms can help harvest energy rich chemical compounds for potential biofuel production by exploiting this particular microorganism and the like [43].
A paradigm shift is also required in the microbiome and resource management as shown in Figure 2. Current practice is to collect environmental samples separately for analyzing the genomics, transcriptomics, and proteomics and metabolomics individually and then analyze the data to construct a microbiome. This needs to change. To develop robust and resilient energy-positive and resource recovering wastewater systems, a five step formula is necessary.
Step 1 - Environmental samples need to be collected to represent over spatial and temporal variations within the wastewater systems and if possible from different geographic locations, i.e. different wastewater treatment plants.
Step 2 - Process the environmental samples through various omics methods to generate the meta-data and consider simultaneous and single (no sample splitting or different samples for different omics analysis) sample analysis via integrated omics approaches.
Step 3) Evaluate the width and depth of the meta data that generates functional Operational Taxonomic Units (OTUs) and associated environmental response hypotheses
Step 4 - Test hypotheses developed in Step 3 in pilot scale or real wastewater systems
Step 5 -Develop robust biotechnological tools for application in environmental remediation with resource recovery benefits in wastewater systems. These tools can be applied to the existing and emerging resource recovery facilities.
Two most promising technologies for providing energy-positive wastewater treatment that require rapid and significant developments in microbial ecology and environmental biotechnology tools are anaerobic digestion (AD) and microbial fuel cells (MFCs) which will be discussed in later sections.
Anaerobic digestion
Anaerobic digestion technology has been referred to as an ideal biological process for removing organic pollutants from wastewaters. The two distinct advantages of this technology over activated sludge process are low sludge production without aeration requirements and its potential to produce biogas, a valuable energy source. Although the application of this technology is mainly focused in stabilization of sludge in the wastewater treatment plants, it is now widely applied in low strength to high strength industrial wastewaters, complex wastewater with persistent and recalcitrant chemical compounds and with above-normal temperatures [44]. Even with several decades of practical knowledge with the anaerobic digestion systems in various applications, the microbiology and the involved biochemical reactions were not completely understood. Earlier studies used the first generation molecular biology techniques namely Sanger sequencing, fluorescence in situ hybridization (FISH), terminal restriction-fragment length polymorphism (T-RFLP), and chemical or temperature denaturing gradient gel electrophoresis (DGGE) to target the 16S rRNA gene of bacteria and archaea to analyze the phylogenetic profiles of the microbial communities involved in anaerobic digestion [45,46] but with low-throughput meaning that the width and breadth of the microbiology of the samples was not captured in detail. 16 sRNA gene clones have revealed various prokaryotic taxa such as the phyla Proteobacteria (mainly in the class Deltaproteobacteria), Chloroflexi, Firmicutes, Spirochaetes, and Bacteroidetes in the domain Bacteria. Similarly, clones in the classes Methanomicrobia, Methanobacteria, and Thermoplasmata in the domain Archaea are those of typical phylotypes found in such sludges [47]. In addition to these relatively known taxa, phylotypes belonging to a variety of uncultured candidate phyla (or classes) (known as ‘clone cluster’) were often detected in these sludges [47-53]. Many uncultured taxonomic groups were also identified. Finding key (or dominant) populations that belong to such uncultured lineages at various taxonomic levels (from species to phylum levels) is one of the major advances in the microbiology of anaerobic digestion processes in the past few years which helped increase their efficiency [44].
Advanced molecular biology techniques (High-throughput shotgun metagenomic sequencing and amplicon sequencing of the 16S rRNA gene) have been applied to understand the microbial communities involved in anaerobic digestion [54,55], while metatranscriptomic [56] and metaproteomic [57,58] analyses have begun to be employed in the past two years. However, the application of all three meta-omics methods in parallel to systematically query the metabolism of the microbial community under different conditions has yet to be widely reported. There are many questions that need attention as we try understand the response of the anaerobic microbial ecosystem and its responses to the environmental stressors. This is crucial to develop anaerobic systems that produce high methane yields with minimal process failures. This could be achieved by integrating the different “omics” methods which may potentially address all the dynamics involved in a microbial community simultaneously. Integrated meta-omics approach including metagenomics, metatranscriptomics, and metaproteomics may help to query the molecular responses at the DNA, RNA and protein levels, respectively, of biomethane-producing microbial communities under normal and stress conditions. In general, the mechanism by which members within a microbial community respond to external stresses is not well understood, and anaerobic digestion can serve as a model system to enhance our understanding of the characteristics of anaerobic microbial communities. Ultimately, knowledge gained in this study could be extrapolated to other microbial communities present in anaerobic environments.
