Research Article Open Access
A Hybrid Cnn-Rf Method for Electron Microscopy Images Segmentation
Guibao Cao1, Shuangling Wang1, Benzheng Wei2, Yilong Yin1* and Gongping Yang11 School of Computer Science and Technology, Shandong University, Jinan, China
2College of Science and Technology, Shandong University of Traditional Chinese Medicine, Jinan, China
- Corresponding Author:
- Yilong Yin
School of Computer Science and Technology
Shandong University, Jinan, China
E-mail: ylyin@sdu.edu.cn
Received date August 16, 2013; Accepted date October 05, 2013; Published date October 15, 2013
Citation: Cao G, Wang S, Wei B, Yin Y, Yang G (2013) A Hybrid Cnn-Rf Method for Electron Microscopy Images Segmentation. J Biomim Biomater Tissue Eng 18:114. doi:10.4172/1662-100X.1000114
Copyright: © 2013 Cao G, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biomimetics Biomaterials and Tissue Engineering
Abstract
To get new insights into the function and structure of the brain,neuroanatomists need to build 3D reconstructions of brain tissue from electron microscopy (EM) images. One key step towards this is to get automatic segmentation of neuronal structures depicted in stacks of electron microscopy images. However, due to the visual complex appearance of neuronal structures, it is challenging to automatically segment membranes in the EM images. Based on Convolutional Neural Network (CNN) and Random Forest classifier (RF), a hybrid CNN-RF method for EM neuron segmentation is presented. CNN as a feature extractor is trained firstly, and then well behaved features are learned with the trained feature extractor automatically. Finally, Random Forest classifier is trained on the learned features to perform neuron segmentation. Experiments have been conducted on the benchmarks for the ISBI2012 EM Segmentation Challenge, and the proposed method achieves the effectiveness results: The Rand error, Warping error and Pixel error attains to 0.109388991, 0.001455688 and 0.072129307, respectively.
Keywords
Biomedical image processing; Membrane segmentation; Feature learning; Convolutional neural network; Random forest
Introduction
Neuroanatomists face the tough task of reconstructing neuronal structure with synaptic resolution in order to gain insights into the functional connectivity of brain. Currently, EM is the main imaging tool which can provide sufficient resolution for studying connections at the neuron level. This imaging device produces amounts of image data. In order to understand the patterns, the image data should be segmented according to structural and functional modules. For human neuroanatomist, segmentation of neuro-images is a trivial task but, unfortunately, it is very time consuming [1]. Therefore, accurate algorithms for automatic neuronal segmentation are indispensable for large scale geometric reconstruction of densely interconnected neuronal tissue. The neuron images have the characteristics of medical image imaging and they have its own characteristics with respect to general medical image as well. Nevertheless, its structure is complexity, such as intricate topology, various cell interference inside, and noisy textures. In addition, the poor quality of the imaging tools causes the border lacking and fuzzy. Those problems make the automatic segmentation of neuronal electron microscopy images very difficult. Therefore, the accurate segmentation needs more distinctive features in detail naturally.
Supervised machine learning methods have been proved to be effective for detecting membranes in EM images in recent years [2]. The main research works that based on feature extraction is explicit or implicit can be broadly divided into two categories: engineered features based methods and deep learning based methods.
The former methods are the traditional method, and image features are predefined before classifier is learned. A supervised learning approach to detect the cell membranes was proposed in Venkataraju et al. [3]. In that paper, the classifier was trained using Adaboost on local and context features with the feature vectors were more than 100 dimensions. The literature [4] proposed a hierarchical segmentation procedure based on statistical learning and topology-preserving grouping. In the process of voxel classification, in order to decide for each voxel whether it represented a membrane, 63 hand-designed features were computed to train a Random Forest. By finding global dense correspondence between two sections, Random forest was trained with 1878 engineered pixel features, including the features from the neighboring section [5]. Burget et al. [6] presented a segmentation using local-level and segment-level features and machine learning algorithms. A novel method that utilizes a hierarchical structure and boundary classification for 2D neuron segmentation was proposed [7]. A set of 141 features extracted from the two merging regions to train a boundary classifier for the purpose of making decisions in a merge tree. For these methods, the hand-designed feature extractor needs elaborately designed features or even applies different types of features to achieve a better accuracy for different problem. In a certain degree, the hand-designed features are difficult to design and problem specific, when the problem changed, then the feature must be redesigned accordingly, so that algorithm has poor portability. In addition, engineered features only capture low-level edge information.
