A Comprehensive Bioinformatics Analysis of Sarcopenia and Covid-19
Received: 06-Jun-2022 / Manuscript No. bcp-22-67393 / Editor assigned: 09-Jun-2022 / PreQC No. bcp-22-67393 / Reviewed: 20-Jun-2022 / QC No. bcp-22-67393 / Revised: 25-Jun-2022 / Manuscript No. bcp-22-67393 / Published Date: 30-Jun-2022 DOI: 10.4172/2168-9652.1000384
Abstract
Although more and more evidence has supported that the interaction between sarcopenia and Corona Virus Disease 2019(COVID-19) could be bidirectional and may form a vicious circle, the common mechanism of its occurrence is still not fully elucidated. Our study reveals the common pathogenesis and potential therapeutic targets of sarcopenia and COVID-19. These common pathways and potential drugs may provide new ideas for further mechanism research.
Keywords
Sarcopenia; COVID-19; Bioinformatic gene analysis; Therapeutic agents
Introduction
Sarcopenia is a clinical syndrome characterized by continuous loss of skeletal muscle mass, muscle strength or function [1]. It is also closely associated with metabolic diseases and cognitive dysfunction, resulting in serious adverse effects on people’s quality of life and inflicting a heavy socio-economic burden [2]. Corona Virus Disease 2019(COVID-19) caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is a novel respiratory infectious disease. Since the outbreak and epidemic, the majority of patients have been elderly. Fever caused by virus infection, loss of appetite, hypoxemia, and a long time in bed, lack of exercise, not only not conducive to the recovery of the disease, but also cause sarcopenia, weakness [3].
It is now clear that survivors of COVID-19 are at increased risk of acute sarcopenia [4], with worsening muscle dysfunction in many cases [5]. Similarly, patients with sarcopenia have a higher susceptibility to COVID-19 [6]and sarcopenia has a potential impact on the severity of COVID-19 infection [7]. Sarcopenia shares several pathogenic mechanisms with disease severity of COVID-19 infection, including intrinsic (systemic inflammation, oxidative stress, tissue hypoxia) and extrinsic (physical inactivity, malnutrition, comorbidity) factors [8, 9]. This study attempted to identify the biological pathways of sarcopenia and COVID-19 and their interrelationships. We analyzed two gene expression datasets (GSE1428 for sarcopenia and GSE154998 for COVID-19) downloaded from the Gene expression omnibus (GEO) database. First, we identified differentially expressed genes (DEGs) for GSE1428 and GSE154998, and the common differentially expressed genes were the basis of the whole study. Comprehensive bioinformatics and enrichment analysis were used to determine the common DEGs. Subsequently, protein interaction networks are formed to identify HUB genes from DEGs and search for potential therapeutic agents.
Material and Methods
Data source GEO (http://www.ncbi.nlm.nih.gov/geo) [10] is a public database containing a large number of high-throughput sequencing and microarray data sets submitted by research institutes worldwide. We searched for related gene expression datasets using “sarcopenia” and “COVID-19 infection” as keywords and the test specimens included should be from humans. Finally, GSE1428 dataset [11] and GSE154998 dataset [12] were downloaded from it. The GSE1428 dataset, which contains 12 sarcopenia samples and 10 normal samples, was performed using the GPL96 (Affymetrix Human Genome U133 Array) platform. The GSE154998 dataset, which contains 7 COVID-19 positive samples and 7 COVID-19 negative samples, was performed using the GPL18573 NextSeq 500 platform (Figure 1).