Microbial fuel cells (MFCs)
MFCs exploit the energy metabolism of microbes that transfer the electrons to the conductive surfaces called electrodes [59]. Microorganisms responsible for electron release, transfer and acceptance are mostly populated on the electrodes in the form of biofilms. Biofilms also grow on all the components of the MFCs either naturally or artificially. This is facilitated by either direct contact of the cell with the electrode surface via electroactive biofilm or by the mediator species that serve as electron shuttles. Microbes within a biofilm will enzymatically extract electrons from organic components in the surrounding media and transfer them to the electrode, which serves as an electron acceptor for biological respiration and/or maintenance. Completion of the MFC reaction takes place in a physically separate but electronically and ionically linked compartment where different bacterial biofilms use the cathode electrode as a source for energy during the reduction of oxidants such as nitrate, sulfate, fumarate, or oxygen [60-62]. Although MFCs research has over a hundred years of history, research in this was reincarnated in 1960s and it was discovered that indigenous microorganisms (exoelectrogenic or electroactive) are able to produce electrons from electron donors such as wastewater and other waste sources and transfer them to the electrodes with the help of soluble mediators such ferricyanide. However, it was realized that this mediated process is not feasible for practical viability of the MFC technology leaving scope for deepening our understanding of the microbial reactions and their interactions especially in the electron release, transfer and acceptance process.
Our understanding of the microbial ecology of the MFCs is in its early stages of evolution. Direct transfer of electrons by means of cell membrane bound redox-proteins (extracellular electron transfer) was studied as a major pathway for Shewanella oneidensis, Geobacter sulfurreducens, and Rhodoferax ferrireducens extensively [63]. Effective extracellular electron transfer is one of the hallmark physiological features of Geobacter species [64]. It was also reported that these microorganisms are able to develop electrically conductive pilus-like filaments, also known as bacterial nanowires [65]. This leads to the hypotheses that the microbial biofilms on the MFC electrodes are able to achieve electron transfer from cell to cell and then to the electrode by means of these bacterial nanowires. However, the exact pathways of the electron transfer and the importance of specific proteins is still under investigation [66]. Concerning mixed microbial communities, recent MFCs studies using different susbstrates (acetate, ethanol, lactate, propionate, butyrate, formate, succinate, glucose, chocolate-; domestic-; winery-; and dairy manure-wastewaters) have reported abundance of the following anode biofilm populations as exoelectrogenic: Geobacter sulfurreducens, Pelobacter propionicus, Thauera aromatic, Azoarcus sp., Desulfuromonas sp., Bacillus sp., and Clostridium sp. [67]. It should be noted that the patterns of microbial film formation and their composition varied with different substrates. In all of these studies, a major portion of the biofilm samples remained unidentified. In some cases, a small portion of the microbial community on the biofilms was identified to have significant or most contribution in the electricity generation. Also, the role of non-electroactive microorganisms in electron transfer is not known to date. To gain better understanding on the dynamics of the microbial populations in electrode biofilms, a close monitoring of the evolution of microbial populations in the biofilms (both anode and biocathode) and correlate the process performance to the environmental stressors and the microbial ecology using integrated meta-analyzes seems to be an important and interesting approach.
While understanding the fundamental electron transfer causes is crucial to the development of MFCs, even more important is to develop knowledge on the functional dynamics of this microbial system. To develop reliable MFC operations for sustainable electricity production, microbial communities with stable metabolic function over time, despite the unavoidable perturbations and disturbances that occur in real world wastewater systems should be investigated. Three important microbial ecology factors that play critical roles in maintaining a stable and robust community function in MFC biofilms are thus (1) functionality; (2) evenness in structure; and (3) population dynamics. Functionality refers to having diverse microbial community capable of diverse metabolic pathways while evenness in microbial diversity structure provide robust metabolic function and the population dynamics are important to adjust to the environmental stressors and disturbances. This can be achieved by implementing the MiRM steps indicated in Fig. 2. Since knowledge of the electron transfer mechanisms by exoelectrogenic bacteria alone is not adequate to develop robust systems, cell to cell interactions, community-wide synergism, and ecological networks and communication matrix can only be comprehensively studied using the integrated omics approach as explained earlier.
