A Case Report and Literature Review on Rare LMO7-ALK Rearrangement in Lung Adenocarcinoma
Received: 14-Jan-2024 / Manuscript No. cmb-24-125077 / Editor assigned: 16-Jan-2024 / PreQC No. cmb-24-125077 / Reviewed: 23-Jan-2024 / QC No. cmb-24-125077 / Revised: 29-Jan-2024 / Manuscript No. cmb-24-125077 / Published Date: 31-Jan-2024
Abstract
Background: In non-small cell lung cancer (NSCLC), anaplastic lymphoma kinase (ALK) gene rearrangement is a critical therapeutic biomarker. Partner LMO7 gene was discovered in patients with NSCLC fused to ALK gene as reported (L15: A20), (L16: A20). Investigating the relationship between various breakpoints and prognosis was significant.
Method: The Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST 1.1) was used to evaluate therapeutic responses. Next-generation sequencing (NGS) detected the genomic data.
Case presentation: A 59-year-old male with a 40-pack year smoking history was diagnosed with lung adenocarcinoma. The patient was clinically classified as stage χ A (cT1bN3M1b) NSCLC. A novel LMO7-ALK fusion breakpoint (L13: A20) was identified in a biopsy sample that underwent NGS for gene mutation. Crizotinib was chosen as the first-line therapy, and the patient experienced a partial response. However, significant disease-progression was confirmed at the fourth follow-up at the neighbourhood hospital, which was done five months later. The pneumocystis jiroveci infection the patient had unfortunately led to severe obstructive pneumonia and his death.
Conclusion: LMO7-ALK rearrangement in advanced NSCLC is a potentially sensitive fusion mutation. The poor prognosis in this patient suggested that the novel breakpoint (L13: A20) LMO7-ALK rearrangement may be a hyperprogressive marker.
Keywords
Non-small cell lung cancer; ALK; LMO7; Rearrangement
Background
Anaplastic lymphoma kinase (ALK) gene rearrangement is presented as driving oncogenesis and crucially therapeutic biomarker in non-small cell lung cancer (NSCLC) since most patients harbouring typical ALK fusion responded well to ALK tyrosine kinase inhibitors (ALK-TKIs). The impact is constantly on the focus that of various fusion breakpoints and independent fusion partners perform on ALK-TKIs [1,2]. In the picture regarding the ALK fusion companies, the EML4 gene was the dominant one, with a total of 16 variant types detected and independently predicted various overall survivals in patients receiving ALK-TKIs [3,4]. Other fusion companies such as TFG, KIF5B, STRN, HIP1, and BIRC6 were screened and presented potential oncogenesis [5]. Partner LMO7 gene was a fibrous actinbinding protein widely expressed in normal bronchial and alveolar epithelial cells, which was lately found in patients with NSCLC fused to ALK gene as reported (L15: A20), (L16: A20) [6,7]. Compared with patients harbouring LMO7-ALK fusion with reported breakpoints, clinical prognosis in this case was totally different. Whether breakpoints in LMO7-ALK fusion play important part on enhancing or weakening therapeutic response needs more discussion. Since LMO7-ALK fusion is an extremely rare mutation among ALK fusion companies, it would be worthwhile to explore the importance of different breakpoints and the clinical conditions [8].
This paper reported a case with a novel breakpoint (L13: A20) of LMO7-ALK rearrangement in advanced NSCLC. Literature with ALK rearrangements identified from 2007 was comprehensively described. Based on this, we provide valuable information on ALK rearrangements in NSCLC to clinicians and scientists who concentrate on the field perpetually [9].
Methods
According to the Response Evaluation Criteria in Solid Tumors
version 1.1 (RECIST 1.1), evaluation of therapeutic responses to targeted therapy measured by computed tomography (CT) screen, were sorted to complete response (CR), partial response (PR), stable disease (SD), progressive disease (PD). Progression-free survival (PFS) was calculated from the date of the initial treatment to a radiologic or clinical observation of the disease progression. Overall survival (OS) was calculated from a randomized date to death for any reason. Genomic DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tumor tissue, analysed by next-generation sequencing (NGS) detection (Amoy Diagnostics, Xiamen, China) [10-15].