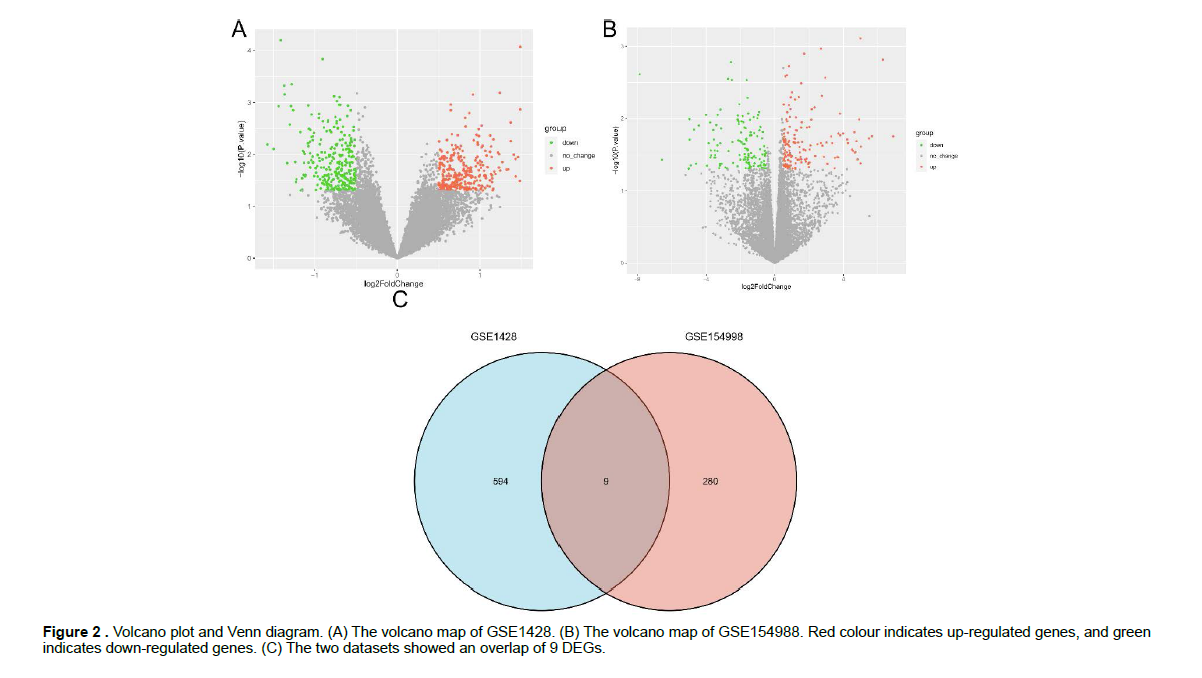
Identification of differentially expressed genes (DEGs) To assess differential expression, using the “limma” package of R software [13]. Normalized expression values according to the “normalizeBetweenArrays” function of the package so that the expression values have similar distribution. A gene was defined as a DEG when the P value was < 0.05 and |log2 FC| ≥ 0.5, which were visualized as Volcano plots. The web tool (http://bioinfogp.cnb. csic.es/tools/venny/index.html) was used to achieve universal gene identification between the DEGs of the GSE1428 and GSE154998 datasets (Figure 2).
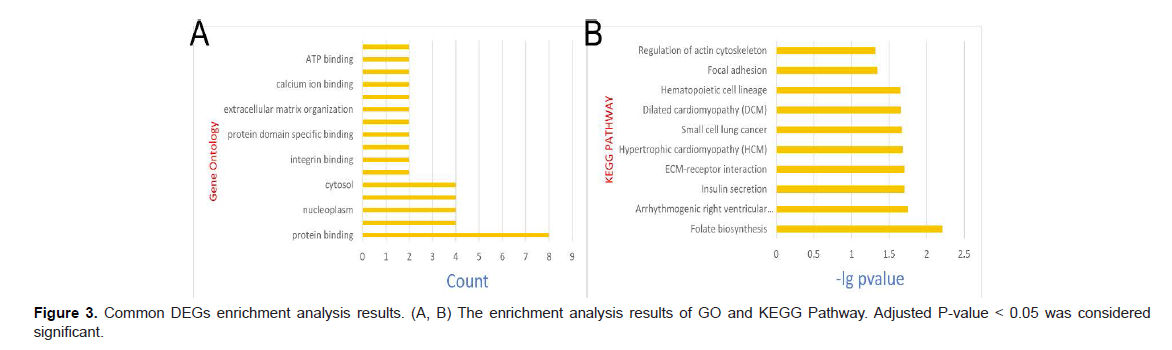
Functional and pathway enrichment analyses of DEGs Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology Based Annotation System (KOBAS) (http://kobas. cbi.pku.edu.cn) [14] is a Web server for gene/protein functional annotation and functional enrichment developed by Peking University, which collects 4325 species functional annotation information. The enrichment analysis results of Gene Ontology (GO) and KEGG were obtained from the KOBAS 3.0 database. P value < 0.05 was considered significant (Figure 3).
