Research Article Open Access
A Comparison of 5P12-vMIP-II and vMIP-II as HIV-1 Entry Inhibitors
| Patricia J LiWang*, Jie Xue, Nai-Wei Kuo and Megan Schill | |
| Molecular Cell Biology, University of California Merced, 5200 North Lake Road, Merced, CA 95343, USA | |
| *Corresponding Author : | Patricia J LiWang Molecular Cell Biology, University of California Merced 5200 North Lake Road, Merced, CA 95343, USA Tel: 209-228- 4568 Fax: 209-724-4459 E-mail: pliwang@ucmerced.edu |
| Received February 22, 2013; Accepted April 23, 2013; Published April 26, 2013 | |
| Citation: LiWang PJ, Xue J, Kuo NW, Schill M (2013) A Comparison of 5P12-vMIPII and vMIP-II as HIV-1 Entry Inhibitors. Biochem Physiol S2:005. doi:10.4172/2168-9652.S2-005 | |
| Copyright: © 2013 LiWang PJ, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Biochemistry & Physiology: Open Access
Abstract
vMIP-II (viral macrophage inflammatory protein-II) is a chemokine analog expressed by human herpesvirus-8 that has the unique ability to bind multiple human chemokine receptors, including CCR5 and CXCR4, representative receptors of two major chemokine subfamilies. This broad binding ability gives vMIP-II powerful anti-inflammatory properties, which have been demonstrated in vitro and in vivo. In addition, vMIP-II is of great interest due to its ability to inhibit HIV infection by both major HIV strains: R5 (strains that enter the host cell using CCR5 as a co-receptor), and X4 (strains that use CXCR4). We have made a vMIP-II variant, “5P12-vMIP-II” in which the N-terminal amino acids of vMIP-II have been replaced by 10 amino acids that have been shown to greatly enhance the anti-HIV potency of the chemokine RANTES for R5 HIV strains. This 5P12-vMIP-II is shown by NMR to be fully folded and similar in structure to wild type vMIP-II. Both vMIP-II and 5P12-vMIP-II showed the ability to inhibit multiple strains of HIV, including several R5 strains and an X4 strain. While the 5P12 N-terminus did not improve the potency of the protein, our results suggest that vMIP-II does not bind CCR5 in the same way as human chemokines. Rather, vMIP-II has sacrificed some binding ability to particular chemokine receptors in order to obtain the ability to bind a broader array of receptors.
| Keywords |
| HIV entry inhibitor; Anti-inflammation; Chemokine analog; vMIP-II; Chemokine; 5P12 RANTES; NMR; CCR5 and CXCR4 |
| Introduction |
| Chemokines (chemotactic cytokines) are small proteins that mediate chemotaxis and activation of immune system cells, with importance both in immune development and inflammation. In addition, some chemokine receptors have been shown to be “coreceptors” for HIV-1 (human immunodeficiency virus type 1) entry, making the chemokine system relevant for both its role in human health and in disease. The protein vMIP-II (viral Macrophage Inflammatory Protein-II) is a chemokine analog produced by human herpesvirus 8 (HHV-8) that has high sequence identity to many chemokines as well as nearly identical tertiary structure [1-3]. However, this protein is unique among chemokines in its ability to bind but not activate receptors of chemokines from multiple sub-families, including the receptors CCR5, CXCR4, CCR1, and CCR2 [4,5]. These properties allow vMIPII to effectively compete with the natural chemokine ligands of these receptors, including MCP-1 (monocyte chemoattractant protein-1; CCL2), MIP-1α (macrophage inflammatory protein-1 alpha; CCL3) and RANTES (regulated on activation normal T cell expressed and secreted; CCL5), and have elicited interest in vMIP-II both as an antiinflammatory agent and for its ability to inhibit HIV [6]. This 71 amino acid protein is also an agonist (having the ability to bind and to cause an intracellular response) for the receptor CCR3 [4,7,8]. |