Concluding Remarks
Wastewater systems are defined to be stable when they have a very diverse microbial community or ecology which enhances their ability to respond to various environmental stressors by optimizing the metabolic pathways. This helps maintain the mass and energy transfer within the system. However, the diversity of the microbial system also poses some major challenges for the environmental scientists and engineers who strive to engineer and manage these systems for environmental remediation. The recent developments in molecular biology tools created exciting opportunities and enhanced our ability to understand these complex microbial communities. With the advent of next generation sequencing and meta-data analyzes, our knowledge on the functional dynamics of the microbial populations has been significantly improved. With increasing expectations on the performances of these engineered systems especially for wastewater systems for their potential role in becoming energy-positive systems via biofuel production and resource recovery systems such as nutrients and high quality effluents in the near future, our use of these advanced technologies through integrated approaches needs to expand over new vistas where these meta-omic technologies can be exploited simultaneously to develop stable and robust environmental systems for reliable operations that provide for sustainable energy and resource recovery.
Acknowledgements
This work was supported by the Office of Research and Economic Development (ORED), Bagley College of Engineering (BCoE), and the Department of Civil and Environmental Engineering (CEE) at Mississippi State University.
References
- Singh R, Paul D, Jain RK (2006) Biofilms: implications in bioremediation. Trends Microbiol 14: 389-397.
- Ritt mann BE (2006) Microbial ecology to manage processes in environmental biotechnology. Trends Biotechnol 24: 261-266.
- Verstraete W (2007) Microbial ecology and environmental biotechnology. ISME J 1: 4-8.
- Logan BE (2009) Exoelectrogenic bacteria that power microbial fuel cells. Nat Rev Microbiol 7: 375-381.
- Verstraete W, Wittebolle L, Heylen K, Vanparys B, De Vos P, et al. (2007) Microbial resource management: the road to go for environmental biotechnology. Engineering in Life Sciences 7: 117-126.
- Gude VG (2015) Energy and water autarky of wastewater treatment and power generation systems. Renewable and Sustainable Energy Reviews 45: 52-68.
- Wagner M, Loy A (2002) Bacterial community composition and function in sewage treatment systems. CurrOpinBiotechnol 13: 218-227.
- Wagner M, Loy A, Nogueira R, Purkhold U, Lee N, et al. (2002) Microbial community composition and function in wastewater treatment plants. Antonie Van Leeuwenhoek 81: 665-680.
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, et al. (2004) Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37-43.
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, et al. (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66-74.
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, et al. (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59-65.
- Ottesen EA, Young CR, Eppley JM, Ryan JP, Chavez FP, et al. (2013) Pattern and synchrony of gene expression among sympatric marine microbial populations. ProcNatlAcadSci U S A 110: E488-497.
- McMahon KD, Martin HG, Hugenholtz P (2007) Integrating ecology into biotechnology. CurrOpinBiotechnol 18: 287-292.
- Koch C, Müller S, Harms H, Harnisch F (2014) Microbiomes in bioenergy production: from analysis to management. CurrOpinBiotechnol 27: 65-72.
- Wang X, Hu M, Xia Y, Wen X, Ding K (2012) Pyrosequencing analysis of bacterial diversity in 14 wastewater treatment systems in China. Appl Environ Microbiol 78: 7042-7047.
- Myrold DD, Zeglin LH, Jansson JK (2014)The potential of metagenomic approaches for understanding soil microbial processes. Soil Science Society of America Journal 78: 3-10.
- Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, et al. (2011) Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331: 463-467.
- Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, et al. (2012) Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337: 1661-1665.
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, et al. (2012) Ultra-high-throughput microbial community analysis on the IlluminaHiSeq and MiSeq platforms. ISME J 6: 1621-1624.
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, et al. (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. ProcNatlAcadSci U S A 108 Suppl 1: 4516-4522.
- Claesson MJ, Wang Q, O'Sullivan O, Greene-Diniz R, Cole JR, et al. (2010) Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic acids Res 38:e200.
- Pedrós-Alió C (2007) Ecology. Dipping into the rare biosphere. Science 315: 192-193.
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, et al. (2006) Microbial diversity in the deep sea and the underexplored "rare biosphere". ProcNatlAcadSci U S A 103: 12115-12120.
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, et al. (2008) Microbial community gene expression in ocean surface waters. ProcNatlAcadSci U S A 105: 3805-3810.
- Lesniewski RA, Jain S, Anantharaman K, Schloss PD, Dick GJ (2012) The metatranscriptome of a deep-sea hydrothermal plume is dominated by water column methanotrophs and lithotrophs. ISME J 6: 2257-2268.