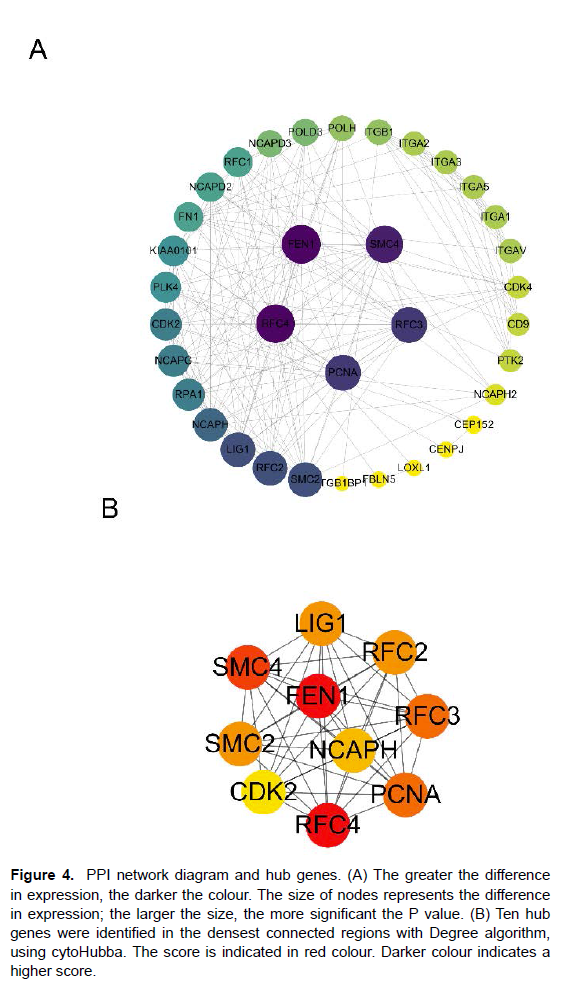
PPI network construction and selection of hub genes The Protein-protein interaction (PPI) network was analyzed using the Search Tool for the Retrieval of Interacting Genes (STRING; http:// string-db.org). Analysis of functional interactions between proteins was performed in order to elucidate the mechanisms of osteogenesis and development [15]. An interaction with a combined score > 0.4 was selected and used to construct a PPI network with Cytoscape software.
Cytoscape (version 3.9.1) is an open source bioinformatics software platform for visualizing molecular interaction networks [16, 17]. The top 10 genes were obtained by Degree algorithm with Cytoscape’s plug-in cytoHubba.
TF-gene interactions and TF-miRNA coregulatory network NetworkAnalyst (https://www.networkanalyst.ca/) is a webbased platform for performing gene comparisons, quantification, and differential gene expression analysis for numerous species [18].
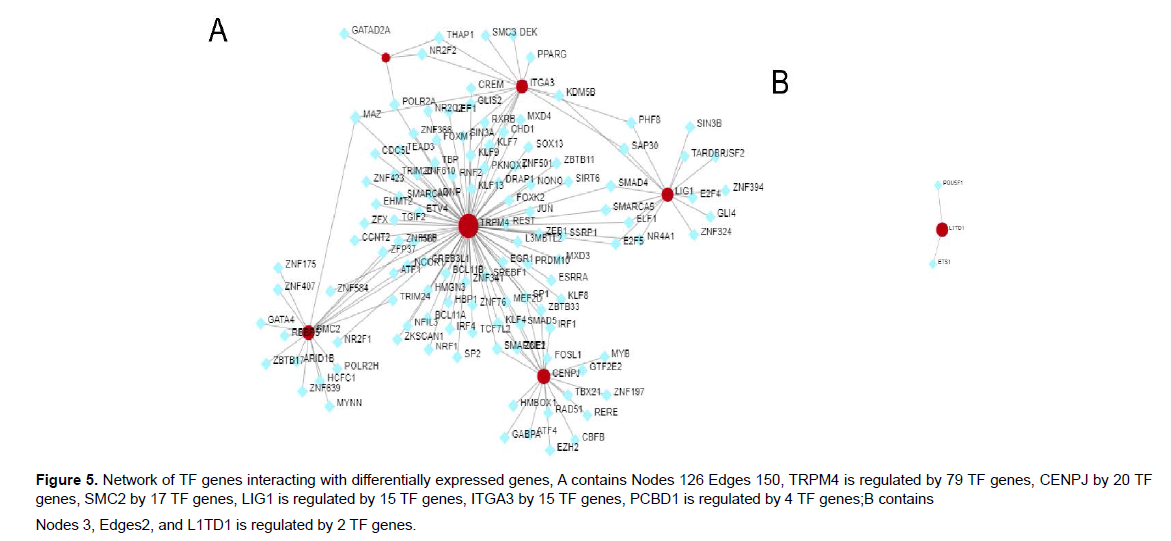
Interaction of transfer factor (TF) with identified common DEGs as a result of TF on functional pathways and gene expression levels was evaluated[19]. Identification of TF gene interactions with common genes via NetworkAnalyst platform. The TF-gene interaction network was obtained from the ENCODE (https://www.encodeproject.org/)) database which in the platform. Also, TF-miRNA coregulatory network was obtained through the analysis of differential genes through the platform (Figure 4) (Table 1).
| Name | Score |
|---|---|
| FEN1 | 19 |
| RFC4 | 19 |
| SMC4 | 18 |
| PCNA | 17 |
| RFC3 | 17 |
| RFC2 | 16 |
| SMC2 | 16 |
| LIG1 | 16 |
| NCAPH | 15 |
| CDK2 | 14 |
Table 1: 10 HUB genes and their scores.