| Regarding its role in inflammation, vMIP-II has shown significant promise as an anti-inflammatory agent in animal models, including prolonging cardiac and corneal allograft survival [9,10]. vMIP-II also effectively reduced damage and neurological deficit in a spinal cord injury model [11-13] and could be safely injected into the mouse brain in order to attenuate inflammation and thereby reduce injury from cerebral ischemia [14]. |
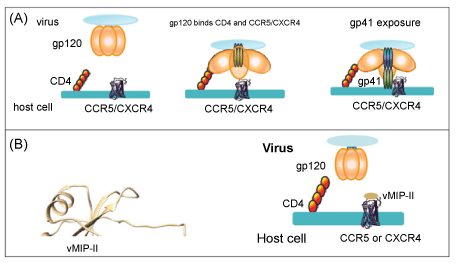
| The process of HIV infection begins with the interaction of HIV surface protein gp120 with human cell surface protein CD4 (Figure 1A). This interaction allows conformational change in gp120, leading to the binding by gp120 of the co-receptor on the cell surface, CCR5 or CXCR4. These co-receptors are chemokine receptors that normally play a role in immune activation and cell chemotaxis. The role of vMIP-II as an anti-HIV agent is based upon its unique ability to act as an antagonist of both chemokine receptors CCR5 and CXCR4 [4,6] (Figure 1B). Human chemokines, by contrast, are able to bind only receptors from one sub-class, so that MIP-1α, MIP-1β (macrophage inflammatory protein-1 beta; CCL4), and RANTES tightly bind the CCR5 receptor (and are able to inhibit infection with HIV strains that use this receptor), but have no effect on the CXCR4 receptor [15-17]. This receptor is bound by the natural ligand SDF-1 (CXCL-12), which in turn has no ability to bind CCR5 [18,19]. |
| Chemokine variants have been developed that are among the most potent HIV entry inhibitors known, with much of the work having been done on the chemokine RANTES. In particular, chemical synthesis at the N-terminus of the protein has allowed the potent RANTES variants AOP-RANTES and PSC-RANTES to be developed, revealing nanomolar activity in vitro [20,21] and, for PSC-RANTES, effectiveness in protection from HIV in the macaque in vivo [22-24]. N-terminal modification of the chemokine has been shown to be transferrable, as AOP-MIP-1α was produced and also shown to be a potent HIV inhibitor [25]. |
| The chemokine variant that currently has the highest potential for clinical use is undoubtedly 5P12-RANTES. This protein was obtained by random mutagenesis at the N-terminus of RANTES, replacing the wild type 9 amino acids with an N-terminus containing 10 amino acids. There are no further synthetic additions, so this protein can be made by recombinant techniques [26]. In addition to being highly potent (with antiviral activity in the sub-nanomolar range for most strains tested), this protein neither activates nor internalizes the CCR5 receptor, alleviating concerns about immune activation, which is not desirable in the context of HIV inhibition [26]. Later work also demonstrated that viral escape from 5P12-RANTES is extremely difficult, unlike many small molecule inhibitors [27], solidifying the position of 5P12- RANTES as a potential therapeutic that could be expected to remain potent despite a large variation in viral sequence. However, despite the nearly uniform positive properties of 5P12-RANTES, one drawback is its inability to bind the CXCR4 co-receptor, leading to inactivity against HIV viral strains that use this receptor (so-called X4 HIV strains). |
| Here, we sought to clarify two major issues regarding vMIP-II. First, could its anti-HIV properties against R5 virus be improved by substituting the N-terminus with the “5P12” sequence, while retaining inhibition against X4 virus; and the related second question, namely whether vMIP-II binds CCR5 using the same mechanism as that used by the human chemokine RANTES. The N-terminus of CC chemokines is largely unstructured until it binds the receptor, and as mentioned vMIP-II shares significant sequence identity and structural properties with RANTES, MIP-1β, and MIP-1α [3,28-30]. Therefore, this strategy could lead