- Baker BJ, Lesniewski RA, Dick GJ (2012) Genome-enabled transcriptomics reveals archaeal populations that drive nitrification in a deep-sea hydrothermal plume. ISME J 6: 2269-2279.
- Shi Y, Tyson GW, DeLong EF (2009) Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column. Nature 459: 266-269.
- Shi Y, Tyson GW, Eppley JM, DeLong EF (2011) Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J 5: 999-1013.
- VerBerkmoes NC, Denef VJ, Hettich RL, Banfield JF (2009) Systems biology: functional analysis of natural microbial consortia using community proteomics. Nature Reviews Microbiology 7: 196-205.
- Armengaud J (2013) Microbiology and proteomics, getting the best of both worlds! Environ Microbiol 15: 12-23.
- Denef VJ, VerBerkmoes NC, Shah MB, Abraham P, Lefsrud M, et al. (2009) Proteomics-inferred genome typing (PIGT) demonstrates inter-population recombination as a strategy for environmental adaptation. Environmental microbiology 11: 313-325.
- Raes J, Bork P (2008) Molecular eco-systems biology: towards an understanding of community function. Nat Rev Microbiol 6: 693-699.
- Morgan XC, Segata N, Huttenhower C (2013) Biodiversity and functional genomics in the humanmicrobiome. Trends in genetics 29: 51-58.
- Daims H, Taylor MW, Wagner M (2006) Wastewater treatment: a model system for microbial ecology. Trends Biotechnol 24: 483-489.
- Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, et al. (2006) Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440: 790-794.
- Kuai L, Verstraete W (1998) Ammonium removal by the oxygen-limited autotrophic nitrification-denitrification system. Applied and Environmental Microbiology 64: 4500-4506.
- Pynaert K, Smets BF, Wyffels S, Beheydt D, Siciliano SD, et al. (2003) Characterization of an autotrophic nitrogen-removing biofilm from a highly loaded lab-scale rotating biological contactor. Applied and Environmental Microbiology 69: 3626-3635.
- Vlaeminck SE, Terada A, Smets BF, De Clippeleir H, Schaubroeck T, et al. (2010) Aggregate size and architecture determine microbial activity balance for one-stage partial nitritation and anammox. Applied and environmental microbiology 76: 900-909.
- Wilmes P, Andersson AF, Lefsrud MG, Wexler M, Shah M, et al. (2008) Community proteogenomics highlights microbial strain-variant protein expression within activated sludge performing enhanced biological phosphorus removal. The ISME journal 2: 853-864.
- Yu K, Zhang T (2012) Metagenomic and metatranscriptomic analysis of microbial community structure and gene expression of activated sludge. PLoS One 7: e38183.
- Muller EE, Glaab E, May P, Vlassis N, Wilmes P (2013) Condensing the omics fog of microbial communities. Trends Microbiol 21: 325-333.
- Sheik AR, Muller EE, Wilmes P (2014) A hundred years of activated sludge: time for a rethink. Front Microbiol 5: 47.
- Muller EE, Pinel N, Gillece JD, Schupp JM, Price LB, et al. (2012) Genome sequence of “CandidatusMicrothrixparvicella” Bio17-1, a long-chain-fatty-acid-accumulating filamentous actinobacterium from a biological wastewater treatment plant. Journal of bacteriology 194: 6670-6671.
- Narihiro T, Sekiguchi Y (2007) Microbial communities in anaerobic digestion processes for waste and wastewater treatment: a microbiological update. CurrOpinBiotechnol 18: 273-278.
- Rittmann BE, Krajmalnik-Brown R, Halden RU (2008) Pre-genomic, genomic and post-genomic study of microbial communities involved in bioenergy. Nat Rev Microbiol 6: 604-612.
- Rivière D, Desvignes V, Pelletier E, Chaussonnerie S, Guermazi S, et al. (2009) Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J 3: 700-714.
- Sekiguchi Y, Kamagata Y (2004) Microbial community structure and functions in methane fermentation technology for wastewater treatment. Strict and Facultative Anaerobes: Medical and Environmental Aspects 361-384.
- Sekiguchi Y (2006) Yet-to-be cultured microorganisms relevant to methane fermentation processes. Microbes and Environments 21: 1-15.