The other methods usually deployed deep learning paradigm. Such methods can retrieve features directly from raw images and have an excellent capability of feature learning. The features learned with such methods gain higher levels of representation and represents more and more abstract functions of the raw input easily [8]. The work of Jain et al. [9] automatically segmented the SBFSEM data set by means of a CNN with more than 34,000 adjustable parameters. In Jurrus et al. [2], a framework to detect neuron membranes that integrates information from the original image together with contextual information by learning a series of artificial neural networks (ANN) was proposed. This makes the network much easier to train because the classifiers in the series are trained one at a time and in sequential order [10] and the experiment results show the advantages over previous membrane detection methods. The latest study in Ciresan et al. [11] described a method that using a special type of deep artificial neural network as a pixel classifier based on GPU implementation, although, the method achieved best result in segmenting the EM neuron images, this approach, however, requires more memory and works with specialized hardware, and it is therefore much more difficult to apply in one’s work. Implicit methods usually deployed deep learning paradigm, in which raw pixel intensities are often directly used as the input to train artificial neural network (ANN) or its variants. The deep architectures have advantages in learning features at multiple levels, but not always optimal for classification.
Feature extraction is one of the key factors in the success of a recognition system. It requires that features should have the most distinguishable characteristics. Firstly, although these supervised methods above achieve a promising segmentation result, the methods based on hand-designed feature extraction require elaborately designed features and cannot process raw images as well as it need a deep understanding for a specific problem [12,13], making it uneasy to adapt to other domain. In addition, the hand-designed features only capture low-level edge information and it is difficult to design features that effectively capture mid-level cues (e.g. edge intersections) or highlevel representation (e.g. object parts) [8], which is very important for neuron images. Also in some other applications, one may not have this knowledge that can be used to develop feature extractors. What’s more, recent developments in machine learning, known as "Deep Learning", have shown how hierarchies of features can be learned directly from data and automatic extraction methods become a tendency in the image processing. Furthermore, a better classifier, distinguishable characteristics combination can generate the highest accuracy for classification.
Inspired by these particular works, this paper presents a hybrid method for the neuron segmentation: combining the capability of distinguishable features learning of CNN and the advantages of Random Forest classifier. This method automatically retrieves features based on the CNN architecture, and recognizes the unknown pattern using the Random Forest recognizer. Experimental results demonstrate the promising performance of our approach.
Related algorithms
Convolutional neural network: The Convolutional Neural Network [14] is a special multilayer neural network, and it is composed of input layers, hidden layers and output layers. The neuron is the basic information processing unit of a CNN which consists of a set of synapses or connecting links, each link characterized by a weight W1, W2,..., Wm, an adder function (linear combiner Eq. (1)) which computes the weighted sum of the inputs
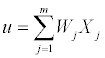 (1)
(1)
and activation function f() for limiting the amplitude of the output of the neuron. The model of the neuron can be viewed as Eq. (2)
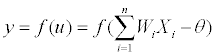 (2)
(2)
Where Xi(i=1,2,3,...,n) is the input vector. Wi represents the weights between two connective neurons. θ is the threshold. f() is the activations function, the commonly used function is sigmoid function Eq. (3)
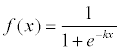 (3)
(3)
y is the desired output.
In CNN architecture, all neurons in a feature map share the same weights (but not the biases). Replicating units in this way allows for features to be detected regardless of their position in the visual field. Additionally, weight sharing offers a very efficient way to greatly reduce the number of free learning parameters. By controlling model capacity, CNNs tend to achieve better generalization on vision problems [14].
The CNN architecture can be viewed as the composition of two parts: an automatic feature extractor and a trainable classifier. The feature extractor contains feature map layers and retrieves discriminating features from the raw images via two operations: convolutional filtering and down sampling. The classifier and the weights learned in the feature extractor are trained by a back-propagation algorithm [15]. Convolutional neural network with its local value shared by the special structure has unique advantages in image processing.
Random forest: Random Forest [16] is a general term for ensemble methods using tree-type classifiers {h(x, βk), k=1…} for classification and regression, where the {βk} are independent identically distributed random vectors and x is an input pattern. In training, the Random Forest algorithm creates multiple CART-like trees each trained on a bootstrapped sample of the original training data, and searches only across a randomly selected subset of the input variables to determine a split (for each node). Each tree is grown as follows: sample N (the number of cases in the training set) cases at random with replacement from the original data; This sample will be the training set for growing the tree; At each node, m predictors are randomly selected out of the M input variables (m<M) and the best split on these m predictors is used to split the node; Each tree is grown to the largest extent possible; There is no pruning. For classification, each tree in the Random Forest casts a unit vote for the most popular class at input x. The output of the classifier is determined by a majority vote of the trees.