to a dual-acting HIV entry inhibitor that would be highly effective against both R5 and X4 strains of HIV. In addition, such work could add to our understanding of the manner in which vMIP-II is able to bind chemokine receptors. Our results show that while 5P12- vMIP-II retained activity against both R5 and X4 HIV strains, the activity was not improved by the 5P12 N-terminus compared to the wild type N-terminus. This result suggests that vMIP-II does not bind the chemokine receptor in the same manner as wild type chemokines, a possibility that has been previously suggested [31]. |
| Materials and Methods |
| Construction of 5P12-vMIP-II |
| The gene for vMIP-II was constructed and cloned into pET32a(+) as described [32]. 5P12-vMIP-II was made by replacing the first ten amino acid of vMIP-II (protein sequence: LGASWHRPDK) with the first ten amino acids of 5P12-RANTES (protein sequence: QGPPLMATQS). The replacement was carried out using oligonucleotide primers and PCR to replace the sequence coding for the N-terminal amino acids. |
| Protein production |
| vMIP-II and 5P12-vMIP-II were expressed in the pET32a(+) vector with a thioredoxin fusion tag (Novagen, Madison, WI). The plasmids were transformed into Escherichia coli BL21(DE3) (Novagen, Madison, WI) cell and expressed in 15N minimal medium using 15NH4Cl as the sole nitrogen source. The protein induction was carried |
| out with 1 mM IPTG (Isopropyl β-D-1-thiogalactopyranoside) as final concentration when the O.D.600 reached 0.6-0.8 at 37°C. The minimal medium was then shifted to 16°C and shaking continued for 16 hours. The cells were harvested by centrifugation at 6000×g for 10 min. The cell pellet was resuspended in 50 mM Tris, 50 mM NaCl, 3 mM EDTA, 5M Guanidinium/HCl, pH 8 with fresh 10 mM benzamidine and then passed through a French Press twice at 16,000 psi. β-mercaptoethanol was then added to a final concentration of 5 mM, and the resulting solution was incubated at room temperature with stirring for 1 hour. Then, the solution was centrifuged at 20,000×g for 1 hour. The supernatant containing the denatured protein was added dropwise to 10X volume of low salt buffer (50 mM Tris, 50 mM NaCl, pH8) with fresh 5 mM β-mercaptoethanol and stirring. Then, the solution was incubated at room temperature without stirring for 2 hours. The solution was then centrifuged at 20,000×g for 30 min and the supernatant was dialyzed against 4 L high salt buffer (20 mM Tris, 500 mM NaCl, 5 mM Imidazole, pH 8) twice overnight at 4°C. After dialysis, the solution was applied to a Nickel chelating column and eluted with a gradient of imidazole (50 mM to 500 mM). The resulting purified protein was dialyzed against 4 L 20 mM Tris, 50 mM NaCl, 2 mM CaCl2, pH 7.4 overnight at 4°C. The solution was made to 0.02% NaN3 to inhibit bacterial growth. To remove the thioredoxin fusion tag, 10 units of recombinant enterokinase (Novagen, Madison, WI) were added. The solution was incubated at room temperature for 3-5 days to allow protease cleavage. Precipitated material was removed by adjusting the solution to contain 10% acetonitrile and 0.1% trifluoroacetic acid followed by centrifugation at 15,000×g for 30 min. The supernatant was purified by a C4 reversed-phase chromatography column (Vydac, Hesperia, CA) using the Akta purification system (GE Healthcare, Pittsburgh, PA). The protein was dried to powder using the Labconco freeze dry system (Labconco Corporation, Kansas City, MO). |
| NMR |
