Research Article Open Access
A Do-it-Yourself Protocol for Making Desired Mutant Proteins
Xue-Ying He1, Carl Dobkin2 and Song-Yu Yang1,3*1Developmental Biochemistry, NYS Institute for Basic Research in Developmental Disabilities, 1050 Forest Hill Road, Staten Island, NY 10314, USA
2Depatment of Human Genetics, NYS Institute for Basic Research in Developmental Disabilities, 1050 Forest Hill Road, Staten Island, NY 10314, USA
3Department of Neuroscience, Graduate School of the City University of New York, 365 Fifth Avenue, New York, NY 10016, USA.
- *Corresponding Author:
- Song-Yu Yang
Department of Developmental Biochemistry
NYS Institute for Basic Research in Developmental Disabilities
1050 Forest Hill Road, Staten Island, NY 10314, USA
Tel: +1-718494-5317
Fax: +1-718698-7916
E-mail: songyu.yang@csi.cuny.edu
Received date: September 4, 2015; Accepted date: October 14, 2015;Published date: October 21, 2015
Citation: He XY, Dobkin C, Yang SY (2015) A Do-it-Yourself Protocol for Making Desired Mutant Proteins. J Chem Biol Ther 1:101. doi: 10.4172/2572-0406.1000101
Copyright: © 2015 He XY, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Chemical Biology & Therapeutics
Abstract
The generation of a mutant protein is often crucial to the elucidation of the pathogenesis of a genetic disease. This requires site-directed mutagenesis of an expression vector containing the gene of interest. Current site-directed mutagenesis procedures are derivatives of either a restriction site elimination method or a PCR-based mutagenesis method, which involve a commercial kit manufactured by any one of a number of biotechnology companies. A new efficient economical site-directed mutagenesis procedure is described here to provide researchers an additional choice that does not to rely on a commercial kit. This novel method has been validated, and requires only basic molecular biological techniques and reagents available in most laboratories even in developing countries. The mutation efficiency of this new method is usually greater than 80%. In case several different mutants of a protein are required for elucidation of the protein structure-function relationships, this method has distinct advantages since multiple mutations can be derived from the initial single strand DNA template.
Keywords
Site-directed mutagenesis; Protein structure-function relationship; Fatty acid oxidation enzymes; HSD10
Introduction
The etiology of most genetic diseases has been found to be a mutation in a gene’s coding region. For example, various mutations have been identified on the HSD17B10 gene, resulting in 17β- hydroxysteroid dehydrogenase type 10 (HSD10) deficiency and mental retardation, choreoathetosis, abnormal behavior (MRXS10) [1].
To elucidate the pathogenesis of many genetic diseases, it is necessary to synthesize mutants of their protein gene products. Site directed mutagenesis is not only essential for this goal but is also critical to advances in biological science, especially for protein engineering and improved understanding of protein structurefunction relationships.
Site specific mutagenesis was first achieved in 1970s [2]. Thereafter, the polymerase chain reaction (PCR) technique simplified the specific alteration of DNA sequences, and the unique restriction digestion raised the mutagenesis efficiency. Site-directed mutagenesis kits on the market are relatively expensive because of the proprietary DNA polymerases and competent cells they contain.
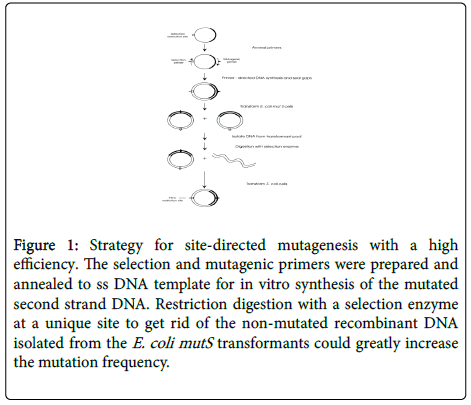
Here we report a do-it-yourself protocol for making desired mutant proteins. This efficient economical method could make a desired mutation at virtually any position in a target gene (Figure 1).
Figure 1: Strategy for site-directed mutagenesis with a high efficiency. The selection and mutagenic primers were prepared and annealed to ss DNA template for in vitro synthesis of the mutated second strand DNA. Restriction digestion with a selection enzyme at a unique site to get rid of the non-mutated recombinant DNA isolated from the E. coli mutS transformants could greatly increase the mutation frequency.