Random forest is a good candidate for software quality prediction, especially for large-scale systems, as it is reported to be consistently accurate when compared with current classification algorithms [17].
Hybrid CNN-RF method
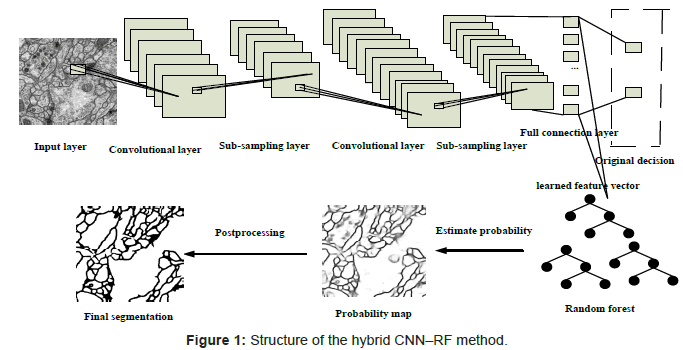
The hybrid CNN–RF method is shown in Figure 1. The proposed approach consists of preprocessing, feature extractor training based on CNN, segmentation based on Random Forest and post processing.
Preprocessing: It is common practice to perform several simple preprocessing steps before attempting to generate features from data. In this work, images were preprocessed with histogram equalization and Gaussian filter, with the purpose of making the intensity more uniform and improving the contrast of the membranes.
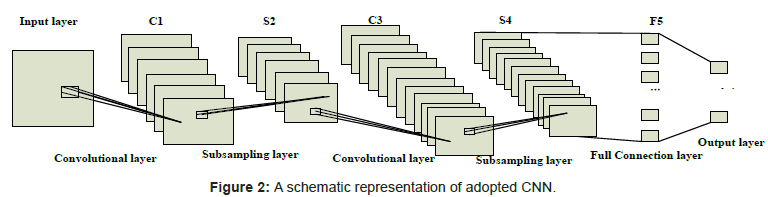
Feature extractor training based on CNN: This section gives a more detailed description of the architecture of the CNN we used in our work, as is shown in Figure 2.
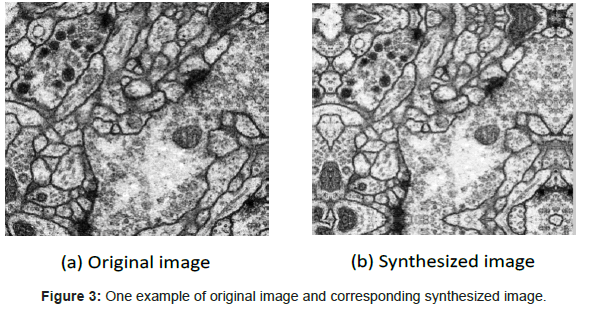
The net of CNN is composed of 7 layers, counting the input and output layers, all of which contain trainable parameters (weights). The input layer is a matrix of the normalized pattern with size 512 by 512 raw pixel images. In order to process the pixels on the border of the image, in this paper, these pixels are synthesized by mirroring the pixels in the actual image across the boundary as shown in Figure 3. Layer C1 is a convolutional layer with 6 feature maps. Layer S2 is a subsampling layer with 6 feature maps. Layer C3 is a convolutional layer with 12 feature maps. Layer S4 is a sub-sampling layer with 12 feature maps. The layer F5 is fully connected, implementing a general purpose classifier over the features extracted by the earlier layers.
Feature map layers are used to compute the features, with different resolutions. Each neuron on a feature map connects with its previous layers, and they are defined by the 5 by 5 convolutional filtering kernel (known as the ‘‘receptive field’’). All the neurons in one feature map share the same kernel and connecting weights (known as the ‘‘sharing weights’’). With a kernel size of 5, and a subsampling ratio of 3, each feature map layer reduces the feature size from the previous feature size. And the CNN learning rate was set to 1. Considering the time cost, the training procedure was stopped after 150 epochs, the consuming time is about one week.