| All NMR samples were prepared in a buffer of 20 mM potassium phosphate, 10 μM DSS, 5% D2O, and 0.02% NaN3 at pH 5.5 or pH 7. All 2D HSQC experiments were carried out on a Bruker 600 MHz AVANCE III spectrometer equipped with a TCI cryoprobe at 25°C. HSQC experiments were run with carrier positions of 4.75 ppm for 1H and 119.3 ppm for 15N, sweep widths of 9615.385 Hz (15.9 ppm) for 1H and 1938.672 Hz (31.8) ppm for 15N with 672* (* complex points) points in 1H and 128* points in 15N. All chemical shifts were referenced to internal DSS (2,2-dimethyl-2- silapentane-5-sulfonic acid). Data were processed and viewed using nmrDraw, PIPP [33,34] and Sparky [35]. The chemical shift changes compared to wild type protein were calculated as described previously [32]. |
| Viral reagents |
| Viral plasmids containing the env gene from HIV-1 were obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH as follows: PVO, clone 4 (SVPB11), AC10.0, clone 29 (SVPB13) was from Dr. David Montefiori, Dr. Feng Gao and Dr. Ming Li [36]; pWITO4160 clone 33 (SVPB18) was from Drs. B. H. Hahn and J. F. Salazar-Gonzalez [36]; DU156.12 (SVPC3) was from Drs. D. Montefiori, F. Gao, S. Abdool Karim and G. Ramjee [37,38]; CAP210.2.00.E8 (SVPC17) was from Drs. L. Morris, K. Mlisana and D. Montefiori [37]; ZM109F.PB4 (SVPC13) was from Drs. E. Hunter and C. Derdeyn [39]; pHxB2-env was from Dr. Kathleen Page and Dr. Dan Littman [40]; pSG3Δenv was from Drs. John C. Kappes and Xiaoyun Wu [41,42]. |
| Single-round HIV infection assays |
| Single-round infection assays were performed as described [43]. Briefly, TZM-bl cells stably expressing CD4, CCR5 and CXCR4 coreceptors were maintained in DMEM (Dulbecco’s Modified Eagle’s Medium) with 10% FBS (fetal bovine serum). The cells were seeded in 96-well plate and serial dilutions of vMIP-II, 5P12-vMIP-II and 5P12- RANTES were added from the top row to the bottom row, as follows: 20 μl ligand of different concentration was added into the first row and mixed well with culture media. Then 20 μl media was removed and added into the second row, and so on. Virus was then added into each well containing different ligands. The cells were incubated at 37°C for 24 hours, at which time the medium was changed, and the cells were then incubated for another 24 hours. Infection with the pseudovirus allows expression of a β-galactosidase reporter gene. PBS containing 0.5% NP-40 was used to lyse the cells and substrate chlorophenol red- D-galactopyranoside (CPRG, Calbiochem, CA) was added into each well. The absorbance signal was measured at 570 nm and 630 nm. The ratio of 570/630 for each well was calculated. EC50 values were determined using a linear equation fitted between two data points surrounding 50% inhibition. For presentation purposes, data shown in figure 4 were plotted and fitted as curves using a four-parameter logistic equation in KaleidaGraph (Synergy Software, Reading, PA). |
| Results |
| Construction and structure of 5P12-vMIP-II |
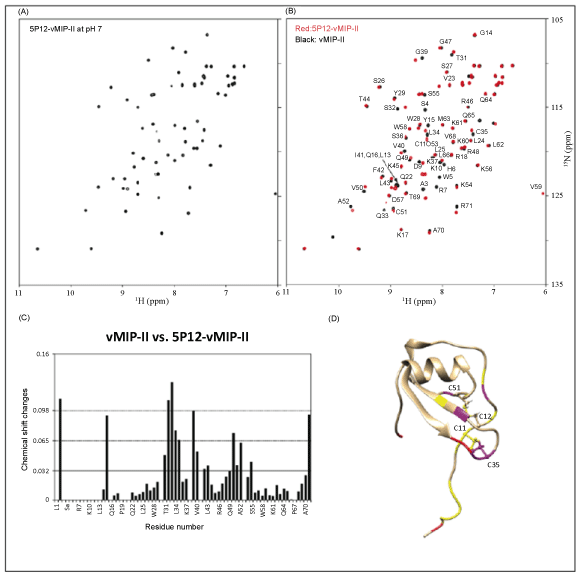
| The “5P12” chemokine N-terminus consists of a 10 amino acid sequence directly before the conserved Cys-Cys of the chemokine: QGPPLMATQS (Figure 2). Thermocycling techniques with overlapping oligonucleotide primers were used to replace the existing N-terminus of vMIP-II with DNA encoding these amino acids. The gene was placed into a pET32a(+) expression vector with an N-terminal thioredoxin fusion tag, and the protein was produced and purified as described in Methods. Figure 3 shows the 15N-1H correlation spectra of 15N labeled 5P12-vMIP-II. At pH 7, the protein showed good chemical shift dispersion and homogeneous peak height, indicating a nicely behaved protein that is not likely experiencing multiple conformations (Figure 3A). Chemical shift assignments of wild type vMIP-II have been carried out at pH 2.5 [3], pH 3.25 [44] and pH 5.4 [32], so spectra were also obtained of 5P12-vMIP-II at pH 5.5, and a comparison is shown in figure 3B. The assignments were adapted from Zhao and LiWang [32]. An overlay of vMIP-II and 5P12-vMIP-II at pH 7 is shown in the Supplementary Material (Figure S1). As seen in figure 3, 5P12- vMIP-II shows similarity to