A Genome-wide Association Study in the Diabetic Patients Finds the 13q35.43-35.46 Locus Associated with Estimated Glomerular Filtration Rate: The Japan Multi-Institutional Collaborative Cohort Study
Received: 15-Jul-2018 / Accepted Date: 19-Jul-2018 / Published Date: 27-Jul-2018
Abstract
Objective: To conduct a genome-wide association studies (GWAS) to find genetic variations that affected renal function in the diabetic patients in Japan.
Research design and methods: In a Japanese population of 955 patients with type 2 diabetes mellitus (T2D), extracted from 14,091 participants appropriate for GWAS from the Japan Multi-Institutional Collaborative Cohort (JMICC) study. Genotyping was performed at a central laboratory with use of a HumanOmniExpressExome-8 v1.2 BeadChip array. Genotype imputation was conducted with use of SHAPEIT, followed by Minimac3 software (with the 1000 Genomes phase 3 as the reference panel). We calculated the estimated glomerular filtration rate (eGFR) for each patient according to Matsuo et al. Association for the imputed variants with eGFR was performed by linear regression analysis with adjustments for age and sex.
Results: We found 77 SNVs upstream of the NBEA genes that were significantly associated with eGFR in T2D participants with P values <5 × 10-8. This gene was reported as participatory in several metabolic functions and was associated with some disease conditions. However, no previous reports implied that the gene was related to nephropathy in diabetes.
Conclusion: We found the 13q35.43-35.46 locus upstream of the NBEA gene was significantly associated with eGFR in participants with T2D in a Japanese population.
Keywords: Genome-wide association study; Diabetes mellitus; Estimated glomerular filtration rate; Chronic kidney disease
Introduction
Diabetic nephropathy is the most common cause of chronic kidney disease (CKD) in developed countries [1]. The clinical characteristics do not fully predict development of nephropathy in diabetes patients. Epidemiological findings suggested genetic background plays an important role in the development of this renal disease [2,3].
There have been several genome-wide association studies (GWAS). In a recent meta-analysis by Pattaro et al. on associations of estimated glomerular filtration rate (eGFR) based on serum creatinine (Scr), cystatin C, and CKD (defined as eGFR based on Scr <60 ml/min/1.73 m2) with about 2.5 million autosomal single-nucleotide variations (SNVs) in 133,413 individuals of European ancestry in the stage 1 discovery analysis, 29 previously identified loci were confirmed and 48 independent novel loci newly identified [4]. In their trans-ethnic analyses in 42,296 Asian participants, 7 out of 24 newly identified loci with eGFR based on Scr achieved direction-consistent significance. If the GWAS discovery analysis had been started in an Asian population, some different SNVs may have been found. Here we found 77 SNVs upstream of NBEA that affected renal function in a Japanese population with type 2 diabetes mellitus (T2D).
Research Design and Methodology
Study population
A cross-sectional GWAS was performed in participants at ages from 35 to 69 years in the Japan Multi-Institutional Collaborative Cohort (JMICC) study. The participants in the J-MICC study were invited from 12 different locations in Japan between 2004 and 2013 (Number of participants: 14,539). The J-MICC study is a cohort study started in 2005 to examine interactions between gene and environment factors in lifestyle-related diseases. Minutes of the J-MICC Study were published elsewhere [5,6]. Briefly, participants answered a questionnaire regarding lifestyle and medical information, and gave a blood specimen at the time of the baseline study. The J-MICC study participants encompasses community citizens, first-visit patients to a cancer institution and health checkup examinees. Written informed consent was obtained from all participants in this study, and the study protocol was approved by the Ethics Committees of Aichi Cancer Center, Nagoya University Graduate School of Medicine, and the other participating institutions in the J-MICC study. Our present study was conducted in keeping with to the principles expressed in the World Medical Association Declaration of Helsinki.
Out of 14,539 participants, 448 were excluded by the GWAS screening described below. Of the remaining 14,091 participants, we identified 1,109 with T2D by the criteria described below. Of the 1,109 participants with T2D, 154 were excluded due to missing data. As a result, we analyzed the data of 955 participants with T2D.
Questionnaire and measurements
The J-MICC study questionnaire consisted of questions on medical history, weight, height, smoking and alcohol drinking habits. Measurements of participants’ body and blood drawing were done as part of the health examination or for research purposes at the J-MICC study participating institutions [5]. Weight and height were measured to the nearest 0.1 kg and 0.1 cm, respectively. Body mass index (BMI) was calculated by dividing body weight in kilograms by the square of height in meters. The HbA1c percentage was obtained with use of a latex aggregation immunoassay (Japan Diabetes Society [JDS] value). The HbA1c percentage was estimated as the National Glycohemoglobin Standardization Program (NGSP) equivalent percentage obtained according to the following formula: HbA1c (NGSP (%))=1.02 × HbA1c (JDS [%])+0.25% [7]. We defined T2D as a fasting blood glucose concentration ≥ 126 mg/dL, or ≥ 200 mg/dL if less than 8 h after meals, or NGSP HbA1c ≥ 47 mmol/mol (6.5%), or participants diagnosed as having DM. Scr was measured enzymatically. The eGFR of each participant was obtained based on SCr, age, and sex with use of the following Japanese eGFR equation proposed by the Japanese Society of Nephrology: eGFR (mL/min/1.73 m2)=194 × Scr (mg/dL)-1.094 × age-0.287 (× 0.739 if female) [8]. Semi-quantified dip-stick urine protein data were available in a part of the patients (N=516) that were classified as 0, 15, 30, 100, or ≥ 250 mg/dL.