- Collins G, O'Connor L, Mahony T, Gieseke A, de Beer D, et al. (2005) Distribution, localization, and phylogeny of abundant populations of Crenarchaeota in anaerobic granular sludge. Appl Environ Microbiol 71: 7523-7527.
- Chouari R, Le Paslier D, Daegelen P, Ginestet P, Weissenbach J, et al. (2005) Novel predominant archaeal and bacterial groups revealed by molecular analysis of an anaerobic sludge digester. Environmental Microbiol 7: 1104-1115.
- Chouari R, Le Paslier D, Dauga C, Daegelen P, Weissenbach J, et al. (2005) Novel major bacterial candidate division within a municipal anaerobic sludge digester. Applied and environmental microbiology 71: 2145-2153.
- Godon JJ, Morinière J, Moletta M, Gaillac M, Bru V, et al. (2005) Rarity associated with specific ecological niches in the bacterial world: the 'Synergistes' example. Environ Microbiol 7: 213-224.
- Leclerc M, Delgènes JP, Godon JJ (2004) Diversity of the archaeal community in 44 anaerobic digesters as determined by single strand conformation polymorphism analysis and 16S rDNA sequencing. Environmental microbiol 6: 809-819.
- Wirth R, Kovács E, Maróti G, Bagi Z, Rákhely G, et al. (2012) Characterization of a biogas-producing microbial community by short-read next generation DNA sequencing. Biotechnol Biofuels 5: 41.
- Wong MT, Zhang D, Li J, Hui RKH, Tun HM, et al. (2013)Towards a metagenomic understanding on enhanced biomethane production from waste activated sludge after pH 10 pretreatment. Biotechnol Biofuels 6: 38.
- Zakrzewski M, Goesmann A, Jaenicke S, Jünemann S, Eikmeyer F, et al. (2012) Profiling of the metabolically active community from a production-scale biogas plant by means of high-throughput metatranscriptome sequencing. Journal of biotechnology 158: 248-258.
- Hanreich A, Schimpf U, Zakrzewski M, Schlüter A, Benndorf D, et al. (2013)Metagenome and metaproteome analyses of microbial communities in mesophilic biogas-producing anaerobic batch fermentations indicate concerted plant carbohydrate degradation. Systematic and applied microbiology, 36: 330-338.
- Heyer R, Kohrs F, Benndorf D, Rapp E, Kausmann R, et al. (2013) Metaproteome analysis of the microbial communities in agricultural biogas plants. N Biotechnol 30: 614-622.
- Bretschger O1, Osterstock JB, Pinchak WE, Ishii S, Nelson KE (2010) Microbial fuel cells and microbial ecology: applications in ruminant health and production research. MicrobEcol 59: 415-427.
- He Z, Angenent LT (2006) Application of bacterial biocathodes in microbial fuel cells. Electroanalysis 18: 2009-2015.
- Clauwaert P, Rabaey K, Aelterman P, de Schamphelaire L, Pham TH, et al. (2007) Biological denitrification in microbial fuel cells. Environ SciTechnol 41: 3354-3360.
- Chen GW, Choi SJ, Lee TH, Lee GY, Cha JH, et al. (2008) Application of biocathode in microbial fuel cells: cell performance and microbial community. ApplMicrobiolBiotechnol 79: 379-388.
- Gorby YA, Yanina S, McLean JS, Rosso KM, Moyles D, et al. (2006) Electrically conductive bacterial nanowires produced by Shewanellaoneidensis strain MR-1 and other microorganisms. Proceedings of the National Academy of Sciences 103: 11358-11363.
- Lovley DR, Ueki T, Zhang T, Malvankar NS, Shrestha PM, et al. (2011) Geobacter: the microbe electric's physiology, ecology, and practical applications. AdvMicrobPhysiol 59: 1-100.
- Logan BE1, Regan JM (2006) Electricity-producing bacterial communities in microbial fuel cells. Trends Microbiol 14: 512-518.
- Rosenbaum M, Angenent LT (2010)Genetically modified microorganisms for bioelectrochemical systems. Bioelectrochemical Systems: From Extracellular Electron Transfer to Biotechnological Application 101-117.
- Kiely PD, Cusick R, Call DF, Selembo PA, Regan JM, et al. (2011) Anode microbial communities produced by changing from microbial fuel cell to microbial electrolysis cell operation using two different wastewaters. Bioresource technology 102: 388-394.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 16830
- [From(publication date):
August-2015 - Jul 15, 2025] - Breakdown by view type
- HTML page views : 12035
- PDF downloads : 4795