Case Presentation
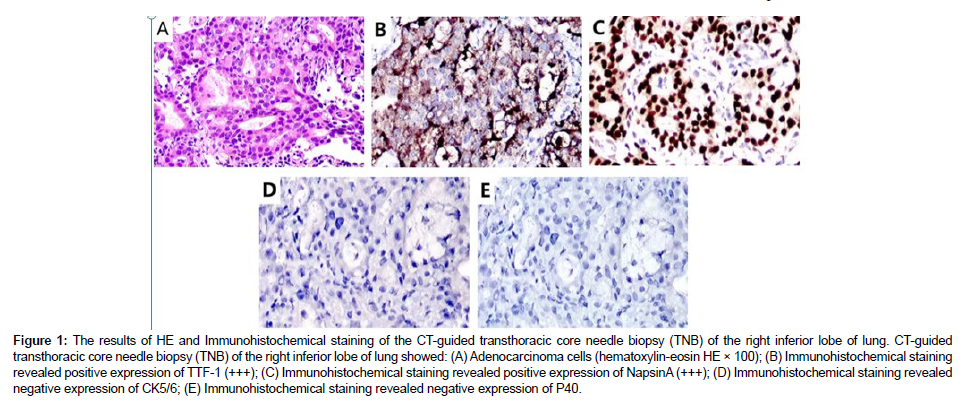
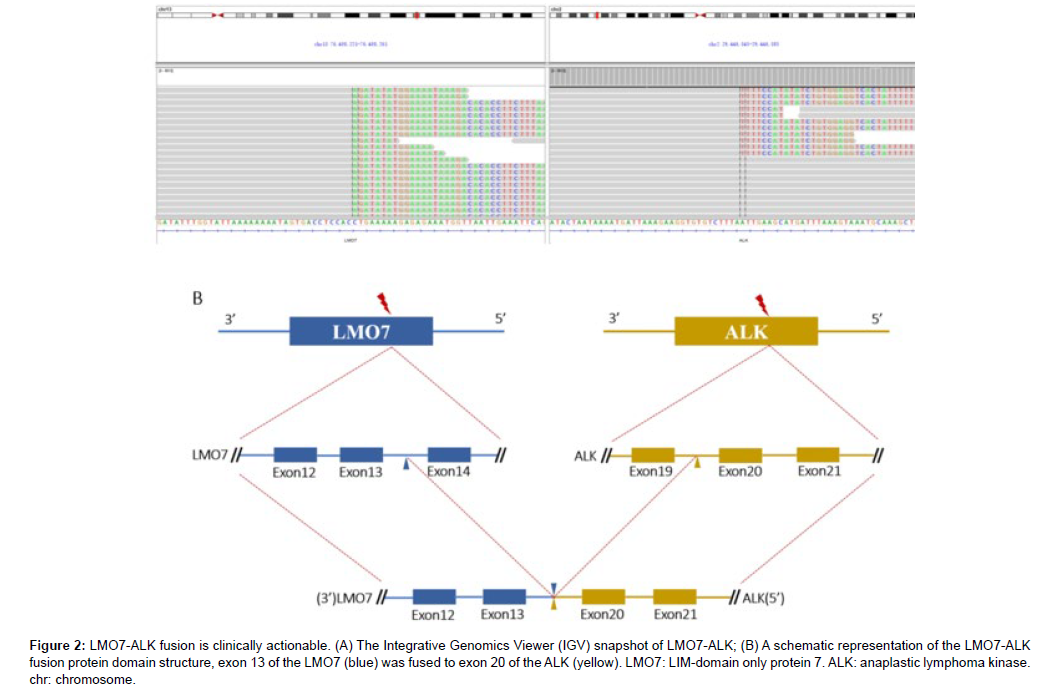
A 59-year-old man with a 40-pack-year smoking history was hospitalized due to a nodule in the right inferior lobe of the lung that had an unknown potential for malignancy. One month before admission to this hospital, CT screen revealed the lung nodule (12.7mm×10mm) during his regular health check, without experiencing any specific symptom. Then 18F-FDG positron-emission tomography-CT (PETCT) was performed to further report that multiple lymph node metastases of the mediastinum (3P, 4R, 4L, 7, 10R) and left cervix (β,χ) developed. Furthermore, no brain metastasis was observed by cranial magnetic resonance [16]. He underwent a CT-guided transthoracic core needle biopsy (TNB) of the specific lung tissue after being admitted to our hospital in order to develop an effective treatment plan. TTF-1, Napsin A and ALK (D5F3) were shown to be expressed positively by immunohistochemistry (IHC) analysis, while CK5/6 and P40 were found to be negatively expressed. PD-L1 was detected to be positively expressed (Figure 1). Subsequently, gene mutation was performed on the biopsy sample by NGS test, whereby a novel LMO7- ALK fusion Breakpoint (L13: A20) was firstly signified (abundance: 3.07%) based on a 448-gene panel on Illumina Novaseq6000/Nextseq 500 (AMOYDX, Xia Men, China) (Figure 2A) [17-20].
Figure 1: The results of HE and Immunohistochemical staining of the CT-guided transthoracic core needle biopsy (TNB) of the right inferior lobe of lung. CT-guided transthoracic core needle biopsy (TNB) of the right inferior lobe of lung showed: (A) Adenocarcinoma cells (hematoxylin-eosin HE × 100); (B) Immunohistochemical staining revealed positive expression of TTF-1 (+++); (C) Immunohistochemical staining revealed positive expression of NapsinA (+++); (D) Immunohistochemical staining revealed negative expression of CK5/6; (E) Immunohistochemical staining revealed negative expression of P40.
Figure 2: LMO7-ALK fusion is clinically actionable. (A) The Integrative Genomics Viewer (IGV) snapshot of LMO7-ALK; (B) A schematic representation of the LMO7-ALK fusion protein domain structure, exon 13 of the LMO7 (blue) was fused to exon 20 of the ALK (yellow). LMO7: LIM-domain only protein 7. ALK: anaplastic lymphoma kinase. chr: chromosome.