PPI network diagram and hub genes. (A)The greater the difference in expression, the darker the colour. The size of nodes represents the difference in expression; the larger the size, the more significant the P value. (B)Ten hub genes were identified in the densest connected regions with Degree algorithm, using cytoHubba. The score is indicated in red colour. Darker colour indicates a higher score.
Screening of candidate drugs
The drug-gene interaction database (DGIdb, http://www.dgidb. org/) is an online resource that presents drug-gene interactions from different sources such as databases and web resources [20]. Candidates selected from detected drug-gene interactions were drugs that had two or more data sources and/or PubMed literature supporting druggene interactions and were approved by the U.S. Food and Drug Administration (FDA) (Figure 5).
Results
Identification of DEGs Gene expression levels of GEO series that have been standardized and the results of pre- and post- standardized were presented in (Figure 1). A total of 603 DEGs with |log2 FC| ≥ 0.5 in sarcopenia samples compared with normal samples was identified, including 314 up-regulated genes and 289 down-regulated genes. A total of 289 DEGs with |log2 FC| ≥ 0.5 in COVID-19 positive samples compared with COVID-19 negative samples was identified, including 154 up-regulated genes and 135 down-regulated genes. Volcano plot of DEGs enrolled in subsequent analyses was showed in Figure 2A,2B. A total of 603 collected sarcopenia genes and 289 COVID-19 genes were compared, and a total of 9 common genes were identified for expression (LIG1 、ITGA3、CENPJ、SMC2、L1TD1、PCBD1、TRPM4、FBLN5 、SLC12A8). The common DEGs between the two datasets were visually compared by the Venn diagram in Figure 2C.
Analysis of the Functional Characteristics of DEGs In order to analyze the biological functions and pathways involved in the 9 common DEGs, GO and KEGG Pathway enrichment analysis were performed. GO analysis results show that these genes were mainly enriched in protein binding, nucleoplasm, cytosol (Figure 3A). In terms of KEGG Pathway, the enrichment pathways are folate biosymthesis (Figure 2B).
PPI network construction and selection of hub genes The PPI network of DEGs and most dense connected regions (35nodes, 186 edges) were obtained by Cytoscape (Figure 4A). Through the Degree algorithms of plug-in cytoHubba, we have calculated the top 10 hub genes, including FEN1、RFC4、SMC4、PCNA、RFC3、 RFC2、SMC2、LIG1、NCAPH、CDK2 (Figure 4B). The 10 hub genes score were shown in Table 1 (Figure 6).
TF gene interactions and TF-miRNA coregulatory network TF-gene interactions and TF-miRNA co-regulatory network were generated using NetworkAnalyst. Differential genes (LIG1、ITGA3、 CENPJ、SMC2、L1TD1、PCBD1、TRPM4、FBLN5、SLC12A8) were screened for TF gene identification(Figure 5). TRPM4 is regulated by 79 TF genes, CENPJ by 20 TF genes, SMC2 by 17 TF genes, LIG1 is regulated by 15 TF genes, ITGA3 by 15 TF genes, PCBD1 is regulated by 4 TF genes, L1TD1 is regulated by 2 TF genes, and these TF genes regulate over one common differential gene in the network, indicating a high degree of interaction between TF genes and differential genes.
The analysis of the TF-miRNA co-regulatory network provided the interaction of miRNAs and TF with the common DEG. This interaction may be responsible for the regulation of DEGS expression. The network created by TF-miRNA co-regulatory network consists of 141 Nodes,143 Edges, consisting of 44 TF genes and 89 miRNAs interacting with differential genes. Figure 6 shows the TF-miRNA coregulatory network.