Segmentation based on random forest: Once the feature extractor is trained based on the CNN, the fully connected layer of CNN was replaced by a Random Forest classifier to predict labels of the input patterns. 108 values from the trained CNN network were used as a new feature vector to represent each input pattern, and were fed into the Random Forest for learning and testing. Once the Random Forest classifier has been well trained, it performs the recognition task and makes new decisions on testing images with such automatically extracted features. In the experiments, the Random Forests was trained with default parameters (the number of trees are 500).
Postprocessing: There are two steps in the postprocessing procedure. The first step is auto-threshold methods [18] provided by Fiji to membrane probability map returned by Random Forest, for the purpose of improving membrane continuity. The second step is iteratively region removing, performed by a series of threshold operations based on region properties such as Area, Euler Number, Solidity and Eccentricity.
Experiments
Dataset
The dataset we used in this paper was provided by the organizers of the ISBI 2012 EM Segmentation Challenge (http://www.biomedicalimaging.org/2012/index.php). The data set consists of training data and testing data of the Drosophila first instar larva ventral nerve cord, which is provided in the form of EM stack. The training data which was labeled by an expert human neuroanatomist is a set of 30 sections from a serial section Transmission Electron Microscopy (ssTEM) data set. The test data (ground truth unknown to the authors) is another volume from the same Drosophila first instar larva ventral nerve cord as the training dataset.
Evaluation metrics
Segmentation result is evaluated through an automated online system; the system computes three error metrics in relation to the hidden ground truth: pixel error, warping error and the Rand error.
- Pixel error: defined as 1 - the maximal F-score of pixel similarity or squared Euclidean distance between the original and the result labels [19].
- Warping error: The warping error is segmentation metric that tolerates disagreements over boundary location, penalizes topological disagreements, and can be used directly as a cost function for learning boundary detection [19].
- Rand error: The Rand error metric is based on the Rand index, defined as 1 - the maximal F-score of Rand index, a measure of similarity between two clusters or segmentations. It has a more intuitive interpretation, but completely disregards non-topological errors [19-21].
Experimental results
This section presents the results obtained by the proposed methods on the publicly-available dataset provided by the organizers of the ISBI 2012 EM Segmentation Challenge. To train the classifier, we use all available slices of the training stack, i.e., 30 images with a 512*512 resolution. For each slice, an automatic representative sample selection method based on superpixel [22] is used in our paper, 10000 samples for each image. This amounts to 300000 training examples in total. Experimental results are shown in Table 1.
| Method | Error Metrics | ||
|---|---|---|---|
| Rand error | Warping error | Pixel error | |
| second human observer | 0.026546995 | 0.000344086 | 0.066553289 |
| simple thresholding | 0.449664478 | 0.017141342 | 0.225194944 |
| DenseETH method | —— | 0.00062 | 0.079264809 |
| Burget’s method | 0.139038440 | 0.002641296 | 0.102285508 |
| CNN | 0.131017450 | 0.001152420 | 0.073262207 |
| CNN-RF | 0.109388991 | 0.001455688 | 0.072129307 |
Note: The first two rows report the performance of the second human observer and of a simple threshold approach.
‘—— ‘the author does not provide the rand error in the article.
Table 1: Results of our approach and competing algorithms.
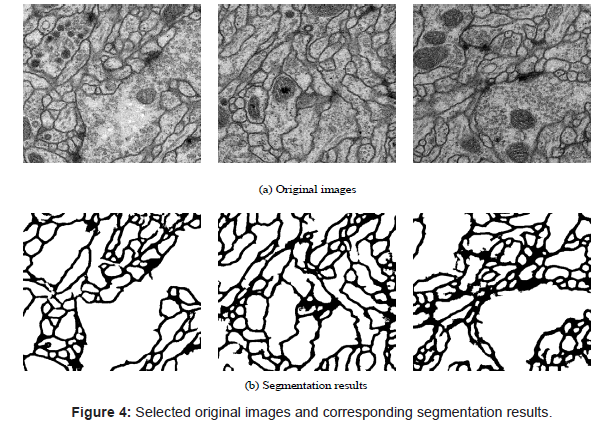
Two popular approaches [5,6] are compared with the proposed method. One of these approaches, DenseETH method described in Laptev et al. [5] constructs a dense correspondence between the neighboring sections and it uses features that are evaluated in all the corresponding pixels for classification. 1848 corresponding handdesigned pixels features are used to train a Random Forest classifier, while only 108 dimensions learned features are used in our method; finally Graph Cut is deployed based on the probability map returned by Random Forest. The second approach is the Burget’s method presented in Burget et al. [6]. In this approach, a segmentation using local-level and segment-level features and machine learning algorithms was used. Firstly, several different transformations with several different parameters were used to get the local features. In order to optimize the parameters of the transforms used, a genetic algorithm optimization was adopted. And then a support vector machine was trained with these features; at last, segment-level features were extracted to train a decision tree for the purpose of removing these unwanted objects in the resulting images from support vector machine. Experiment on the pure CNN is executed to verify the effectiveness of the use of Random Forest classifier. According to the information given in the table, we can find that the results of the proposed method are appeared to be competitive to theirs and their solution was weaker in rand error and pixel error. Some selected segmentation results are shown in Figure 4.