Genotyping and quality control filtering
Buffy coat fractions were prepared from blood specimen and stored at −80°C at the central office for J-MICC Study. DNA was prepared from all buffy coat fractions with use of a BioRobot M48 Workstation (Qiagen Group, Tokyo, Japan) at the central study office. For the specimen from two locations (Fukuoka and Kyushu-KOPS), DNA was prepared locally from specimen of whole blood with use of an automatic nucleic acid isolation system (NA-3000, Kurabo, Co., Ltd, Osaka, Japan). The 14,539 study participants from the 12 locations of the J-MICC study were genotyped at RIKEN Center for Integrative Medicine Sciences with use of a HumanOmniExpressExome-8 v1.2 BeadChip array (Illumina Inc., San Diego, CA, USA). Inconsistent sex information in 26 participants between an estimate from genotyping and the questionnaire was detected, and these participants were excluded. The identity-by-descent method supplemented in the PLINK 1.9 software [9,10] identified 388 close relationship pairs (pihat> 0.1875) and one sample from each pair of the 388 was deleted. Principal component analysis (PCA) [11,12] with a 1000 Genomes reference panel (phase 3) [13,14] found 34 participants whose estimated ancestries were outside the Japanese population [15]. These 34 participants were excluded. In the remaining 14,091 participants, SNVs with a Hardy-Weinberg equilibrium exact test P-value <1 × 10-6, and/or a genotype call rate <0.98, a departure from the allele frequency computed from the 1000 Genomes Phase 3 EAS samples, or a low minor allele frequency (MAF) <0.01, were excluded. These filtering resulted in 14,091 individuals and 570,162 SNVs.
Genotype imputation
Genotype imputation was conducted with use of SHAPEIT [16], followed by Minimac3 [17] software (with the 1000 Genomes phase 3 as the reference panel [13]). Strict quality control filters were applied after genotype imputation; i.e., variants with an R2<0.3 were excluded, resulting in 12,617,547 variants. Finally 4, 233,797 variants with MAF <0.01 in T2D patients were removed, resulting in 8,383,750 variants for the analysis. We used Dosage Convertor software [18] to convert dosage files in VCF format from Minimac3 to PLINK formats.
Association analyses between genetic variants and eGFR in T2D
Associations between all imputed variants and eGFR were analyzed with adjustments for age and sex with use of PLINK 1.9 software. Variants achieving genome-wide significance (P<5 × 10-8) were regarded to be eGFR-associated variants. An R package for creating QQ plot, GWASTools was used [19]. For scatter plots of P values derived from genome-wide scan results for eGFR in T2D, a software, Haploview was used [20]. To visualize regions of interest, we used the LocusZoom program [21].
We performed replication analysis for previously-reported SNVs with use of the J-MICC samples [4,22].
Confounding factor adjustment
In 519 patients with urine protein data, the following additional association analyses were performed. Model 1: associations between imputed variants in chromosome 13 and eGFR were tested with adjustment for age and sex with use of PLINK 1.9 software. Model 2: analysis with model 1 variables plus urine protein data. Model 3: model 1 variables plus PCA components 1 to 3, which were obtained in 14,091 participants with the use of Eigensoft 6.0.1 [10,11]. This analysis was performed since the structure of Japanese population was reported as not homogenous [15]. Model 4: model 1 variable plus BMI, alcohol consumption (g/day), and smoking (current, or past smoking vs. never smoking).
Functional annotations
Genomic locations of variants found in this study were examined based on the UCSC [23] and Ensembl [24] genome browsers. CiseQTL pairs of variants and genes were obtained from the GTEx [25].
Results
Baseline characteristics
Baseline characteristics of the participants with T2D are shown in Table 1. The mean age of the participants was 59.1 ± 7.4 y and the percentage of women and prevalence of current smokers were 34% and 25%, respectively. Mean of BMI, Scr concentration, HbA1c, eGFR and alcohol consumption were 24.8 ± 4.1 kg/m2, 0.76 ± 0.30 mg/dL, 55 ± 11 mmol/mol (7.2 ± 1.5%), 89.3 ± 27.1 ml/min/1.73 m2, and 17.7 ± 27.5 g/day, respectively. Baseline characteristics of the participants with T2D who had urine protein data are also shown in Table 2. The mean age of the participants was 58.5 ± 7.6 y and the percentage of women and prevalence of current smokers were 33% and 24%, respectively. Mean of BMI, Scr concentration, HbA1c, eGFR, alcohol consumption and urinary protein were 25.4 ± 4.0 kg/m2, 0.76 ± 0.22 mg/dL, 53 ± 10 mmol/mol (7.1 ± 1.4%), 88.8 ± 29.5 ml/min/1.73 m2, 18.4 ± 29.5 g/day and 10.3 ± 34.1 mg/dL, respectively.