Candidate drugs screening In the present study, eleven FDA-approved medications that possibly target the protein products of the four key genes were identified using DGIdb. Results show that the promising targets for potential drugs include the CDK2 (55.6%, 7/11), PCNA(22.2%, 2/11), FEN1, and LIG1 (11.1%, 1/9) genes (Table 2).
| Gene | Drug | Interaction types | Sources | Score |
|---|---|---|---|---|
| LIG1 | BLEOMYCIN | inhibitor | TdgClinicalTrial|TEND | 9.76 |
| PCNA | CAPSAICIN | NA | NCI | 2.75 |
| PCNA | PENTOXIFYLLINE | NA | NCI | 1.21 |
| CDK2 | ERIBULIN | NA | CIViC | 0.34 |
| CDK2 | RALTITREXED | NA | NCI | 0.29 |
| CDK2 | LOVASTATIN | NA | NCI | 0.2 |
| CDK2 | PACLITAXEL | NA | NCI | 0.09 |
| FEN1 | METHOXSALEN | NA | NCI | 0.08 |
| CDK2 | ACETAMINOPHEN | NA | NCI | 0.05 |
| CDK2 | CARBOPLATIN | NA | CIViC | 0.03 |
| CDK2 | DEXAMETHASONE | NA | NCI | 0.02 |
Table 2: Top drug compounds.
Discussion
Sarcopenia is an age-limited disease that causes a gradual loss of skeletal muscle mass and a decline in muscle strength and function [21]. It is a key risk factor for increasing the vulnerability of older adults to COVID-19 [22] and, in patients with COVID-19, preexisting sarcopenia is associated with progression of disease severity, manifested as multiple drug therapy, multiple organ failure, intensive care unit (ICU) admissions, increased need for mechanical ventilation, and mortality [23, 24]. Angiotensin converting enzyme 2(ACE2) is a receptor for coronavirus 2 and is present in skeletal muscle. It is hypothesised that long-term immune changes caused by COVID-19 lead to a reduction in the body’s ability to synthesize muscle, with the long-term sequelae of acute sarcopenia. The main mechanism of impaired immune function in patients with sarcopenia refers to abnormal actin proteins, such as interleukin-15, IL-17 and IL-6, which regulate the proliferation and function of innate and adaptive immune cells [25]. The inflammatory response to COVID-19, particularly the cytokine storm of interferon A, interferon G, IL-6, Tumor necrosis factor-A, C-reactive protein, and monocyte chemotaxis protein-1 observed in severe infections, refers to metabolic stress and increased muscle catabolic metabolism [26].
In order to further study the molecular mechanism of the relationship between the two diseases, we performed an in-depth analysis using bioinformatics methods. First, common DEGs (LIG1 、ITGA3、CENPJ、SMC2、L1TD1、PCBD1、TRPM4、FBLN5 、SLC12A8) were identified in GSE1428 and GSE154998 datasets. After identification, nine common DEGs were identified. In the following studies, GO analysis, KEGG pathway analysis, PPI, TF-gene interaction, TF-miRNA co-regulatory network and candidate drug determination were carried out.
From the results of GO analysis, it can be seen that the biological composition and function of sarcopenia and COVID-19 are roughly the same KEGG showed that common DEGs were enriched in pathway of folate biosymthesis. Folate is an essential water-soluble micronutrient that plays a key role in the nucleic acid synthesis and normal cellular function. Several studies have suggested that folate may have a positive effect on skeletal muscle development [27, 28]. Folate can increase the differentiation of skeletal muscle myoblasts, especially affecting the differentiation process and myotube morphology [29]. The activation of Akt is major mechanism that contributes to folic acid-stimulated myogenesis [29]. In one study of older mice, folate deficiency was found to have significant impact on muscle health, in the form of reduced power generation and fatigue resistance, as well as impaired physical activity [30]. In addition, folate deficiency induces an increase in plasma homocysteine (Hcy) [31], and an increase in Hcy leads to skeletal muscle weakness [32]. Meisel et al. found reduced folate levels in 38 (11.4%) of 333 patients diagnosed with COVID-19 infection [33]. Folate in SARS-COV-2-infected cells is significantly reduced, raising the possibility that host folate metabolism is hijacked to meet the viral subgenomic RNA replication needs [34]. Moreover, SARS-CoV-2 activates folate metabolism at the post-transcriptional level in newly infected cells to provide substantial requirements for ribonucleotide synthesis [34]. Collectively, we can hypothesis that the over-consumption of folate in the COVID-19 patients results in folate deficiency, which affects the normal physiological function of skeletal muscle.