the wild type protein in most regions of the spectrum, but the spectrum contains no matching peaks for the N-terminal residues, as expected. However, seven new peaks could be observed in the 5P12-vMIP-II spectrum (red) that were not close to any assigned peaks from the wild type vMIP-II spectrum (black). These likely correspond to the 7 non-Pro amino acids that are different between the variant and wild type vMIP-II (Figure 2 and Figure 3B). Moreover, peaks in the variant spectrum near the resonances in the wild type spectrum for G2, Q33, and K37 could be observed when the contour level is lowered indicating that these are likely the analogous resonances of the variant (data not shown). Finally, the 5P12 variant contains two additional glutamines, Q1 and Q9; in the HSQC spectrum two additional pairs of side-chain peaks are observed, indicating the presence of Q1 and Q9 in 5P12- vMIP-II. Therefore, the HQSC spectrum is fully consistent with a folded 5P12-vMIP-II. |
| While a comparison of the wild type and variant spectra indicate an overall similar structure, some regions with significant changes compared to the wild type protein were observed in 5P12-vMIP-II. Specifically, nine out of 71 residues (residues: 2, 15, 32, 33, 34, 35, 39, 50, and 71) showed chemical shift changes that were bigger than 2 standard deviations from the average. One of these (residue 2) is in the mutated region and another (residue 71) is at the C-terminus. Others include amino acids near Cys 35 (that participates in a disulfide bond with Cys 11 that is adjacent to the mutated N-terminus) and amino acids near Cys 51 (that participates in a disulfide bond with Cys 12). Figure 3C shows chemical shift changes at each residue in 5P12-vMIPII compared to wild type vMIP-II. Figure 3D maps these onto the structure of the wild type protein and shows the structure of the wild type protein. Most significant changes occur near these N-terminusadjacent disulfide bonds. Overall it can be concluded that 5P12- vMIPII is a folded protein that is quite similar in tertiary structure to wild type vMIP-II. |
| Anti-HIV function of vMIP-II and 5P12-vMIP-II |
| While the anti-HIV properties of vMIP-II have been known for some time, few studies have reported assays against multiple viral strains. For this work we used single-round HIV infection assays, in which a viral particle lacking key elements of the HIV genome (but having a functional HIV envelope) is used to infect target cells. The viral particle is therefore the same size and has the same surface proteins (and is presumed to have a similar ability to infect the host cell) as replication-competent HIV, but is not able to replicate. This type of assay is therefore highly useful as an efficient, accurate test of inhibition of HIV entry into the host cell, with each different “pseudovirus” defined by the different sequences of the envelope proteins (i.e., gp120) on the surface [45]. |
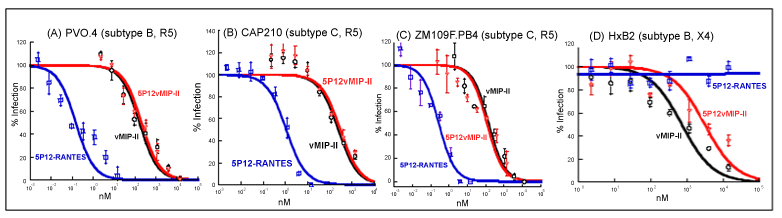
| We tested wild type vMIP-II against seven strains of pseudotyped HIV: R5 strains PVO.4, AC10.0 and pWITO4160 clone 33 (SVPB18) (all from subtype B, which tends to be common in North America and Europe) [36]; R5 strain ZM109F.PB4, DU156.12 and CAP210.2.00. E8 (all from subtype C, which tends to be common in Asia and Africa) [37]; and X4 strain HxB2 (subtype B). Strain PVO.4 also has been categorized as a “Tier 3” HIV strain, indicating low sensitivity to antibody-mediated neutralization, and adding significance to the search for inhibitors for such “difficult to inhibit” viruses [46]. Strain ZM109F.PB4 is also particularly significant because it has shown orders of magnitude less sensitivity than most strains to another major type of HIV inhibitor, namely the lectin griffithsin [47]. |