Analysis of experimental results
We have implemented the CNN-RF method presented in previous sections and successfully used it to perform the segmentation from the data set. Based on experiments on the data set, it has shown that the hybrid CNN-RF method yields a better performance improvement over the competing algorithms. Comparison with the pure CNN and the state of the art methods would verify the overall performance of the proposed method. Compared with the competing methods, one of the main advantages of proposed method is that distinguishable features can be learned directly from raw images instead of being designed manually.
The advantage of the CNN classifier is that it automatically extracts the salient features of the input image. These learned features have a deeper characterization for the input image, and can collect more representative and relevant information form original images. On the contrary, the hand-designed feature extractor needs elaborately designed features and only capture low-level edge information; furthermore, CNN uses the receptive field concept successfully to obtain local visual features to describe the topology of the images which is more important for neuronal structures. Since the theoretical learning method of CNN is the same as that for the MLP, it is an extension model of the MLP. A limitation of MLP is that it tends to assign a high value (nearly +1) to one neuron at the output layer whereas all the remaining neurons have a low value (nearly -1). This causes difficulties in rejecting errors in real applications [15]. But the Random Forest classifier calculates the estimated probability in the classification decision. This probability information provides a more reliable rank list of label predictions. Beside, using those probability values can help us to design an efficient rejection mechanism.
The Random Forest approach should be of great interest for classification since the approach is not only nonparametric [23], but it also provides a way of estimating the importance of the individual variables (data channels) in the classification and can handle high dimensional data while maintaining high computational efficiency. Even in presence of many noisy features, Random Forest works well, so it is unnecessary to perform feature selection procedure. The most important is that it is reported to be consistently accurate when compared with current classification algorithms [17]. As a result, it can improve the classification accuracy of the hybrid method after replacing the output units in the CNN.
Conclusion and Future Work
In this paper, a hybrid method CNN-RF is proposed for EM neuron segmentation. CNN works as a trainable feature extractor to automatic extract features from raw pixels and Radom Forest performs as a recognizer. The method combines the capability of distinguishable features learning of CNN and the advantages of Radom Forest classifier. Using these distinguishable features learned by CNN to train the Random Forest classifier, we get the effectiveness results for the segmentation of neuronal structures in EM stacks. Comparisons with existing methods demonstrate the superiority of the approach.
Future studies might consider building larger architectures together with optimized parameters for the training of the network so as to further improve the recognition accuracy.
Acknowledgements
The work is supported by NSFC Joint Fund with Guangdong under Key Project U1201258, the Research Found for the Doctoral Program of Higher Education under Grant No. 20110131130004, the Natural Science Foundation of Shandong Province (NO.ZR2011FQ033) and A Project of Shandong Province Higher Educational Science and Technology Program NO.J13LN23.
References
- Jurrus E, Whitaker R, Jones B W, Marc R, Tasdizen T (2008) An optimal-path approach for neural circuit reconstruction, Biomedical Imaging: From Nano to Macro, 2008. ISBI 2008. 5th IEEE International Symposium on. IEEE, 1609-1612.
- Jurrus E, Paiva AR, Watanabe S, Anderson JR, Jones BW, et al. (2010) Detection of neuron membranes in electron microscopy images using a serial neural network architecture. Med Image Anal 14: 770-783.
- Venkataraju KU, Paiva ARC, Jurrus E, Tasdizen T (2009) Automatic markup of neural cell membranes using boosted decision stumps, In: proc. of Biomedical Imaging: From Nano to Macro, 2009. ISBI '09. IEEE International Symposium on, Boston (ISBI), MA: 2009, 1039-1042.