| T2D | T2D with UP data | |
|---|---|---|
| N | 955 | 516 |
| Age (y) | 59.1 ± 7.4 | 58.5 ± 7.6 |
| Women (%) | 34% | 33% |
| BMI (kg/m2) | 24.8 ± 4.1 | 25.4 ± 4.0 |
| Creatinine (mg/dL) | 0.76 ± 0.30 | 0.76 ± 0.22 |
| HbA1c (mmol/mol [%]) | 55 ± 11 [7.2 ± 1.5] | 53±10 [7.1 ± 1.4] |
| eGFR (ml/min/1.75) | 89.3 ± 27.1 | 88.8 ± 26.6 |
| Alcohol intake (g/day) | 17.7 ± 27.5 | 18.4 ± 29.5 |
| Current smoking (%) | 25% | 24% |
| Urine protein (mg/dL) | 10.3 ± 34.1 | |
| T2D=Type 2 Diabetes Mellitus, UP=Urine Protein, BMI=Body Mass Index, HbA1c=Glycated Hemoglobin, eGFR=Estimated glomerular filtration rate based on serum creatinine. | ||
Table 1: Background characteristics of the study participants.
| SNP | Chr | Gene | Position | EA | NEA | EA FRQ | β | SE | P |
|---|---|---|---|---|---|---|---|---|---|
| rs9599756 | 13 | NBEA (upstream) | 35435744 | C | T | 0.197 | 6.917 | 1.212 | 1.55E-08 |
| rs77176728 | 13 | NBEA (upstream) | 35439902 | A | T | 0.192 | 7.096 | 1.230 | 1.08E-08 |
| rs9599763 | 13 | NBEA (upstream) | 35440514 | A | G | 0.198 | 6.926 | 1.214 | 1.56E-08 |
| rs74575233 | 13 | NBEA (upstream) | 35447752 | T | C | 0.196 | 6.808 | 1.220 | 3.17E-08 |
| rs143794932 | 13 | NBEA (upstream) | 35450075 | G | A | 0.196 | 6.779 | 1.217 | 3.35E-08 |
| rs77941208 | 13 | NBEA (upstream) | 35451176 | T | C | 0.196 | 6.776 | 1.217 | 3.35E-08 |
| rs76112805 | 13 | NBEA (upstream) | 35451557 | G | A | 0.196 | 6.776 | 1.217 | 3.37E-08 |
| rs146196432 | 13 | NBEA (upstream) | 35452275 | T | C | 0.196 | 6.776 | 1.217 | 3.41E-08 |
| We found 77 SNVs upstream of the NBEA gene that were significantly associated with eGFR in participants with T2D with P<5E-08. A representative 8 SNVs are shown. Information for all 77 SNVs is shown in the supporting information (Table S1). T2D=Type 2 Diabetes Mellitus, SNP=Single Nucleotide variant, Chr=Chromosome, Position=Chromosomal Position (GRCh37/hg19), EA=Effect Allele, NEA=Non-effect Allele, FRQ=Frequency, β=Regression Coefficient for EA, SE=Standard error of effect estimate, P=Association test p-value. | |||||||||
Table 2: SNVs associated with eGFR in T2.
Genome-wide association study
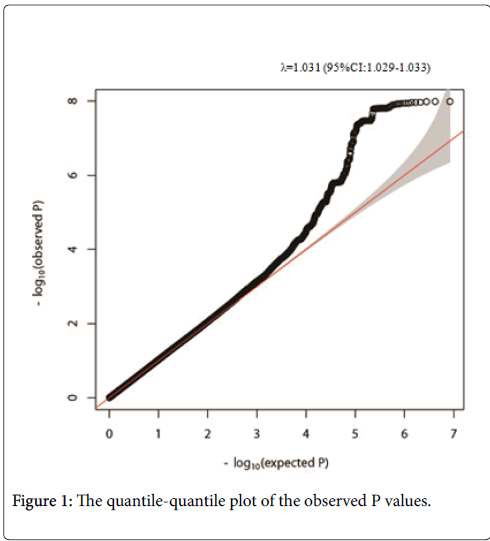
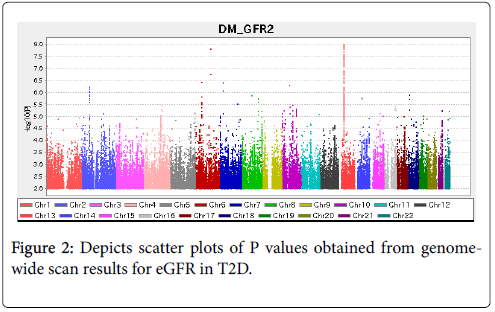
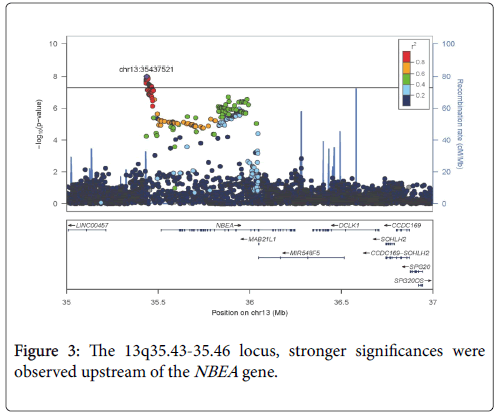
Among the 8,383,750 variants, genome-wide analyses with adjustment for age and sex, we found 77 SNVs upstream of the NBEA gene that were significantly associated with eGFR in participants with T2D with P values <5 × 10-8. A representative 8 SNVs are shown in Table 2. Information for all 77 SNVs is shown in the Supplementary Information (Table S1). The quantile-quantile plot of the observed P values is depicted in Figure 1. The inflation factor of the genome-wide scan was 1.031, which indicates that the population structure was welladjusted. Figure 2 depicts scatter plots of P values obtained from genome-wide scan results for eGFR in T2D, which found that 77 close SNVs at 13q35.43-35.46 met significance (P<5 × 10-8). In the 13q35.43-35.46 locus, stronger significances were observed upstream of the NBEA gene (Figure 3).