| As shown in figure 4 and table 1, vMIP-II was able to inhibit these strains at high nanomolar or low micromolar levels. We then tested 5P12-vMIP-II against these strains. It also showed effectiveness at high nanomolar or low micromolar levels, but did not show improvement over the wild type protein (Table 1). As a control, 5P12- RANTES was tested, and it showed excellent potency against R5 strains as expected, but no effectiveness against the X4 strain (Figure 4 and Table 1). |
| Discussion |
| Since its discovery in 1996-1997 [4-6], vMIP-II has been the focus of a variety of studies, based on its ability to bind multiple chemokine receptors. Most of these studies dealt with the potential of this virallyencoded chemokine homolog to inhibit inflammation. Indeed, vMIPII has been shown in various animal models to attenuate cellular infiltration after spinal cord injury [11-13], to protect the brain against focal cerebral ischemia [14], and to protect allograft survival in both the heart and the cornea [9,10]. However, from the earliest reports, it was also clear that vMIP-II possessed the ability to inhibit HIV infection. Further, the unique ability of this chemokine analog to tightly bind both of the main HIV co-receptors (CCR5 and CXCR4) gives it the potential to be more broadly acting than any of the current chemokine-based inhibitors, which are limited by their ability to bind receptors from only one family (i.e., either CCR5 or CXCR4). This is particularly significant given that initial HIV infection occurs with viruses using CCR5 (socalled R5 strains), while progression to AIDS is often correlated with the switch to X4 virus in a patient [48]. |
| Structural studies have been reported on full length vMIP-II [1-3,44,49] and on peptide fragments of vMIP-II [1]. Other work has focused on peptides derived from vMIP-II in order to generate smaller molecules that retain anti-HIV properties, including the use of D-peptides [50-53]. Interestingly, in studies investigating the binding of vMIPII to human chemokine receptors, data indicate that vMIP-II does compete with natural chemokines but does not necessarily bind CCR5 analogously. In these studies, it was found that while mutating negatively charged positions in CCR5 affected binding by both vMIPII and RANTES, vMIP-II maintained the ability to bind CCR5 when extracellular loop 2 of CCR5 was mutated, while binding by natural chemokines was abrogated [31]. This “partial cross-competition” as described by the authors is supported by our ongoing work that demonstrates the importance of basic residues in the ability of vMIP-II to bind receptors (which is likely important in other chemokines) but the lack of importance of a hydrophobic residue at the 13th position of vMIP-II (unpublished). The “F13/Y13” position is critical for natural chemokine ligands such as RANTES in the binding of CCR5 [54,55]. While these experiments show differences between vMIP-II and chemokines, they also provide evidence for one of the most important facets of vMIP-II action, namely the manner in which it is uniquely able to bind multiple, diverse chemokine receptors, a function not shared with human chemokines, despite these proteins sharing as much as 40% sequence identity and an essentially identical tertiary structure. |
| The chemokine RANTES has been improved as an HIV inhibitor by a variety of changes to the N-terminus, the most promising of which is the “5P12” mutation used in the present work. Previous research by others also included chemical modifications at the amino terminus [21,56]. These have been shown in at least some cases to be transferrable to other chemokines [25]. 5P12-RANTES has been found to retain the ability to tightly bind the receptor CCR5, while having a greatly increased potency against R5 HIV with EC50s usually in the picomolar range. The inhibitor is also very robust to HIV mutation with “virus escape” being rare and requiring a switch to CXCR4 coreceptor use [27]. 