- Andres B, Köthe U,Helmstaedter M,Denk W, Hamprecht FA (2008) Hamprecht. Segmentation of SBFSEM Volume Data of Neural Tissue by Hierarchical Classification. Pattern Recognition. In: Rigoll G (Ed.), Springer Berlin Heidelberg 5096: 142-152.
- Laptev D, Vezhnevets A, Dwivedi S, Buhmann J (2012) Anisotropic ssTEM Image Segmentation Using Dense Correspondence across Sections. In: Ayache N, Delingette H, Golland P, Mori K (Eds.), proc. of Medical Image Computing and Computer-Assisted Intervention ��? MICCAI. Springer Berlin Heidelberg 7510: 323-330.
- Burget R, Uher V, Masek J (2012) Trainable Segmentation Based on Local-level and Segment-level Feature Extraction. In: Proc. of IEEE International Symposium on Biomedical Imaging: From nanoto Macro.
- Liu T, Jurrus E, Seyedhosseini M, Ellisman M, Tasdizen T (2012) Watershed Merge Tree Classification for Electron Microscopy Image Segmentation. In: proc. of the 21th conference of the International Association for Pattern Recognition ICPR. Tsukuba.
- Bengio Y (2009) Learning Deep Architectures for AI. Found. Trends Mach. Learn ® in Machine Learning. 2: 1-127.
- Jain V, Murray JF, Roth F, Turaga S, Zhigulin V, et al. (2007) Supervised Learning of Image Restoration with Convolutional Networks,"Computer Vision, 2007. ICCV 2007. IEEE 11th International Conference on, 8: 14-21.
- Seyedhosseini M, Kumar R, Jurrus E, Giuly R, Ellisman M, et al. (2011) Detection of Neuron Membranes in Electron Microscopy Images Using Multi-scale Context and Radon-Like Features. Med Image Comput Comput Assist Interv 14: 670-677.
- Ciresan DC, Giusti A, Gambardella LM, Schmidhuber J (2012) Deep neural net-works segment neuronal membranes in electron microscopy images. In: proc. Neural Information Processing Systems (NIPS). Lake Tahoe, Nevada, United States.
- Kumar R, Va´zquez-Reina A, Pfister H (2010) Radon-Like features and their application to connectomics. In: proc. of Computer Vision and Pattern Recognition Workshops (CVPRW). IEEE Computer Society Conference on, 186-193.
- Smith K, Carleton A, Lepetit V (2009) Fast Ray features for learning irregular shapes. Computer Vision, 2009 IEEE 12th International Conference on. 397-404.
- LeCun Y, Bottou Y, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proceedings of the IEEE 86: 2278-2324.
- Niu XX, Suen CY (2012) A novel hybrid CNN��?SVM classifier for recognizing handwritten digits. Pattern Recognition 45: 1318-1325.
- Breiman L (2001) Random forests.Machine learning45.1: 5-32.
- Guo L, Yan M, Cukic B, Singh H (2004) Robust prediction of fault-proneness by random forests[C]//Software Reliability Engineering, 2004. ISSRE 2004. 15th International Symposium on. IEEE 417-428.
- Kittler J, Illingworth J (1986) "Minimum error thresholding", Pattern Recognition 19: 41-47.
- Jain V, Bollmann B, Richardson M, Berger DR, Helmstaedter MN, et al. (2010) Boundary Learning by Optimization with Topological Constraints. In: proc. of Computer Vision and Pattern Recognition (CVPR), IEEE Conference on, 2488-2495.
- Rand WM (1971) Objective Criteria for the Evaluation of Clustering Methods. J Am Stat Assoc 66: 846-850.
- Unnikrishnan R, Pantofaru C, Hebert M (2007) Toward objective evaluation of image segmentation algorithms. IEEE Transactions on Pattern Analysis and Machine Intelligence 29: 929-944.
- Achanta R, Shaji A, Smith K, Lucchi A, Fua P, et al. (2012) SLIC Superpixels Compared to State-of-the-Art Superpixel Methods. Pattern Analysis and Machine Intelligence, IEEE Transactions on, 3: 2274-2282.
- Gislason PO, Benediktsson JA, Sveinsson JR (2006) Random Forests for land cover classification. Pattern Recognit Lett 27: 294-300.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 25109
- [From(publication date):
November-2013 - Apr 19, 2025] - Breakdown by view type
- HTML page views : 20177
- PDF downloads : 4932