Confounding factor adjustment
The results of confounding factor adjustment analysis for the representative 8 SNVs in 519 T2D patients with urine protein data are shown in Table 3. Compared to the results in all 955 T2D patients, P values were more significant and β was larger in the age sex adjusted analysis in 519 T2D patients with urine protein data (model 1). With the addition of urine protein in model 1, P values were attenuated very slightly (model 2). With the addition of PCA components to model 1, P values were somewhat attenuated (model 3). With the addition of BMI, smoking, and alcohol consumption to model 1, P values were slightly attenuated (model 4) (Table 3).
| Model 1 | Model 2 | Model 3 | Model 4 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables adjusted | Age, sex | Age, sex, urine protein | Age, sex, PCA | Age, sex, BMI, smoking, alc | ||||||||||||
| Position | Gene | EA | NEA | EA FRQ | β | SE | P | β | SE | P | β | SE | P | β | SE | P |
| 35435744 | rs9599756 | C | T | 0.209 | 9.34 | 1.55 | 2.96E-09 | 9.24 | 1.54 | 3.95E-09 | 8.92 | 1.54 | 1.13E-08 | 9.28 | 1.56 | 5.41E-09 |
| 35439902 | rs77176728 | A | T | 0.206 | 9.35 | 1.56 | 3.76E-09 | 9.26 | 1.56 | 5.02E-09 | 8.93 | 1.55 | 1.47E-08 | 9.29 | 1.58 | 7.24E-09 |
| 35440514 | rs9599763 | A | G | 0.209 | 9.35 | 1.55 | 2.92E-09 | 9.26 | 1.55 | 3.88E-09 | 8.94 | 1.54 | 1.11E-08 | 9.30 | 1.56 | 5.32E-09 |
| 35447752 | rs74575233 | T | C | 0.211 | 8.90 | 1.55 | 1.62E-08 | 8.80 | 1.55 | 2.25E-08 | 8.53 | 1.54 | 4.99E-08 | 8.85 | 1.57 | 2.81E-08 |
| 35450075 | rs143794932 | G | A | 0.212 | 8.86 | 1.55 | 1.78E-08 | 8.76 | 1.55 | 2.48E-08 | 8.49 | 1.54 | 5.43E-08 | 8.80 | 1.56 | 3.06E-08 |
| 35451176 | rs77941208 | T | C | 0.212 | 8.86 | 1.55 | 1.78E-08 | 8.75 | 1.55 | 2.48E-08 | 8.48 | 1.54 | 5.44E-08 | 8.80 | 1.56 | 3.06E-08 |
| 35451557 | rs76112805 | G | A | 0.212 | 8.86 | 1.55 | 1.79E-08 | 8.75 | 1.555 | 2.49E-08 | 8.48 | 1.54 | 5.46E-08 | 8.80 | 1.56 | 3.07E-08 |
| 35452275 | rs146196432 | T | C | 0.215 | 8.81 | 1.55 | 2.02E-08 | 8.70 | 1.54 | 2.83E-08 | 8.44 | 1.53 | 6.04E-08 | 8.75 | 1.56 | 3.52E-08 |
| The results of confounding factor adjustment analysis on the representative 8 SNVs are shown. T2D=Type 2 Diabetes Mellitus, PCA=Principal Component Adjustment (components 1-3), BMI=Body Mass Index, alc=Alcohol Consumption, Position=Chromosomal position in chromosome 13 (GRCh37/hg19), EA=Effect Allele, NEA=Non-effect Allele, FRQ=Frequency, β=Regression Coefficient for EA, SE=Standard error of effect estimate, P=Association test p-value. | ||||||||||||||||
Table 3: Confounding factor adjustment analyses in SNVs in chromosome 13 associated with eGFR in T2D with urine protein data.