Materials and Methods
Anneal primer(s)
The single strand (ss) DNA template containing the target gene fadB [3] cloned into an M13 vector was prepared according to the published procedure [4,5]. It was then primed with one or more 5’phosphorylated mutagenic primer(s) and a 5’phosphorylated selection primer (5’-CAGGCATGCACGCGTGGCGTAATC-3’) with a Mlu I restriction site instead of a Hind III site. The mismatched nucleotide(s) were underlined, and had to be flanked by at least 10 matched nucleotides on each side. The selection primer was designed to change a unique restriction site (Figure 1). The nucleotide sequence of mutagenic primer used in the current study was 5’- TGCGCTGGGCTTTGGCTGCGAA-3’. 0.1 μg of ssDNA template was mixed with appropriate primers (0.1 μg each), 2 μl of 10x annealing buffer (200 mM Tris-HCl, pH 7.5, 100 mM MgCl2, and 500 mM NaCl) and water to a total volume 20 μl. After incubation at 100°C for 1 min the annealing mixture was transferred into a 40°C water bath for 3 min, and then put on ice for 5 min.
Synthesis of second strand DNA
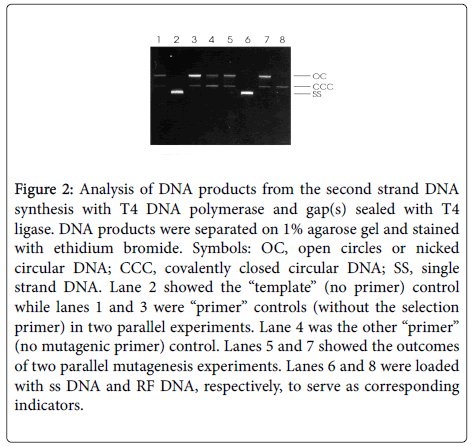
In order to extend the primers and to seal the gaps in the newly synthesized second strand, 2 μl of 10x synthesis buffer (100 mM Tris- HCl, pH 7.5, 5 mM each dNTP, 10 mM ATP and 20 mM dithiothreitol), 1 μl T4 DNA polymerase (3units). 1 μl T4 DNA ligase (6 units) and 5 μl H2O were added into the annealing mixture, and then incubated at 37°C for 2 h. The in vitro synthesis reaction was stopped by heating the mixture at 70°C for 5 min. Controls were treated with the same procedures except that no primer was added (template control) or only one primer was included (primer control). Thereafter 5 μl of the reaction mixture and 5 μl of each control were analyzed by electrophoresis on a 1% agarose gel (Figure 2).
Figure 2: Analysis of DNA products from the second strand DNA synthesis with T4 DNA polymerase and gap(s) sealed with T4 ligase. DNA products were separated on 1% agarose gel and stained with ethidium bromide. Symbols: OC, open circles or nicked circular DNA; CCC, covalently closed circular DNA; SS, single strand DNA. Lane 2 showed the “template” (no primer) control while lanes 1 and 3 were “primer” controls (without the selection primer) in two parallel experiments. Lane 4 was the other “primer” (no mutagenic primer) control. Lanes 5 and 7 showed the outcomes of two parallel mutagenesis experiments. Lanes 6 and 8 were loaded with ss DNA and RF DNA, respectively, to serve as corresponding indicators.
Transformation of E. coli mutator strain
When it was determined that in vitro synthesis was satisfactory as shown in Figure 2, only those reaction mixtures containing the desired mutant DNAs were used to transform the E. coli mutS competent cells prepared with a one-step method [6]. 100 μl competent BMH 71-18 mutS cells were gently mixed with 10 μl of the synthesis reaction mixture, and incubated on ice for 20 min. After a heat shock at 42°C for 1 min, the cells were gently mixed with 0.5 ml LB medium and incubated at 37°C for 45 min as a recovery period. The transformants were amplified by adding 5 ml LB medium, and incubated at 37°C overnight with shaking at 200 rpm.
Digestion with selection enzyme
The replicative form (RF) DNA was isolated from the transformant pool using a standard miniprep procedure [7], and then digested with the selection enzyme as follows: 0.1 μg RF DNA was mixed with 1 μl of the appropriate restriction enzyme, 10 units of Hind III (Promega, WI) in the current study, 2 μl of 10x digestion buffer, and water to a total volume of 20 μl, and incubated at 37°C for 2 h. Then, an additional 1 μl of the same restriction enzyme was added and incubated at 37°C for 1h. The total enzyme used should not be more than 1/10 volume of the digestion system to avoid any inhibition caused by glycerol. The linearization of parental DNA decreased its transformation efficiency more than a hundred-fold.