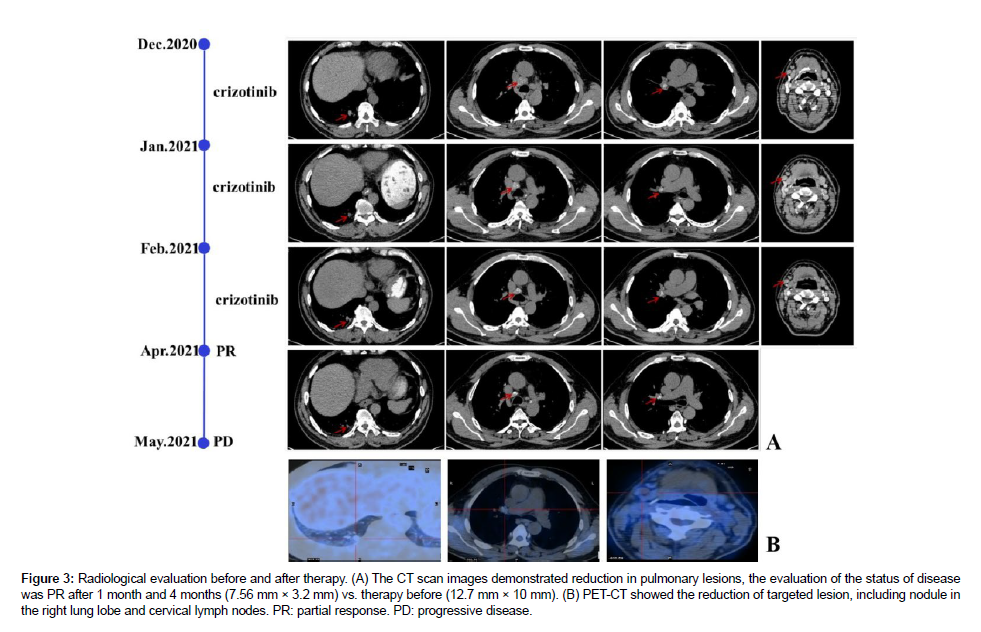
The diagnosis was a poorly differentiated adenocarcinoma with a strong expression of the ALK fusion protein at stage IV A (cT1bN3M1b, 8thUICC/AJCC). The response of the fusion variant obtaining novel breakpoint (L13: A20) to ALK-TKIs remained unclear. the patient accepted Crizotinib orally at a dosage of 250mg twice daily as the standard first-line targeted treatment ever since disease identification in the real-world study. The target lesion was reduced after four weeks of Crizotinib treatment [21-24]. Since then, a chest CT scan was routinely evaluated and conducted every four weeks. The lung lesion and cervix lymph nodes decreased with a partial response (PR) 3 months later after Crizotinib therapy according to the RECIST1.1 (Figure 3). During the targeted treatment period, the patient suffered from diarrhea, which was finally cured by an antidiarrheic treatment, without any additional symptoms. However, significant disease progression on Chest CT scans was confirmed at the fourth follow-up five months later at a local hospital. Unfortunately, he developed severe obstructive pneumonia caused by pneumocystis jiroveci infection and lost his life [25-30].
Figure 3: Radiological evaluation before and after therapy. (A) The CT scan images demonstrated reduction in pulmonary lesions, the evaluation of the status of disease was PR after 1 month and 4 months (7.56 mm × 3.2 mm) vs. therapy before (12.7 mm × 10 mm). (B) PET-CT showed the reduction of targeted lesion, including nodule in the right lung lobe and cervical lymph nodes. PR: partial response. PD: progressive disease.
Discussion
LIM-domain only protein 7 (LMO7), a fibrous actin-binding protein, is a member of PDZ and LIM domain-containing protein family, which is widely expressed in adults’ tissues including normal bronchial and alveolar epithelial cells. LMO7 gene is considered as a molecule that facilitates the formation and maintenance of epithelial architecture via remodelling of the actin cytoskeleton.6 LMO7 gene is located in chromosome 13q22 and circumferentially in the plasma membrane of adenocarcinoma cells [31]. The decreased expression of LMO7 was significantly correlated with tumor progression and a poor prognosis in patients with lung adenocarcinoma, but it remains unclear whether LMO7 may be a candidate for molecular-targeting therapy.5 More than 90 atypical non-EML4 ALK fusion partners have been reported in NSCLC since 2007. The effect that diverse fusion breakpoints have on ALK-TKIs is inconsistent [32].
In light of this, we have compiled a list of the many ALK fusion partners that have been reported since 2007 along with related occurrences, namely breakpoints during rearrangement, the reaction to ALK-TKIs, and observed PFS (Tables 1 and 2). ALK-rearranged information was provided as rich as possible to guide clinician prognosis and therapy. The ALK-rearranged information was made available as richly as possible to assist physician prognosis and treatment [33]. PFS of the table listed visibly showed remarkable therapeutic efficacy, where 45 cases (45/52, 86.54%) achieved six-month PFS at least, 26 cases (26/52, 50%) reached twelve-month PFS at least, and PFS over eighteen months reached the point of 16 cases (16/52, 30.77%) [34]. The PFS of the patients in the table clearly demonstrated the treatment efficacy: PFS over six months was obtained in 45 cases (45/52, 86.54%), PFS over twelve months was reached in 26 cases (26/52, 50%), and PFS over eighteen months was attained in 16 cases (16/52, 30.77%). ALK-TKIs significantly improved the survival of patients in NSCLC harbouring ALK rearrangements [35].