Functional annotations for SNVs associated with eGFR in T2D in the 13q35.43-35.46 locus
We examined the expression quantitative trait loci (eQTL) relationship between the 77 significant SNVs and NBEA gene. From the GTEx database [25], 31 eQTL hits were found, of which expression levels were significantly associated with the 13q35.43-35.46 variants (P<1.0E-04; Table 4 and Supplementary Information (Table S1)).
| SNP | Chr | Position | EA | NEA | Gene(s) | Tissue | β | P |
|---|---|---|---|---|---|---|---|---|
| rs9599756 | 13 | 35435744 | C | T | No hit | |||
| rs77176728 | 13 | 35439902 | A | T | No hit | |||
| rs77176728 | 13 | 35440514 | A | G | No hit | |||
| rs74575233 | 13 | 35447752 | T | C | NBEA | Artery-Tibial | 0.59 | 7.6E-06 |
| rs143794932 | 13 | 35450075 | G | A | NBEA | Artery-Tibial | 0.57 | 4.3E-05 |
| rs77941208 | 13 | 35451176 | T | C | NBEA | Artery-Tibial | 0.59 | 7.8E-06 |
| rs76112805 | 13 | 35451557 | G | A | NBEA | Artery-Tibial | 0.59 | 7.6E-06 |
| rs146196432 | 13 | 35452275 | T | C | NBEA | Artery-Tibial | 0.59 | 7.6E-06 |
| Functional annotations for the representative 8 SNVs are shown. Information for all 77 SNVs is shown in the supporting information (Table S1). Chr=Chromosome; Position (GRCh37/hg19); EA=Effect Allele; NEA=Non-effect Allele; β=effect size. | ||||||||
Table 4: Functional annotations for SNVs associated with eGFR in T2D in the 13q35.43-35.46 locus.
Replication of previously reported SNVs in the Asian population
We performed a replication study on the 7 out of 24 newly identified loci associated with eGFR based on Scr that achieved directionconsistent significance in the trans-ethnic analyses of Asian participants in the meta-analysis conducted by Pattaro et al. [4]. Only one SNP (rs163160) at the intron of the KCNQ1 gene was statistically significant (P=0.032, EA FRQ=0.132), however, the direction was inconsistent. We could not replicate nine SNVs at the ELMO1 gene that were reported previously in a Japanese population [22]. However, 8 SNVs at the ELMO1 gene in the present study showed significant association with eGFR (P<0.025) (Table 5).
| SNP | Chr | Position | Gene (variant type) | EA | NEA | EA FRQ | β | SE | P | Effect direction |
|---|---|---|---|---|---|---|---|---|---|---|
| Replication of the study by Pattaro et al. | ||||||||||
| rs4014195 | 11 | 65506822 | AP5B1 (intergen) | G | C | 0.156 | -0.153 | 1.344 | 0.909 | + |
| rs2712184 | 2 | 217682779 | IGFBP5 (intron) | A | C | 0.496 | -1.477 | 0.999 | 0.140 | + |
| rs9682041 | 3 | 170091902 | SKIL (intron) | T | C | 0.940 | 2.340 | 2.025 | 0.248 | - |
| rs7956634 | 12 | 115321194 | PTPRO (intron) | C | T | 0.369 | 0.813 | 1.033 | 0.432 | + |
| rs11666497 | 19 | 38464262 | SIPA1L3 (intron) | T | C | 0.094 | 1.810 | 1.679 | 0.281 | - |
| rs163160 | 11 | 2789955 | KCNQ1 (intron) | G | A | 0.132 | 3.064 | 1.429 | 0.032 | - |
| rs10277115 | 7 | 1285195 | UNCX (intergen) | T | A | 0.313 | -1.284 | 1.096 | 0.242 | + |
| Replication of the study by Shimazaki et al. | ||||||||||
| rs4723596 (A/G) | 7 | 36951044 | ELMO1 (intron) | C | T | 0.240 | -1.955 | 1.138 | 0.086 | - |
| rs11983698 (A/G) | 7 | 36948547 | ELMO1 (intron) | C | T | 0.288 | -1.225 | 1.087 | 0.260 | - |
| rs4723593 (C/T) | 7 | 36947292 | ELMO1 (intron) | A | G | 0.231 | -2.173 | 1.168 | 0.063 | - |
| rs28496648 (C/T) | 7 | ?36938446 | ELMO1 (intron) | A | G | 0.322 | -1.186 | 1.084 | 0.274 | - |
| rs7799004 (A/G) | 7 | 36928964 | ELMO1 (intron) | T | C | 0.716 | 1.731 | 1.109 | 0.119 | - |
| rs741301 (A/G) | 7 | 36917995 | ELMO1 (intron) | T | C | 0.658 | 1.462 | 1.057 | 0.167 | - |
| rs1558688 (G/T) | 7 | 36915185 | ELMO1 (intron) | C | T | 0.665 | 1.342 | 1.062 | 0.207 | - |
| rs3807163 (A/G) | 7 | 36908759 | ELMO1 (intron) | C | T | 0.275 | -1.462 | 1.132 | 0.197 | - |
| rs1541727 (T/G) | 7 | 37482424 | ELMO1 (intron) | C | A | 0.291 | -1.566 | 1.439 | 0.277 | + |
| rs7804092 (A/T) | 7 | 36893232 | ELMO1 (intron) | A | T | 0.275 | -1.582 | 1.129 | 0.162 | + |
| Replication study onELMO1 | ||||||||||
| rs117198906 | 7 | 37194931 | ELMO1 (intron) | A | G | 0.021 | -12.608 | 4.901 | 0.010 | |
| rs192248614 | 7 | 37205346 | ELMO1 (intron) | A | G | 0.010 | -14.437 | 6.118 | 0.019 | |
| rs117874121 | 7 | 37348682 | ELMO1 (intron) | A | G | 0.282 | 3.415 | 1.477 | 0.021 | |