Amplification of mutant DNA
5 μl of the restriction enzyme digest was used to transform 100 μl of competent E. coli MV1190 cells, and the transformants were plated to generate plaques. Several plaques were picked randomly, and each was incubated with 100 μl of overnight culture of MV1190 in 10 ml LB medium supplemented with 5.0 mM MgSO4 at 37°C for 3 h with shaking at 300 rpm. The RF DNA was isolated from the infected E. coli cells by a standard miniprep procedure [7].
Verification of desired mutation(s)
Since the mutation frequency by this method is usually very high, a preliminary screening of mutants by restriction digestion was omitted. The desired mutation(s) was readily identified by DNA sequencing of the coding region of fadB gene.
Overexpression and purification of mutant protein
The mutant expression plasmids were transformed into E. coli BL21(DE3)pLysS according to one-step method [6]. The transformants were grown in 2YT medium to an absorbance of about 1.0 at 600 nm and then induced with 0.5 mM isopropyl-1-thio-β-Dgalactopyranoside (IPTG) for 4 h. Cells were harvested by centrifugation for 10 min at 3000x g, 4°C, and washed twice with icecold 200 mM KPi, pH 8.0. The preparation of cell extract, purification of mutant protein FAOMP (fatty acid oxidation multifunctional protein) G116F, and the characterization of the mutant protein were performed as described previously [8].
Results and Discussion
Efficient in vitro synthesis of the mutated second strand DNA is essential for achieving a high frequency of mutation. As shown in Figure 2, almost all the single DNA template has been converted to double strand DNA either nicked or covalently closed circular DNA in this study (Figure 2). In contrast, the synthesis of the mutated second strand DNA was very poor in the unique site elimination (USE) method [9] because it was severely inhibited by the annealing of the two complementary parental strands. This provides an explanation for the failure of the improved USE method to mutate DNA cloned into some expression plasmid and phagemid vectors [9,10]. The synthesis buffer reported here contains ATP and dithiothreitol, indispensable for activities of T4 ligase and T4 polymerase, respectively. Thus, this economical site-directed mutagenesis method was successfully validated. In contrast to the improved USE method, this new efficient site-directed mutagenesis method has only one selection step rather than two, because (a) the residual ssDNA, if any, has very low transformation efficiency; (b) ssDNA is extremely refractory to restriction digestion; (c) T4 DNA polymerase does not have strand displacement activity; (d) the nicked or covalently closed circular, heteroduplex DNA product, containing only the mutagenic primer but not the selection primer, is a very minor component which is destroyed with the homoduplex parental DNA.
The frequency of the altering of the restriction site in this method is about 90%. For example, conversion of the Hind III site into Mlu I site of an expression vector containing fadB gene was found to be 9/10, 13/16 and 12/12 in three independent experiments. Since the 5’phosphorylated mutagenic primer was annealed with the ssDNA template simultaneously in the same reaction mixture, it would be incorporated into the mutated second strand DNA as readily as the selection primer (Figure 1). The mutagenesis frequency of this method is, therefore, not only much higher than the improved USE method [9] but also comparable to those announced for commercial PCR-based mutagenesis kits in the market, i.e., >80% mutation efficiency for single site mutagenesis. A potentially misleading statement that it is possible "....to introduce a variety of mutations in any vector in a single day" was also found in an announcement of the QuickChange II XL Site- Directed Mutagenesis Kit (Agilent), in which the Dpn I restriction digestion serves as the selection step to eliminate the methylated DNA template. However, if bacterial transformation is necessary for converting the mutated PCR product to a useful preparation of mutated expression vector, any PCR-based site-directed mutagenesis method cannot be achieved in a single day. With regard to introducing a mutation into a sequence, the first stage of this method (Figure 2) could be, indeed, completed within a few hours.