| No. | Fusion Partner | Breakpoint | Published Year | Age | Gender | Tumor Sourse | PFS (months) | Method of detection | FISH/IHC | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | TFG | (T3, A20) | 20075 | NR | NR | Tumor | NT | NR | 5’RACE PCR DNA sequencing | UK/UK |
| 2 | KIF5B | (K24, A20) | 20095 | NR | NR | Tumor | NT | NR | RT-PCR | +/+ |
| (K15, A20) | 20115 | NR | NR | Tumor | NT | NR | 5’RACE PCR DNA sequencing | +/+ | ||
| (K17, A20) | 20125 | NR | NR | Tumor | NT | NR | RT-PCR | +/+ | ||
| (K20, A20) | 20218 | 70 | M | Tumor | NR | 7 | NGS | UK/UK | ||
| 3 | KLC1 | (K9, A20) | 20125 | NR | NR | Tumor | NT | NR | RT-PCR | +/UK |
| 4 | PTPN3 | (P2, A10) | 20129 | NR | NR | Tumor | NT | NR | RT-PCR | UK/UK |
| 5 | STRN | (S3, A20) | 20135 | NR | NR | NR | NR | Targeted RNA sequencing | +/+ | |
| (S3, A20) | 20175 | 59 | M | Plasma | PR to crizotinib | >6 | DNA NGS | UK/UK | ||
| (S3, A20) | 20175 | 51 | M | Lymph node | PD to crizotinib | NR | RNA sequencing | +/+ | ||
| (S3, A20) | 2E+05 | 64 | M | Plasma | PR to alectinib | >19 | Targeted NGS | +/UK | ||
| 6 | HIP1 | (H21, A20) | 20145 | 38 | F | Tumor | PR to crizotinib | 15 | RT-PCR | +/+ |
| (H30, A20) | 20145 | 58 | F | Liver | PR to crizotinib, CR to alectinib | 5, 12 | DNA NGS | +/UK | ||
| (H19, A20) | 2E+05 | 56 | F | lumbar | PD to crizotinib, | / | NGS | UK/+ | ||
| spine | PR to alectinib | >9 | ||||||||
| 7 | TPR | (T15, A20) | 20145 | 60 | M | Tumor | NT, adjuvant chemotherapy | >18 | PCR | +/+ |
| 8 | BIRC6 | (B10, A20) | 20155 | 45 | F | Tumor | PR to crizotinib | >8 | DNA NGS | -/+ |
| 9 | DCTN1 | (D26, A20) | 20155 | NR | NR | Tumor | NR | NR | DNA NGS | +/UK |
| 10 | SQSTM1 | (S5, A20) | 20155 | NR | NR | Tumor | NR | NR | DNA NGS | +/UK |
| 11 | SOCS5 | NR | 2E+05 | NR | NR | Tumor | PD | NR | NGS | -/UK |
| 12 | CLIP4 | NR | 2E+05 | NR | NR | Tumor | PR to crizotinib | 5 | NGS | -/UK |
| UK | 2E+05 | NR | NR | Tumor | NR | NR | NGS | UK/UK | ||
| 13 | SEC31A | (S21, A20) | 20165 | 53 | M | Tumor | NT | NR | NGS | +/+ |
| 14 | CLTC | (C31, A20) | 20165 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 15 | PRKAR1A | (P5, A20) | 20165 | 67 | M | Tumor | PR to crizotinib | 7 | NGS | +/+ |
| 16 | PPM1B | (P1, A20) | 20165 | NR | NR | Tumor | PR to crizotinib | NR | NGS | UK/UK |
| 17 | EIF2AK3 | (E2, A20) | 20165 | 71 | F | Tumor | PR to crizotinib | 28 | NGS | -/- |
| 18 | CRIM1 | NR | 20165 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 19 | PICALM | (P19, A20) | 2E+05 | NR | NR | Tumor | PR to crizotinib | NR | NGS | -/+ |
| 20 | SPTBN1 | (S6, A20) | 2E+05 | 69 | M | Plasma | PD to crizotinib | / | DNA NGS | UK/- |
| 21 | COL25A1 | NR | 2E+05 | 58 | M | Tumor | PR to crizotinib | 6.3 | NGS | UK/UK |
| 22 | FAM179A | NR | 2E+05 | 36 | F | Tumor | PR to crizotinib | 12 | NGS | UK/UK |
| (F1, A19) | 2E+05 | 27 | F | Brain and plasma | PR to lorlatinib | 23 | NGS | UK/+ | ||
| 23 | CEBPZ | (C2, A20) | 20175 | NR | NR | Tumor | PR to crizotinib | NR | NGS | +/+ |
| 24 | CLIP1 | (C22, A20) | 2E+05 | NR | NR | Tumor | UK | NR | NGS | +/+ |
| 25 | BCL11A | (B4, A20) | 20175 | 64 | F | Tumor | PR to crizotinib | >6 | DNA and RNA NGS | UK/UK |
| (B2, A18) | 20195 | 29 | M | Tumor and plasma | PR to crizotinib | 13 | DNA NGS | UK/UK | ||
| 26 | GCC2 | (G12, A20) | 20175 | 28 | F | Tumor | PR to crizotinib | 18 | RT-PCR, NGS | +/+ |
| (G19, A20) | 2E+05 | NR | NR | Tumor | NT | NR | Targeted RNA sequencing | +/+ | ||
| 27 | LMO7 | (L15, A20) | 20176 | NR | NR | Tumor | NR | NR | RT-PCR, NGS | +/+ |
| (L16, A20) | 20217 | 56 | F | Tumor | PR to ensartinib | 18 | NGS | UK/+ | ||
| (L13, A20) | present | 59 | M | Tumor | PR to crizotinib | 4 | NGS | -/+ | ||
| 28 | PHACTR1 | (P7, A20) | 20176 | NR | NR | Tumor | NR | NR | RT-PCR, NGS | +/+ |
| 29 | CMTR1 | (C2, A20) | 20185 | 75 | M | Lymph node | PD to crizotinib | NR | NGS | -/- |
| 30 | VIT | (V7, A20) | 20185 | 64 | F | Tumor | PR to alectinib | 5 | NGS | +/+ |
| 31 | DYSFa | (D10, A20) | 20185 | 44 | M | Pleural effusion | PD(Extracranial PR but intracranial progression to crizotinib) | 23 | Pleural effusion | UK/+ |