| rs12532146 | 7 | 37353302 | ELMO1 (intron) | C | T | 0.135 | -3.510 | 1.433 | 0.014 | |
| rs150130338 | 7 | 37359086 | ELMO1 (intron) | T | G | 0.016 | -11.777 | 5.212 | 0.024 | |
| rs10245068 | 7 | 37365234 | ELMO1 (intron) | A | C | 0.203 | 3.308 | 1.242 | 0.008 | |
| rs10248478 | 7 | 37365649 | ELMO1 (intron) | T | C | 0.181 | 3.311 | 1.312 | 0.012 | |
| rs10243081 | 7 | 37369690 | ELMO1 (intron) | T | C | 0.173 | 3.774 | 1.355 | 0.005 | |
| Chr=Chromosome; Position (GRCh37/hg19); EA=Effect Allele; NEA=Non-effect Allele; FRQ=Frequency; β=effect size; SE=Standard error of effect size. | ||||||||||
Table 5: Replication analysis using the J-MICC samples for previously-reported SNVs.
Discussion and Conclusion
In the present GWAS of participants with T2D in a Japanese population, we found 77 novel SNVs located upstream of the NBEA gene that were associated with eGFR. The NBEA gene involves a member of a large, diverse group of A-kinase anchor proteins that designate the activity of protein kinase A to specific subcellular locations by binding to its type II regulatory subunits. NBEA is expressed in blood cells, the brain, internal systems such as the kidneys and colon, and secretory systems such as the pancreas and adrenal glands [26]. A wide variety of diseases are associated with NBEA including migraine in bipolar disorder [27], idiopathic autism [28], schizophrenia [29], major depression [30], substance abuse [30], and multiple myeloma [31,32]. Olszewski et al. found a significant association for two upstream SNVs in NBEA , rs17775456 and rs7990537 with BMI as a continuous quality and trends for weight among the overweight adult men. They also found that NBEA+/2 mice result in moderately elevated body weight during early adulthood. Increased insulin concentrations are consistent with this phenotype [33]. Despite these, there are no reports indicating an association of this gene with renal function in diabetes or in general. Our findings warrant replication studies and further functional study in addition to support the observed association between SNVs located upstream of the NBEA gene locus and eGFR in T2D.
Proteinuria is a marker of nephropathy in diabetes. Plasma proteins leak into the urine in increased glomerular permeability. These proteins taken up by proximal tubular cells can initiate an inflammatory process and interstitial scarring, ending up in fibrosis [34]. An important role of advanced glycation end products in the pathogenesis of proteinuria and degenerative changes in kidney was recently recognized [35]. In one of our confounding factor adjustment analyses, the addition of urine protein in the model did not attenuate the associations of newly found SNVs located upstream of the NBEA gene with eGFR. This indicated that the gene has an effect on eGFR independently of urinary protein.
The other confounding factor adjustment analyses, adjusting for PCA scores, or for BMI, smoking and alcohol intake did not result in an alteration of the association between the gene and eGFR. The P values were more significant and β was larger in the age sex adjusted analysis in 519 T2D patients with urine protein data compared to the results in all 955 T2D patients, despite a reduction in the number of participants. The reason for this finding is not clear. It is possible that data quality in T2D with urine protein data were better than in T2D without protein urine data.
The KCNQ1 gene was significant in our replication of prior reported SNVs in the Asian population. This was one of 7 replicated loci that were associated with eGFR in the trans-ethnic analyses of Asian participants in the meta-analysis study by Pat taro et al. The KCNQ1 gene involves a voltage-gated potassium channel that is required for the repolarization phase of the cardiac action potential. Disease conditions associated with KCNQ1 include Long QT Syndrome [36-41] and gestational diabetes mellitus [42]. Although we could not replicate the nine SNVs at the ELMO1 gene that were reported previously to be associated with nephropathy in diabetes in a Japanese population by Shimazaki et al. [22], we found 8 SNVs at the ELMO1 gene in the present study that showed significant association with eGFR (P<0.025). Success in replication studies of ELMO1 in non- Japanese populations was reported [43-47].