We construct a complex interaction network to identify key nodes by their common DEGs. This comprehensive bioinformatics approach has been shown to be reliable in a variety of diseases [35]. The analysis of PPIs also arose from LIG1、ITGA3、CENPJ、SMC2、L1TD1 、PCBD1、TRPM4、FBLN5、SLC12A8 genes as these genes are common DEGs. According to the PPI network, FEN1、RFC4、SMC4 、PCNA、RFC3、RFC2、SMC2、LIG1、NCAPH、CDK2 genes were declared as hub genes due to their high interaction rate or degree values. In order to focus on important regions of PPI network, modular analysis of hub genes was performed. The reason for concentration in highly concentrated areas is that more effective drug compounds are recommended.
TF-gene interactions were obtained with the common DEGs. From the network, TRPM4 interacts with other TF genes at a high rate. Among the regulators, the degree of MAZ (Myc-associated zinc finger protein, MAZ) which interacted significantly in TF-gene interaction network was 3. The human MAZ gene is located on chromosome 16p11.2, and it’s regulation of transcription based on the interaction between the GC-rich DNA-binding site and its carboxylterminal zinc finger motif [36]. As an important transcription factor that is widely expressed in most tissues of the human body, it has been identified as a transcriptional regulator of muscle-specific genes in skeletal. Himeda et al. identified a variety of different MAZ motifs in the promoters of key muscle regulators such as Myogenin, MEF2C and Six4, and demonstrated that MAZ can trans-activate MCK promoters in differentiated skeletal muscle cell cultures [37]. Shoemaker et al. created a CA04 (influenza A/California/04/2009 [H1N1], referred to as CA04) differentially regulated host response network through microarrays, suggesting that MAZ had enhanced viral RNA sensing, immune cell signaling, and cell cycle arrest in CA04-infected lungs [38].
Regulatory biomolecules can serve as potential biomarkers for a variety of complex diseases. The activities of miRNAs and TF genes for the regulatory analysis of common DEGs were visualized in a TF-miRNA co-regulatory network. 89 miRNAs and 44 TF-genes are identified in the research. Among the most interacted TFs, E2F1 has the higher degree value of 3. E2F1 is an important transcription factor in normal development of organisms. E2F1 overexpression in myoblasts promotes myoblast proliferation and inhibits myoblast differentiation [39]. Blanchet et al. found that E2F1 has transcriptional regulation of metabolic oxidation reactions in the switch to oxidative muscles fibers [40]. E2F1 has been reported to be upregulated in NSCLC tissues and cell lines, as well as in primary cultured lung fibroblasts from hyperoxia exposed rats [41,42].
According to the DSigDB database, drug molecules are proposed by hub genes. Of all the drug candidates, the current study highlights the top 11 important drugs. Bleomycin (BLM) is the best candidate for COVID-19 and sarcopenia. Seo et al. showed that preconditioning mice with BLM reduced influenza virus-mediated lung inflammation, alleviated influenza virus infection mainly by activating type I interferon(IFN-I)signaling in mice, and stimulated IFN-I secretion in plasmacytoid dendritic cells (pDCs) to enhance resistance to influenza virus infection [43]. However, BLM has not been used for clinical treatment of SARS-COV-2. Since SARS-COV-2 is a new virus, relatively little research has been done so far. In the future, as more research is conducted, the current research will be more effective in the case of a SARS-COV-2 pandemic.
Conclusion
We identified the common DEGs of sarcopenia and COVID-19, and made enrichment and PPI network analysis. We found that sarcopenia and COVID-19 have many common pathogenic mechanisms, which may be mediated by specific central genes. In terms of transcriptome analysis, there are no other studies on sarcopenia and COVID-19 to date. Analysis of sarcopenia and COVID-19 predicted methods of detecting various disease infections. Drug target recommendations are logical because they are made by identifying central genes. These common pathways and potential drugs may provide new ideas for further mechanism research. However, the limitations of this study should be considered. First of all, this is a retrospective study that requires external verification to verify our findings; Secondly, the function of the hub gene needs to be further verified in an in vitro model, which will be the focus of our future work.
Availability of data and materials
GSE1428 dataset for this study is openly available in Gene Expression Omnibus database at https://www.ncbi.nlm.nih.gov/ geo/query/acc.cgi?acc=GSE1428. GSE154998 dataset for this study is openly available in Gene Expression Omnibus database at https:// www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154998.
Competing interest
The authors declare that they have no competing interests.
Acknowledgments
Thanks to each of the authors who contributed to this paper. This work is supported by the Natural Science Foundation of Shandong Province (ZR2019MH044), the TCM Science and Technology Development Plan of Shandong Province (No.2019-0164) and the Xu Zhanwang Shandong Construction Project of Inheritance Studio for Famous and Old TCM Experts ([2019] No. 92). They provided technical guidance in designing, implementing and analyzing data.