| 32 | ITGAVa | (I2, A20) | 20185 | 44 | M | Pleural effusion | NR | 23 | Pleural effusion | UK/+ |
| 33 | PLEKHA7 | (P26, A19) | 20185 | 70 | F | Plasma | PR to alectinib、 osimertinib | 6 | DNA NGS | UK/UK |
| 34 | CUX1 | (C8, A20) | 20185 | NR | NR | Tumor | PR to crizotinib | 20 | NGS | UK/UK |
| 35 | VKORC1L1 | (V1, A20) | 20185 | 54 | M | Plasma | PR to crizotinib alectinib | 24 | NGS | +/UK |
| 36 | FBXO36 | NR | 20185 | NR | NR | Tumor | PR to crizotinib | NR | NGS | UK/+ |
| 37 | EML6b | (E1, A20) | 20185 | 56 | M | Tumor | PR to crizotinib | 10 | NGS | UK/+ |
| 38 | FBXO11b | (F1, A20) | 20185 | 56 | M | Tumor | PR to crizotinib | 10 | NGS | UK/+ |
| 39 | CAMKMT | (C3, A 20) | 20195 | 67 | F | Tumor | NT, adjuvant chemotherapy | / | NGS | +/+ |
| 40 | NCOA1 | (N12, A20) | 20195 | 59 | M | Tumor | PR to crizotinib | >18 | NGS | UK/+ |
| 41 | MYT1L | (M14, A20) | 20195 | 41 | M | Tumor | PR on crizotinib, PD on ceritinib | 4 | NGS | -/UK |
| 42 | SRBD1 | (S20, A20) | 20195 | 56 | F | Tumor | NT | / | NGS | UK/+ |
| (S6, A20) | 2E+05 | 59 | M | Lymph node | PR to crizotinib | >10 | NGS | +/+ | ||
| 43 | between CENPA and DPYSL5 | IGR | 2E+05 | 60 | M | Tumor | PR | UR | NGS | +/+ |
| 44 | SRD5A2 | (S1, A20) | 20195 | NR | NR | Tumor | NT | / | NGS | UK/+ |
| 45 | NYAP2 | (N3, A20) | 20195 | NR | NR | Tumor | NT | / | NGS | UK/- |
| 46 | MPRIP | (M21, A20) | 20195 | 27 | F | Tumor | PR to crizotinib | >11 | RNA sequencing | +/+ |
| 47 | ADAM17 | (A4, A20) | 20195 | NR | NR | Plasma | PR to alectinib | NR | DNA NGS | UK/UK |
| 48 | ALK | (A6, A20) | 20195 | NR | NR | Plasma | NR | NR | DNA NGS | UK/UK |
| 49 | LPIN1 | NR | 20195 | NR | NR | Tumor | PR to crizotinib erlotinib | NR | NR | UK/UK |
| 50 | WDPCP | (W17, A20) | 20195 | 52 | F | Tumor | PR to crizotinib | 11 | DNA NGS | +/+ |
| 51 | CEP55 | (C3, A20) | 20195 | NR | NR | Tumor | NR | NR | DNA NGS | UK/UK |
| 52 | ERC1 | (E15, A20) | 20195 | NR | NR | Tumor | NR | NR | DNA NGS | UK/UK |
| 53 | SLC16A7 | (S1, A20) | 20195 | NR | NR | Tumor | PR to crizotinib | NR | DNA NGS | UK/UK |
| 54 | TNIP2/ABIN2 | (T5, A20) | 20195 | 49 | F | Tumor/Plasma | PR to crizotinib | NR | DNA NGS | UK/+ |
| 55 | ATAD2B | (A1, A20) | 20195 | 46 | M | Tumor/Plasma | NR | NR | DNA NGS | UK/+ |
| 56 | SLMAP | (S12, A20) | 20195 | 73 | M | Tumor/Plasma | UK, adjuvant treatment with crizotinib | / | DNA NGS | +/+ |
| 57 | FBN1 | NR | 20195 | NR | NR | Tumor/Plasma | NR | NR | DNA NGS | UK/UK |
| 58 | SWAP70 | NR | 20195 | NR | NR | Tumor/Plasma | NR | NR | DNA NGS | UK/UK |
| 59 | TCF12 | NR | 20195 | NR | NR | Tumor/Plasma | NR | NR | DNA NGS | UK/UK |
| 60 | TRIM66 | NR | 20195 | NR | NR | Tumor/Plasma | NR | NR | DNA NGS | UK/UK |
| 61 | WNK3 | NR | 20195 | NR | NR | Tumor/Plasma | NR | NR | DNA NGS | UK/UK |
| 62 | AKAP8L | NR | 20195 | NR | NR | Plasma | ensartinib | NR | DNA NGS | UK/UK |
| 63 | SPECC1L | (S9, A20) | 20195 | NR | NR | Tumor | NT | / | DNA NGS | UK/UK |
| 64 | PRKCB | (P2, A19) | 20195 | 44 | M | Tumor plasma | PR to crizotinib | 6 | NGS | UK/UK |
| 65 | CDK15 | (C10, A19) | 2E+05 | 54 | F | Tumor | NR | / | DNA NGS | UK/UK |
| 66 | LCLAT1 | NR | 2E+05 | NR | NR | Tumor | NR | NR | DNA NGS | UK/UK |
| 67 | YAP1 | NR | 2E+05 | NR | NR | Tumor | NR | NR | DNA NGS | UK/UK |
| 68 | MEMO1 | NR | 2E+05 | NR | NR | Tumor | NR | NR | DNA NGS | UK/UK |
| 69 | PLEKHM2 (SCLC) | (P7, A20) | 2E+05 | 63 | F | Tumor | SD to crizotinib and brigatinib | 12,7 | NGS | UK/+ |
| 70 | DCHS1 | NR | 20205 | NR | NR | Tumor | UK,ensartinib | NR | NGS | UK/UK |
| 71 | PPFIBP1 | NR | 20205 | NR | NR | Tumor | UK,ensartinib | NR | NGS | UK/UK |
| 72 | ATP13A4 | (A9, A19) | 20205 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 73 | C12orf75 | (C1, A20) | 20205 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 74 | EPAS1 | (E1, A20) | 20205 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 75 | HMBOX1 | (H4, A20) | 2E+05 | NR | NR | FFPE | UK | UK | NGS | UK/UK |