We should state the limitations of this study. First, we did not make a replication study in a different population. Second, there is not a large number of participants with T2D, because J-MICC is not a study specific for T2D. Third, we only had semi-quantitative urine protein data in a part of the patients, and we lacked important variables such as duration of T2D.
In conclusion, we have discovered that the 13q35.43-35.46 locus is associated with eGFR in T2D participants among a Japanese population. Future studies are needed to examine the biological mechanism that relates the locus and renal function in T2D.
Acknowledgment
We would like to thank all the staff of the Laboratory for Genotyping Development, Center for the Integrative Medical Sciences, RIKEN, and the staff of the BioBank Japan project.
The following grants supported fully or in part this study: Grantsin- Aid for Scientific Research for Priority Areas of Cancer (No. 17015018) and Innovative Areas (No. 221S0001), and JSPS KAKENHI Grants (No. 16H06277 and 15H02524) from the Japanese Ministry of Education, Culture, Sports, Science and Technology, a Grant-in-Aid from the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant B Numbers 24390165, 20390184, 17390186; (Grant-in-Aid for Young Scientists) Numbers 15K19236 and 17K15840, funding for the BioBank Japan Project from the Japan Agency for Medical Research and development since April 2015, and the Ministry of Education, Culture, Sports, Science and Technology from April 2003 to March 2015.
References
- United States Renal Data System (2017) Annual Data Report Volume 1– Chronic Kidney Disease (CKD) in the United States. Chapter 1: CKD in the General Population.
- Nakai S, Shinzato T, Sanaka T, kikuchi K, Kitaoka T, et al. (2002) The current state of chronic dialysis treatment in Japan. J Jpn Soc Dial Ðer 35: 1155-1184.
- Krolewski AS, Waram JH, Rand LI, Kahn CR (1987) Epidemiologic approach to the etiology of type 1 diabetes mellitus and its complications. N Engl J Med 317: 1390-1398.
- Pattaro C, Kottgen A, Teumer A, Garnaas M, Boger CA, et al. (2012) Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet 8: e1002584.
- Hamajima N (2007) The Japan Multi-Institutional Collaborative Cohort Study (J-MICC Study) to detect gene-environment interactions for cancer. Asian Pac J Cancer Prev 8: 317-323.
- Nakagawa-Senda H, Hachiya T, Shimizu A, Hosono S, Oze I, et al. (2018) A genome-wide association study in the Japanese population identifies the 12q24 locus for habitual coffee consumption: The J-MICC Study. Sci Rep 8: 1493.
- Kashiwagi A, Kasuga M, Araki E, Oka Y, Hanafusa T, et al. (2012) International clinical harmonization of glycated hemoglobin in Japan: from Japan Diabetes Society to National Glycohemoglobin Standardization Program values. J Diabetes Investig 3: 39-40.
- Matsuo S, Imai E, Horio M, Yasudaa Y, Tomita K, et al. (2009) Revised equations for estimated GFR from serum creatinine in Japan. AJKD 53: 982-992.
- Purcell S, Neale B, Todd-Brown K, Ðomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559-575.
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, et al. (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4: 7.
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904-909.
- Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2: 190.
- 1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56-65.
- 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, et al. (2015) A global reference for human genetic variation. Nature 526: 68-74.
- Kabata Y, Nakazono K, Takahashi A, Saito S, Hosono N, et al. (2008) Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am J Hum Genet 83: 445-456.
- Delaneau O, Marchini J, Zagury JF (2011) A linear complexity phasing method for thousands of genomes. Nat Methods 9: 179-181.
- Das S, Forer L, Schonherr S, Sidore C, Locke AE, et al. (2016) Next generation genotype imputation service and methods. Nat Genet 48: 1284-1287.
- Gogarten SM, Bhangale T, Conomos MP, Laurie CA, McHugh CP, et al. (2012) GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics 28: 3329-3331.
- Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 15: 263-265.
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, et al. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26: 2336-2337.
- Shimazaki A, Kawamura Y, Kanazawa A, Sekine A, Saito S, et al. (2005) Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes 54: 1171-1178.
- Tyner C, Barber GP, Casper J, Clawson H, Diekhans M, et al. (2017) The UCSC Genome Browser database: 2017 update. Nucleic Acids Res 45: 626-634.
- Aken BL, Achuthan P, Akanni W, Amode MR, Bernsdorff F, et al.(2017) Ensembl 2017. Nucleic Acids Res 45: 635-642.
- Consortium GT (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648-660.
- Jacobsen KK, Nievergelt CM, Zayats T, Greenwood TA, Anttila V, et al. (2015) Genome wide association study identifies variants in NBEA associated with migraine in bipolar disorder. J Affect Disord 72: 453-461.
- Castermans D, Wilquet V, Parthoens E, Huysmans C, Steyaert J, et al. (2003) The neurobeachin gene is disrupted by a translocation in a patient with idiopathic autism. J Med Genet 40: 352-356.
- Camargo LM, Collura V, Rain JC, Mizuguchi K, Hermjakob H, et al. (2007) Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry 12: 74-86.