References
- Rosenberg IH (2011) Sarcopenia: origins and clinical relevance. Clin Geriatr Med. 27:337-339.
- Chen LK, Woo J, Assantachai P, Auyeung TW, Chou MY, et al (2020) Asian Working Group for Sarcopenia: 2019 Consensus Update on Sarcopenia Diagnosis and Treatment. J Am Med Dir Assoc.21:300-307.
- Chen N, Zhou M, Dong X, Qu J, Gong F, et al (2020) Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 395:507-513.
- Disser NP, De Micheli AJ, Schonk MM, Konnaris MA, Piacentini AN et al. (2020) Musculoskeletal Consequences of COVID-19. J Bone Joint Surg Am. 102:1197-1204.
- Bagnato S, Boccagni C, Marino G, Prestandrea C, D'Agostino T et al.(2020) Critical illness myopathy after COVID-19. Int J Infect Dis. 99:276-278.
- Ali AM, Kunugi H (2021) Screening for Sarcopenia (Physical Frailty) in the COVID-19 Era. Int J Endocrinol.21:556-960.
- Molfino A, Imbimbo G, Rizzo V, Muscaritoli M, Alampi D (2021) The link between nutritional status and outcomes in COVID-19 patients in ICU: Is obesity or sarcopenia the real problem? Eur J Intern Med. 91:93-95.
- Lang T, Streeper T, Cawthon P, Baldwin K, Taaffe DR et al. (2010) Sarcopenia: etiology, clinical consequences, intervention, and assessment. Osteoporos Int. 21:543-559.
- Welch C, Greig C, Masud T, Wilson D, Jackson TA (2020) COVID-19 and Acute Sarcopenia. Aging Dis. 11:1345-1351.
- Edgar R, Domrachev M, Lash AE (2002)Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210.
- Giresi PG, Stevenson EJ, Theilhaber J, Koncarevic A, Parkington J, et al. (2005) Identification of a molecular signature of sarcopenia. Physiol Genomics 21:253-263.
- Gill SE, Dos Santos CC, O'Gorman DB, Carter DE, Patterson EK, et al. (2020) Transcriptional profiling of leukocytes in critically ill COVID19 patients: implications for interferon response and coagulation. Intensive Care Med Exp. 8:75-80.
- Diboun I, Wernisch L, Orengo CA, Koltzenburg M (2006) Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics 7:252-273.
- Wu J, Mao X, Cai T, Luo J, Wei L (2006) KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 34:720-724.
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, et al.(2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45:362-368.
- Demchak B, Hull T, Reich M, Liefeld T, Smoot M, et al. (2014) Cytoscape: the network visualization tool for GenomeSpace workflows. F1000Res. 3:151-175.
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498-2504.
- Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, et al. (2019) NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 47:234-241.
- Ye Z, Wang F, Yan F, Wang L, Li B, et al. (2019) Bioinformatic identification of candidate biomarkers and related transcription factors in nasopharyngeal carcinoma. World J Surg Oncol. 17:60-74.
- Cotto KC, Wagner AH, Feng YY, Kiwala S, Coffman AC, et al. (2018) DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 46:1068-1073.
- Ali AM, Kunugi H (2020) Apitherapy for Age-Related Skeletal Muscle Dysfunction (Sarcopenia): A Review on the Effects of Royal Jelly, Propolis, and Bee Pollen. Foods. 9:1362-1376.
- Ali AM, Kunugi H (2021) Propolis, Bee Honey, and Their Components Protect against Coronavirus Disease 2019 (COVID-19): A Review of In Silico, In Vitro, and Clinical Studies. Molecules 26:1232-1236.
- Tehrani S, Killander A, Astrand P, Jakobsson J,Gille-Johnson P et al. (2021) Risk factors for death in adult COVID-19 patients: Frailty predicts fatal outcome in older patients. Int J Infect Dis. 102:415-421.
- Ma Y, Hou L, Yang X, Huang Z, Yang X et al. (2020) The association between frailty and severe disease among COVID-19 patients aged over 60 years in China: a prospective cohort study. BMC. 18:274-276.
- Nelke C, Dziewas R, Minnerup J, Meuth SG, Ruck T (2019) Skeletal muscle as potential central link between sarcopenia and immune senescence. EBio Med. 49:381-388.
- Meftahi GH, Jangravi Z, Sahraei H, Bahari Z (2020) The possible pathophysiology mechanism of cytokine storm in elderly adults with COVID-19 infection: the contribution of "inflame-aging. Inflamm Res. 69:825-839.