5P12-RANTES does not inhibit X4 HIV in vitro, since this chemokine does not significantly interact with the CXCR4 receptor [26]. Previously, in order to overcome this limitation we expressed 5P12-RANTES covalently linked with a peptide that is derived from the C-terminus of HIV gp41 (a C-peptide) that is known to bind to the N-terminal helices of gp41 and inhibit viral entry. This bifunctional chimera strategy was very successful, resulting in potent inhibition of both R5 and X4 viral strains [57]. In the present work, an alternate route to a broadly-acting inhibitor was tested, namely to place the 5P12 N-terminal sequence onto the vMIP-II chemokine homolog. |
| Our results show that 5P12-vMIP-II is a folded protein that is stable over a range of pH values and that likely retains structural similarity to wild type vMIP-II. Unlike 5P12-RANTES, 5P12-vMIP-II is able to inhibit X4 strains of HIV. However, 5P12-vMIPII is much less potent than 5P12-RANTES against R5 HIV, and shows similar effectiveness as wild type vMIP-II. Therefore, while the “5P12” N-terminus did not significantly reduce the effectiveness of vMIP-II, it also clearly does not enable interaction with the receptor in the same way as it does when placed onto RANTES, where it usually engenders subnanomolar inhibition against R5 HIV. This provides evidence that vMIP-II does not bind CCR5 in the same was as RANTES (nor, by extension, the other human chemokines MIP-1β and MIP-1α). Therefore, it appears that vMIP-II’s ability to bind multiple chemokine receptors from multiple families relies on shared structural features among chemokines, and likely on the highly basic surface on vMIP-II [32], but that this comes at the price of not binding these receptors identically to their natural chemokine ligands. |
| Conclusions |
| vMIP-II is a virally expressed chemokine analog that has the unique ability to bind both HIV co-receptors, CCR5 and CXCR4. We have modified vMIP-II at its Nterminus to make 5P12-vMIP-II, based on the N-terminal modification in 5P12-RANTES, which had made RANTES one of the most potent known R5 HIV entry inhibitors. While 5P12-vMIP-II retained anti-HIV activity against both R5 and X4 HIV strains, the 5P12 Nterminus did not improve the inhibitor. This provides evidence that despite high sequence identity and essentially identical structure to human chemokines, vMIP-II does not interact with chemokine receptors in an analogous manner to their natural chemokine ligands. |
| Acknowledgement |
| This work was supported by US Army grant W911NF-11-1-0139. |
References
|
Tables and Figures at a glance
| Table 1 |
Figures at a glance
 |
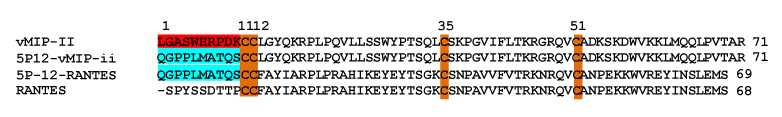 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
Relevant Topics
- Analytical Biochemistry
- Applied Biochemistry
- Carbohydrate Biochemistry
- Cellular Biochemistry
- Clinical_Biochemistry
- Comparative Biochemistry
- Environmental Biochemistry
- Forensic Biochemistry
- Lipid Biochemistry
- Medical_Biochemistry
- Metabolomics
- Nutritional Biochemistry
- Pesticide Biochemistry
- Process Biochemistry
- Protein_Biochemistry
- Single-Cell Biochemistry
- Soil_Biochemistry
Recommended Journals
- Biosensor Journals
- Cellular Biology Journal
- Journal of Biochemistry and Microbial Toxicology
- Journal of Biochemistry and Cell Biology
- Journal of Biological and Medical Sciences
- Journal of Cell Biology & Immunology
- Journal of Cellular and Molecular Pharmacology
- Journal of Chemical Biology & Therapeutics
- Journal of Phytochemicistry And Biochemistry
Article Tools
Article Usage
- Total views: 15152
- [From(publication date):
specialissue-2013 - Jul 09, 2025] - Breakdown by view type
- HTML page views : 10494
- PDF downloads : 4658