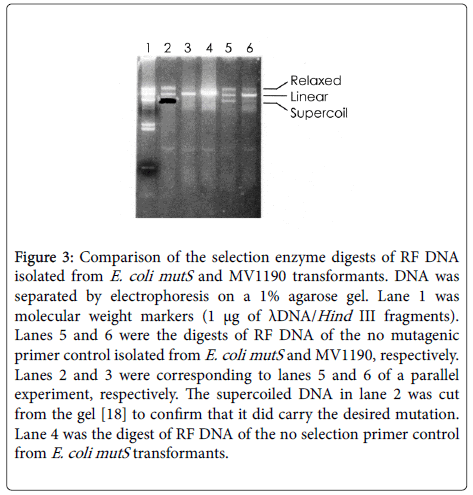
When the restriction enzyme digests of RF DNA isolated from E. coli mutS transformants were analyzed, the supercoiled and relaxed circular DNA resistant to the digestion due to the mutation accounted for about half of the total DNA (see lanes 2 and 5 of Figure 3). In contrast, the wild type MV1190 competent cells cannot serve for this purpose (see lanes 3 and 6 of Figure 3). As shown in Figure 2, the template (no primer) and one primer controls of this method serve to monitor the success in the synthesis of the mutated second strand DNA. This efficient and economical site-directed mutagenesis method is especially suitable for protein engineering and studies on structurefunction relationships of an important protein such as the fatty acid oxidation and related enzymes: hydratase, HADH (3-hydroxyacyl-CoA dehydrogenase), and HADHII (17β-hydroxysteroid dehydrogenase type 10) [11-13]. The mutant protein FAOMPG116F was found to lose both enoyl-CoA hydratase and 3-hydroxyacyl-CoA epimerase activities while retaining 3-hydroxyacyl-CoA dehydrogenase and some of its Δ 3-cis-Δ 2-trans-enoyl -CoA isomerase activities due to the substitution of phenylalanine for glycine at residue 116. If several different mutants need to be made and characterized, the ssDNA template could be repeatedly utilized (Table 1).
Figure 3: Comparison of the selection enzyme digests of RF DNA isolated from E. coli mutS and MV1190 transformants. DNA was separated by electrophoresis on a 1% agarose gel. Lane 1 was molecular weight markers (1 μg of λDNA/Hind III fragments). Lanes 5 and 6 were the digests of RF DNA of the no mutagenic primer control isolated from E. coli mutS and MV1190, respectively. Lanes 2 and 3 were corresponding to lanes 5 and 6 of a parallel experiment, respectively. The supercoiled DNA in lane 2 was cut from the gel [18] to confirm that it did carry the desired mutation. Lane 4 was the digest of RF DNA of the no selection primer control from E. coli mutS transformants.
| Mutagenic primer Nucleotide sequence | Mutant protein | Ratios of specific activities of mutant vs WT (%) | Ref | ||||
|---|---|---|---|---|---|---|---|
| Hydratase | Dehydrogenase | Isomerase | Epimerase | ||||
| 5’-CCGGATCTGCA AATCGGTCTGC-3’ | FAOMPR134Q | 62 | 106 | 155 | 122 | 14 | |
| 5’-CGGTCTGCCGC AAACCAAACT-3’ | FAOMPE139Q | 0 | 115 | 33 | 0 | 14 | |
| 5’-CTGGGTGCAGC GATTATGGG-3’ | FAOMPG322A | 170 | 46 | 103 | NDa | 15 | |
| 5’-CTGCGGGATGG CCTTCTTTAACC-3’ | FAOMPH450A | 63 | 0 | 64 | NDa | 15 | |
| 5’-CGGGATGCAAT TCTTTAACC-3’ | FAOMPH450Q | 45 | 0 | 33 | NDa | 15 | |
| 5’GGTGGCTGCCAATGCGTGCT-3’ | FAOMPE119Q | 0.9 | 62 | 5 | NDa | 16 | |
| 5’GTCGGTGCTAATATCACCGAA-3’ | FAOMPD69N | 9 | 95 | 76 | NDa | 16 | |
| 5’-GCAAGATCGCC AAAGTGATG-3’ | FAOMPD524A | 48 | 11 | 69 | NDa | 17 | |
| 5’ATCTGCTGGCCGTTGTGGG-3’ | FAOMPD542A | 93 | 6 | 32 | NDa | 17 | |
| 5’-GCCGTTGGTAC AAATTATTCGCG-3’ | FAOMPE462Q | 62 | 0.4 | 37 | NDa | 17 | |
| 5’-GCCGTTGGTAG CAATTATTCGCG-3’ | FAOMPE462A | 83 | 1.5 | 47 | NDa | 17 | |
| *A selection primer (5’-CAGGCATGCACGCGTGGCGTAATC-3’) was used for site-directed mutagenesis experiments. In each of these 11 experiments 5 clones were chosen at random for sequence analysis. All clones contained the desired mutations. aND, not done. | |||||||
Table 1: Utilization of a ss DNA template for making various mutant proteins*
Moreover, the production of M13 phage and ssDNA has become much easier [5]. Last but not least, this method requires only basic molecular biological techniques and reagents that are readily available in most laboratories in economically developed as well as in developing countries.
Acknowledgements
This work was supported in part by the New York State Office for People with Developmental Disabilities.
References
- Yang SY, Dobkin C, He XY, Philipp M, Brown WT (2013) A 5-methylcytosine hotspot responsible for the prevalent HSD17B10 mutation. Gene 515: 380-384.