| 76 | FUT8 | (F3, A20) | 20205 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 77 | LIMD1 | (L2, A20) | 20205 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 78 | LINC00327 | (L2, A20) | 20205 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 79 | LOC349160 | (L1, A20) | 20205 | 57 | M | Tumor | SD to crizotinib | NR | NGS | UK/UK |
| 80 | LYPD1 | (L3, A20) | 20205 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 81 | RBM20 | (R1, A20) | 20205 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 82 | TACR1 | (T1, A20) | 20205 | 33 | F | Tumor | PR to crizotinib | 15 | NGS | UK/UK |
| 83 | TANC1 | (T3, A20) | 20205 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 84 | TTC27 | (T12, A20) | 20205 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 85 | TUBBB | (T3, A20) | 20205 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 86 | SMPD4 | (S1, A20) | 20205 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 87 | SORCS1 | (S10, A20) | 20205 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 88 | LINC00211 | IGR | 20205 | 47 | F | CSF | PR to crizotinib alectinib, SD to lorlatinib | 7.6, 8.7 | NGS | UK/+ |
| 89 | SOS1 | (S2, A20) | 20205 | 52 | M | FFPE | PR to crizotinib | >6 | NGS | UK/UK |
| 90 | C9orf3 | (C12, A20) | 20205 | NR | NR | FFPE | NR | NR | NGS | UK/+ |
| 91 | CYBRD1 | (C21, A20) | 20205 | NR | NR | FFPE | NR | NR | NGS | UK/UK |
| 92 | MTA3 | (M6, A20) | 20205 | NR | NR | FFPE | SD to crizotinib | UK | NGS | UK/UK |
| 93 | THADA | (T25, A20) | 20205 | NR | NR | Plasma | SD to crizotinib, PR to ceritinib | UK | NGS | UK/UK |
| 94 | TSPYL6 | (T6, A20) | 20205 | NR | NR | FFPE | PR to crizotinib, SD to alectinib | UK | NGS | UK/UK |
| 95 | WDR37 | (W6, A20) | 20205 | NR | NR | FFPE | PR to crizotinib | UK | NGS | UK/UK |
| 96 | PLEKHH2 | (P6, A20) | 20205 | 54 | F | FFPE | PR to alectinib | 18 | Targeted RNA seqencing | +/+ |
| 97 | CCNYc | (C1, A20) | 2E+05 | 32 | M | Tumor | PR to crizotinib | >6 | DNA NGS | +/+ |
| 98 | ATICc | (A7, A20) | 2E+05 | 32 | M | Tumor | PR to crizotinib | >6 | DNA NGS | +/+ |
| 99 | HPCAL1 | (H1, A19) | 2E+05 | 53 | M | Plasma | PR to crizotinib, PR to alectinib | 5 | NGS | +/+ |
| 100 | NLRC4 | (N6, A20) | 2E+05 | 64 | F | Tumor | UK, adjuvant treatment with crizotinib | >10 | NGS | UK/UK |
| 101 | PNPT1 | (P19, A20) | 2E+05 | 72 | F | FFPE | PR to crizotinib | >13 | NGS | UK/+ |
| 102 | PLB1 | UK | 2E+05 | UK | UK | Tumor | UK | / | NGS | UK/UK |
| 103 | between Linc00308 and D21S2088E | IGR | 2E+05 | 61 | M | FFPE | PR to crizotinib | >6 | NGS | +/+ |
| 104 | SPECC1L | (S8, A20) | 2E+05 | 44 | F | Tumor | PR to crizotinib、bevacizumab | >23 | NGS | +/+ |
| 105 | COX7A2Le | IGR | 2E+05 | 53 | M | Tumor | PR to crizotinib | 12 | NGS | UK/+ |
| 106 | LINC0121e | IGR | 2E+05 | 53 | M | Tumor | PR to crizotinib | 12 | NGS | UK/+ |
| 107 | ATP13A4e | (A9, A19) | 2E+05 | 53 | M | Tumor | PR to crizotinib | 12 | NGS | UK/+ |
| 108 | SLCO2A1e | IGR | 2E+05 | 53 | M | Tumor | PR to ceritinib | 7 | NGS | UK/+ |
| 109 | GPC1 | (G1, A20) | 2E+05 | 65 | F | Tumor | NR | NR | RNA NGS | +/+ |
| 110 | CDCA7d | NR | 2E+05 | 51 | M | Tumor | PR to crizotinib | UK | NGS | UK/+ |
| 111 | FSIP2d | NR | 2E+05 | 51 | M | Tumor | PR to crizotinib | UK | NGS | UK/+ |
| 112 | ERLEC1d | NR | 2E+05 | 51 | M | Tumor | PR to crizotinib | UK | NGS | UK/+ |
| 113 | THUMPD2 | (T6, A20) | 2E+05 | 43 | F | Tumor | PR to crizotinib | >17 | NGS | +/+ |
| 114 | OFCC1 | IGR | 2E+05 | 51 | F | FFPE | NT | / | NGS | UK/+ |
| 115 | RMDN2 | (R1, A15) | 2E+05 | NR | NR | Tumor | NR | NR | NGS | UK/UK |
| 116 | between KLHL31 and LRRC1 | IGR | 2E+05 | 50 | M | Plasma | PR to ensartinib | >6 | NGS | UK/+ |
| 117 | MRPS9 | IGR | 2E+05 | 60 | F | Mediastinal puncture | PR to crizotinib alectinib | 10,>10 | NGS | -/- |
Table 1: Catalog of Fusion Partners in ALK+ NSCLC.
| ALK fusion | Breakpoint of LMO7 | Published Year | Positivity to ALK-TKIs | PFS (months) |
|---|---|---|---|---|
| (L15, A20) | Exon15 | 20176 | Unknown | Unknown |
| (L16, A20) | Exon16 | 20217 | PR to ensartinib | 18 |
| (L13, A20) | Exon13 | 2022 | PR to crizotinib | 5 |
Table 2: Different LMO7-ALK fusion variants published till now.