- Gratacos M, Costas J, De Cid R, Bayes M, Gonzalez JR, et al. (2009) Identification of new putative susceptibility genes for several psychiatric disorders by association analysis of regulatory and non-synonymous SNVs of 306 genes involved in neurotransmission and neurodevelopment. Am J Med Genet B Neuropsychiatr Genet 150: 808-816.
- O'Neal J, Gao F, Hassan A, Monahan R, Barrios S, et al. (2009) Neurobeachin (NBEA) is a target of recurrent interstitial deletions at 13q13 in patients with MGUS and multiple myeloma. Exp Hematol 37: 234-244.
- Nagoshi H, Taki T, Hanamura I, Nitta M, Otsuki T, et al. (2012) Frequent PVT1 rearrangement and novel chimeric genes PVT1-NBEA and PVT1- WWOX occur in multiple myeloma with 8q24 abnormality. Cancer Res 72: 4954-4962.
- Olszewski PK, Rozman J, Jacobsson JA, Rathkolb B, Stromberg S, et al. (2012) Neurobeachin, a regulator of synaptic protein targeting, is associated with body fat mass and feeding behavior in mice and bodymass index in humans. PLoS Genet. 8: e1002568.
- http://www.clevelandclinicmeded.com/medicalpubs/diseasemanagement/ nephrology/diabetic-nephropathy/
- Kumar P A, Chitra PS, Reddy GB (2016) Advanced glycation endproducts mediated cellular and molecular events in the pathology of diabetic nephropathy. BioMol Concepts 7: 293-309.
- Bruce HA, Kochunov P, Paciga SA, Hyde CL, Chen X, et al. (2017) Potassium channel gene associations with joint processing speed and white matter impairments in schizophrenia. Genes Brain Behav 16: 515-521.
- Amin AS, Pinto YM, Wilde AA (2013) Long QT syndrome: beyond the causal mutation. J Physiol 591: 4125-4139.
- Amin AS, Giudicessi JR, Tijsen AJ, Spanjaart AM, Reckman YJ, et al. (2012) Variants in the 3' untranslated region of the KCNQ1-encoded Kv7.1 potassium channel modify disease severity in patients with type 1 long QT syndrome in an allele-specific manner. Eur Heart J 33: 714-723.
- Westaway SK, Reinier K, Huertas-Vazquez A, Evanado A, Teodorescu C, et al. (2011) Common variants in CASQ2, GPD1L, and NOS1AP are significantly associated with risk of sudden death in patients with coronary artery disease. Circ Cardiovasc Genet 4: 397-402.
- Daelemans C, Ritchie ME, Smits G, Abu-Amero S, Sudbery IM, et al. (2010) High-throughput analysis of candidate imprinted genes and allele-specific gene expression in the human term placenta. BMC Genet 11: 25.
- Sudandiradoss C, Sethumadhavan R (2009) In silico investigations on functional and haplotype tag SNPs associated with congenital long QT syndromes (LQTSs). Genomic Med 2: 55-67.
- Wang X, Li W, Ma L, Ping F, Liu J, et al. (2017) Investigation of miRNA-binding site variants and risk of gestational diabetes mellitus in Chinese pregnant women. Acta Diabetol 54: 309-316.
- Pezzolesi MG, Katavetin P, Kure M, Poznik GD, Skupien J, et al. (2009) Confirmation of genetic associations at ELMO1 in the GoKinD collection supports its role as a susceptibility gene in diabetic nephropathy. Diabetes 8: 2698-2702.
- Leak TS, Perlegas PS, Smith SG, Keene KL, Hicks PJ, et al. (2009) Variants in intron 13 of the ELMO1 gene are associated with diabetic nephropathy in African Americans. Ann Hum Genet 73: 152-159.
- Wu HY, Wang Y, Chen M, Zhang X, Wang D, et al. (2013) Association of ELMO1 gene polymorphisms with diabetic nephropathy in Chinese population. J Endocrinol Invest 36: 298-302.
- Craig DW, Millis MP, DiStefano JK (2009) Genome-wide SNP genotyping study with use of pooled DNA to identify candidate markers mediating susceptibility to end-stage renal disease attributed to Type 1 diabetes. Diabet Med 26: 1090-1098.
- Williams WW, Salem RM, McKnight AJ, Sandholm N, Forsblom C, et al. (2012) Association testing of previously reported variants in a large casecontrol metaanalysis of diabetic nephropathy. Diabetes 61: 2187-2194.
Citation: Nakamura Y, Narita A, Hachiya T, Sutoh Y, Shimizu A, et al. (2018) A Genome-wide Association Study in the Diabetic Patients Finds the 13q35.43-35.46 Locus Associated with Estimated Glomerular Filtration Rate: The Japan Multi-Institutional Collaborative Cohort Study. J Clin Diabetes 2: 102.
Copyright: © 2018 Nakamura Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 5569
- [From(publication date): 0-2018 - Nov 27, 2025]
- Breakdown by view type
- HTML page views: 4586
- PDF downloads: 983