- Hwang SY, Kang YJ, Sung B, Jang JY, Hwang NL et al.(2018) Folic acid is necessary for proliferation and differentiation of C2C12 myoblasts. J Cell Physiol. 233:736-747.
- Hwang SY, Kang YJ, Sung B, Kim M, Kim DH et al. (2015) Folic acid promotes the myogenic differentiation of C2C12 murine myoblasts through the Akt signaling pathway. Int J Mol Med. 36:1073-1080.
- Hwang SY, Sung B, Kim ND (2019) Roles of folate in skeletal muscle cell development and functions. Archives of Pharmacal Res. 42:319-325.
- van Dijk M, Dijk FJ, Hartog A, van Norren K, Verlaan S et al. (2018) Reduced dietary intake of micronutrients with antioxidant properties negatively impacts muscle health in aged mice. J Cachexia Sarcopenia Muscle. 9:146-159.
- Linhart HG, Troen A, Bell GW, Cantu E, Chao WH et al. (2009) Folate deficiency induces genomic uracil misincorporation and hypomethylation but does not increase DNA point mutations. Gastroenterology 136:227-235.
- Veeranki S, Winchester LJ, Tyagi SC (2015) Hyperhomocysteinemia associated skeletal muscle weakness involves mitochondrial dysfunction and epigenetic modifications. Biochim Biophys Acta. 1852:732-741.
- Meisel E, Efros O, Bleier J, Beit Halevi T, Segal G et al. (2021) Folate Levels in Patients Hospitalized with Coronavirus Disease 2019. Nutrients 13:134-146.
- Zhang Y, Guo R, Kim SH, Shah H, Zhang S et al. (2021) SARS-CoV-2 hijacks folate and one-carbon metabolism for viral replication. Nat Commun. 12:1676-1764.
- Su W, Zhao Y, Wei Y, Zhang X, Ji J et al.(2021) Exploring the Pathogenesis of Psoriasis Complicated With Atherosclerosis via Microarray Data Analysis. Frontiers in Immunol.12:134-155.
- Zheng C, Wu H, Jin S, Li D, Tan S et al. (2022) Roles of Myc-associated zinc finger protein in malignant tumors. Asia Pac J Clin Oncol.23:765-786.
- Himeda CL, Ranish JA, Hauschka SD (2008) Quantitative proteomic identification of MAZ as a transcriptional regulator of muscle-specific genes in skeletal and cardiac myocytes. Mol Cell Biol. 28:6521-6535.
- Shoemaker JE, Fukuyama S, Eisfeld AJ, Muramoto Y, Watanabe S et al. (2012) Integrated network analysis reveals a novel role for the cell cycle in 2009 pandemic influenza virus-induced inflammation in macaque lungs. BMC Syst Biol. 6:117-213.
- Luo W, Li G, Yi Z, Nie Q, Zhang X (2016) E2F1-miR-20a-5p/20b-5p auto-regulatory feedback loop involved in myoblast proliferation and differentiation. Sci Rep. 6:279-304.
- Blanchet E, Annicotte JS, Lagarrigue S, Aguilar V, Clape C et al.(2011) E2F transcription factor-1 regulates oxidative metabolism. Nat Cell Biol. 13:1146-1152.
- Zhao S, Chen Z, Han S, Wu H (2021) Effects of the p16/cyclin D1/CDK4/Rb/E2F1 pathway on aberrant lung fibroblast proliferation in neonatal rats exposed to hyperoxia. Exp Ther Med.22:1057-1063.
- Meng Q, Liu M, Cheng R (2020) LINC00461/miR-4478/E2F1 feedback loop promotes non-small cell lung cancer cell proliferation and migration. Biosci Rep. 40:456-476.
- Seo SU, Jeong JH, Baek BS, Choi JM, Choi YS et al. (2021) Bleomycin-Induced Lung Injury Increases Resistance to Influenza Virus Infection in a Type I Interferon-Dependent Manner. Front Immunol. 12:697-762.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at , Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar,Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: Su Y, Xin J, Gao S, Song Y, Chen R, et al. (2022) A Comprehensive Bioinformatics Analysis of Sarcopenia and Covid-19. Biochem Physiol 11: 384. DOI: 10.4172/2168-9652.1000384
Copyright: © 2022 Su Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 3189
- [From(publication date): 0-2022 - Dec 07, 2025]
- Breakdown by view type
- HTML page views: 2693
- PDF downloads: 496