ALK fusion partners identification and breakpoints discovery still play an important role. ALK kinase domain is activated through the autophosphorylation involving dimerization and the N-terminal fusion partner provided a promoter that causes constitutive expression of the ALK fusion protein When gene fusion happens.38 In previous reports, LMO7-ALK variants (L16: A20) were reported with PD to alectinib and PR to ensartinib separately. In this fusion variant, exon 13 of LMO7 (NM_015842) is fused to exon 20 of ALK (NM_004304) [36]. The patient showed a partial response to Crizotinib whereafter developed rapidly and lost his life 5 months later, which is never reported before (Figure 2B). The predicted LMO7-ALK protein product contained amino acids comprising the N-terminal amino acid of LMO7 and C-terminal amino acid of ALK, containing the entire tyrosine kinase domain presenting that the fusion variant responded to ALK-TKIs [37].
However, poor prognosis indicated the novel breakpoint (L13: A20) LMO7-ALK rearrangement may be a hyper-progressive marker. In conclusion, the LMO7-ALK rearrangement in advanced NSCLC is identified as a potentially sensitive fusion mutation. Poor prognosis, in this case, indicated the novel breakpoint (L13: A20) LMO7-ALK rearrangement may be a hyper-progressive marker and result in shorter survival time than other breakpoints in LMO7-ALK rearrangement and other ALK fusions. However, the rearrangement was rare that leads to more observation between molecular mechanism and survival prognosis in real world. To signify efficient rearrangements in the molecule aspect and supply more detailed information about ALK fusion variants in NSCLC, we reviewed the ALK fusion partners as well as the relative PFS, response to ALK-TKIs from 2007. According to this research, it is possible that more accurate recommendations for precision medicine will be provided by employing the NGS test to find more fusion partners with the ALK gene [38].
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Funding
This work was supported by the This work has been strongly supported by the External Cooperation of Science and Technology Program of Fujian Province (No. 202210034) and the 900th Hospital of the Joint Logistic Support Force of China: National Science and Technology Fund Incubation Special Program(No. 2023GK04)..
Conflict of Interest Disclosure
The authors report no declarations of interest.
Consent Statement
The follow-up data and images included in this paper have been acquired with the patient’s consent.
Acknowledgment
Thank AMOYDX (Xia Men, China) for providing support in genetic analysis technology. Ping Lin drafted the manuscript and collected present illness. Wencui Kong supplied the clinical data. Ying Chen evaluated disease status and therapeutic response to ALKTKIs. Lijuan Qu identified the pathology of lung cancer. Zongyang Yu modified the manuscript critically for important intellectual content and confirmed final approval of the version to be published. All authors confirmed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Herbst RS, Morgensztern D, Boshoff C (2018) The biology and management of non-small cell lung cancer. Nature 553: 446-454.
- Remon J, Pignataro D, Novello S (2021) Current treatment and future challenges in ROS1- and ALK-rearranged advanced non-small cell lung cancer. Cancer Treat Rev 95: 102-178.
- Soda M, Choi YL, Enomoto M (2007) Identification of the Transforming EML4-ALK Fusion Gene in Non-Small Cell Lung Cancer. Nature 448: 561-6.
- Qiao M, Zhao C, Liu Q (2002) Impact of ALK variants on brain metastasis and treatment response in advanced NSCLC patients with oncogenic ALK fusion. Transl Lung Cancer Res 9: 1452-1463.
- Ou SI, Zhu VW, Nagasaka M (2020) Catalog of 5' Fusion Partners in ALK-positive NSCLC Circa 2020. JTO Clin Res Rep 1: 10-15.
- Noh KW, Lee MS, Lee SE (2017) Molecular breakdown: a comprehensive view of anaplastic lymphoma kinase (ALK)-rearranged non-small cell lung cancer. J Pathol 243: 307-319.
- Li M, An Z, Tang Q (2021) Mixed responses to first-line alectinib in non-small cell lung cancer patients with rare ALK gene fusions: A case series and literature review. J Cell Mol Med 25: 9476-9481.
- Zeng H, Liu Y, Wang W (2021) A rare KIF5B-ALK fusion variant in a lung adenocarcinoma patient who responded to crizotinib and acquired the ALK L1196M mutation after resistance: a case report. Ann Palliat Med 10: 8352-8357.
- Jung Y, Kim P, Jung Y (2012) Discovery of ALK-PTPN3 gene fusion from human non-small cell lung carcinoma cell line using next generation RNA sequencing. Gene Chromosomes Can 51: 590-597.
- Su C, Jiang Y, Jiang W (2020) STRN-ALK Fusion in Lung Adenocarcinoma with Excellent Response upon Alectinib Treatment: A Case Report and Literature Review. Onco Targets Ther 13: 12515-12519.
- Li M, Tang Q, Chen S (2021) A novel HIP1-ALK fusion variant in lung adenocarcinoma showing resistance to Crizotinib. Lung Cancer 151: 98-100.
- Drilon A, Wang L, Arcila ME (2015) Broad, Hybrid Capture-Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches. Clin Cancer Res 21: 3631-3639.
- Yang S, Gong F, Wang G (2020) Anaplastic lymphoma kinase (ALK) partners identified by next-generation sequencing in Chinese patients with solid tumors. J Clin Oncol 38: 3555-3555.
- Li W, Zhang J, Guo L (2017) Combinational Analysis of FISH and Immunohistochemistry Reveals Rare Genomic Events in ALK Fusion Patterns in NSCLC that Responds to Crizotinib Treatment. J Thorac Oncol 12: 94-101.
- Gu FF, Zhang Y, Liu YY (2016) Lung adenocarcinoma harbouring concomitant SPTBN1-ALK fusion, c-Met overexpression, and HER-2 amplification with inherent resistance to crizotinib, chemotherapy, and radiotherapy. J Hematol Oncol 9: 66.
- Cui S, Zhang W, Xiong L (2017) Use of capture-based next-generation sequencing to detect ALK fusion in plasma cell-free DNA of patients with non-small-cell lung cancer. Oncotarget 8: 2771-2780.