- Hutchinson CA, Philipps S, Edgeell MH, Gilliam S, Jahnke P, et al. (1978) Mutagenesis at a specific position in a DNA sequence. J BiolChem 253: 6551-6560.
- Yang XYH, Schulz H, ElzingaM,Yang SY (1991) Nucleotide sequence of the promoter and fadB gene of the fadBA operon and primary structure of the multifunctional fatty acid oxidation protein from Escherichia coli. Biochemistry 30: 6788-6795.
- Yang SY, Elzinga M (1993) Association of both enoyl-CoA hydratase and 3-hydroxyacyl-CoA epimerase with an active site in the amino-terminal domain of the multifunctional fatty acid oxidation protein from Escherichia coli. J BiolChem268: 6588-6592.
- Reddy P, McKenny K (1996) Improved method for the production of M13 phage and single-strand DNA for DNA sequencing. BioTechniques 20: 854-860.
- Chung, CT, Niemela SL, Miller RH (1989) One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. ProcNatlAcadSci USA 86: 2172-2175.
- Serghini MA, RitzenthalerC,Pinck L (1989) A rapid and efficient 'miniprep' for isolation of plasmid DNA. Nucleic Acids Res 17: 3604.
- Yang SY, Li J, He XY, Cosloy SD, Schulz H (1988) Evidence that the fadB gene of the fadAB operon of Escherichia coli encodes 3-hydroxyacyl-coenzyme A (CoA) epimerase, ?3-cis-?2-trans-enoyl-CoA isomerase and enoyl-CoA hydratase in addition to 3-hydroxyacyl-CoA dehydrogenase. J Bacteriol 170: 2543-2548.
- Zhu L, Holtz AE (1993) Improved protocol for the TransformerTM site directed mutagenesis. CLONTECHniques VIII (4): 22-23.
- Wong F, Komaromy M (1995) Site-directed mutagenesis using thermostable enzymes. Biotechniques 18: 1034-1038.
- Yang SY, He XY, Schulz H (2005) 3-Hydroxyacyl-CoA dehydrogenase and short-chain 3-hydroxyacyl-CoA dehydrogenase in human health and disease. FEBS J 272: 4874-4883.
- Yang SY, He XY (1999) Molecular mechanisms of the fatty acid b-oxidation enzyme catalysis:Current Views of Fatty Acid Oxidation and Ketogenesis from Organelles to Point Mutations. Kluwer Academic/Plenum Publishers, New York, NY.
- Yang SY, He XY, Isaacs C, Dobkin C, Miller D, et al. (2014) Roles of 17beta-hydroxysteroid dehydrogenase type 10 in neurodegenerative disorders. J Steroid BiochemMolBiol 143: 460-472.
- Yang SY, He XY, Schulz H (1995) Glutamate 139 of the large a-subunit is the catalytic base in the dehydration of both D- and L-3-hydroacyl-CoA but not the isomerization of ?-3, ?2-enoyl-CoA catalyzed by the multienzyme complex of fatty acid oxidation from Escherichia coli. Biochemistry 34: 6441-6447.
- He XY, Yang SY (1996) Histidine-450 is the catalytic residue of L-3-hydroxyacyl-CoA dehydrogenase associated with the large a-subunit of the multienzyme complex of fatty acid oxidation from Escherichia coli. Biochemistry 35: 9625-9630.
- He XY, Yang SY (1997) Glutamate-119 of the large a-subunit is the catalytic base in the hydration of 2-trans-enoyl-coenzyme A catalyzed by the multienzyme complex of fatty acid oxidation from Escherichia coli. Biochemistry 36: 11044-11049.
- He XY, DengH,Yang SY (1997) Importance of the g-carboxyl group of the large a-subunit for the catalytic function and the stability of the multienzyme complex of fatty acid oxidation from Escherichia coli. Biochemistry 36: 261-268.
- Volgestein B, Gillespie D (1979) Preparative and analytical purification of DNA from agarose. ProcNatlAcad USA 76: 615-619.
Relevant Topics
- Analytical Methods
- Biocatalysis and biotransformation
- Bioinorganic Chemistry
- Biological chemistry
- Bioorganic chemistry
- Chemical Biology of Tetracyclines
- Chemotherapeutic Agents
- Enzyme Inhibitor
- Mechanisms of DNA Damage and Repair
- Nanomaterials For Imaging and Drug Delivery
- Natural Product Biosynthesis
- Nucleic Acid Analogs
- Spectroscopic Probes
Recommended Journals
Article Tools
Article Usage
- Total views: 18997
- [From(publication date):
February-2016 - Jul 13, 2025] - Breakdown by view type
- HTML page views : 17916
- PDF downloads : 1081