- Yan J, Zhou X, Pan D (2020) A case of one lung adenocarcinoma patient harbouring a novel FAM179A-ALK (F1, A19) rearrangement responding to lorlatinib treatment. Lung Cancer 147: 26-29.
- Vendrell JA, Taviaux S, Béganton B (2017) Detection of known and novel ALK fusion transcripts in lung cancer patients using next-generation sequencing approaches. Sci Rep 7: 12510.
- Chen Y, Zhang X, Jiang Q (2020) Lung adenocarcinoma with a novel SRBD1-ALK Fusion responding to crizotinib. Lung Cancer 146: 370- 372.
- Fei X, Zhu L, Zhou H (2019) A Novel Intergenic Region between CENPA and DPYSL5-ALK Exon 20 Fusion Variant Responding to Crizotinib Treatment in a Patient with Lung Adenocarcinoma. J Thorac Oncol 14: 191-193.
- Wen S, Dai L, Wang L (2019) Genomic Signature of Driver Genes Identified by Target Next-Generation Sequencing in Chinese Non-Small Cell Lung Cancer. Oncologist 24: 1070-1081.
- Li T, Zhang F, Wu Z (2020) PLEKHM2-ALK: A novel fusion in small-cell lung cancer and durable response to ALK inhibitors. Lung Cancer 139: 146-150.
- Liu S, Huang T, Liu M (2020) The Genomic Characteristics of ALK Fusion Positive Tumors in Chinese NSCLC Patients. Front Oncol 10: 726.
- Wu X, Zhou H, He Z (2020) Coexistence of a novel CCNY-ALK and ATIC-ALK double-fusion in one patient with ALK-positive NSCLC and response to crizotinib: a case report. Transl Lung Cancer Res 9: 2494-2499.
- Wang R, Qin J, Fan Y (2020) Responses to ALK Inhibitor Treatments in a Patient with Non-Small Cell Lung Cancer Harbouring a Novel HPCAL1- ALK Fusion Variant: A Case Report. Onco Targets Ther 13: 4183-4187.
- Wu X, Wang W, Zou B (2020) Novel NLRC4-ALK and EML4-ALK double fusion mutations in a lung adenocarcinoma patient: A case report. Thorac Cancer 11: 1695-1698.
- Jin L, Wang Y, Li S (2020) Novel PNPT1-ALK fusion variant exerted significant benefit to crizotinib in NSCLC. Lung Cancer 146: 382-384.
- Zhang J, Zou C, Zhou C (2020) A Novel Linc00308/D21S2088E Intergenic Region ALK Fusion and Its Enduring Clinical Responses to Crizotinib. J Thorac Oncol 15: 1073-1077.
- Ma L, Zhang Q, Dong Y (2020) SPECC1L-ALK: A novel gene fusion response to ALK inhibitors in non-small cell lung cancer. Lung Cancer 143: 97-100.
- Cai C, Long Y, Li Y (2020) Coexisting of COX7A2L-ALK, LINC01210-ALK, ATP13A4-ALK and Acquired SLCO2A1-ALK in a Lung Adenocarcinoma with Rearrangements Loss During the Treatment of Crizotinib and Ceritinib: A Case Report. Onco Targets Ther 13: 8313-8316.
- Xiong L, Li X, Chen D (2021) GPC1-ALK: A novel ALK fusion in a patient with pulmonary sarcomatoid carcinoma. Lung Cancer 151: 104-105.
- Zhao G, Chen L, Xiao M (2021) Rare coexistence of three novel CDCA7-ALK, FSIP2-ALK, ALK-ERLEC1 fusions in a lung adenocarcinoma patient who responded to Crizotinib. Lung Cancer 152: 189-192.
- Wang YL, Wu ZZ, Zhang HR (2021) Coexistence of a novel RGS18 downstream intergenic region ALK fusion and a THUMPD2-ALK fusion in a lung adenocarcinoma patient and response to crizotinib. Lung Cancer 154: 216- 218.
- Zhai X, Wu Q, Zeng Z (2021) OFCC1-ALK (Ointergenic: A20): A novel OFCC1 intergenic region-ALK fusion identified from a lung adenocarcinoma patient. Lung Cancer 153: 171-173.
- Jiang L, Chen S, Stinnett V (2021) Concomitance of a novel RMDN2-ALK fusion and an EML4-ALK fusion in a lung adenocarcinoma. Cancer Genet 258-259: 18-22.
- Qiu H, Li Q, Xiao Y (2021) A Novel Intergenic Region Between KLHL31 and LRRC1-ALK Exon 20 Fusion Variant in Advanced Lung Adenocarcinoma and its Remarkable Response to ALK Inhibitor. J Thorac Oncol 16: 21-23.
- Zhou H, Xu B, Xu J (2021) Novel MRPS9-ALK Fusion Mutation in a Lung Adenocarcinoma Patient: A Case Report. Front Oncol 11: 670907.
- Katayama R, Lovly CM, Shaw AT (2015) Therapeutic targeting of anaplastic lymphoma kinase in lung cancer: a paradigm for precision cancer medicine. Clin Cancer Res 21: 2227-2235.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: Lin P, Kong W, Chen Y, Qu L, Yu Z (2024) A Case Report and LiteratureReview on Rare LMO7-ALK Rearrangement in Lung Adenocarcinoma. Cell MolBiol, 70: 311.
Copyright: © 2024 Lin P, et al. This is an open-access article distributed underthe terms of the Creative Commons Attribution License, which permits unrestricteduse, distribution, and reproduction in any medium, provided the original author andsource are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 1088
- [From(publication date): 0-2024 - Mar 04, 2025]
- Breakdown by view type
- HTML page views: 1015
- PDF downloads: 73