Research Article Open Access
2D-PAGE of Cashew Stem Coupled to LC ESI Q-TOF MS/MS Reveals Abundance of Antioxidant Enzymes and Heat Shock Proteins, Compatible with the Crop Adaptation to the Semi-Arid Conditions of Tropical Countries
Darcy MF Gondim1,2, Ilka M Vasconcelos1, Frederico BMB Moreno2, Ana CO Monteiro-Moreira2, Jose H Araújo-Filho1, Jeferson Segalin3, Paulo M Pinto4, Célia RRS Carlini3, Jose E Cardoso5 and Jose TA Oliveira1*1Department of Biochemistry and Molecular Biology, Federal University of Ceará, Brazil
2University of Fortaleza (UNIFOR) – Ceará, Brazil
3Department of Biophysics and Center of Biotechnology, Federal University of Rio Grande do Sul, Brazil
4Institute of Biotechnology, University of Caxias do Sul, Caxias do Sul, RS, Brazil
5Brazilian Agricultural Research Corporation, Tropical Agroindustry Research Center (EMBRAPA), Brazil
- *Corresponding Author:
- Jose TA Oliveira
Department of Biochemistry and Molecular Biology
Federal University of Ceará, Brazil
Tel: +55 85 33669823
Fax: +55 85 33669789
E-mail: jtaolive@ufc.br
Received date: June 17, 2014; Accepted date: July 14, 2014; Published date: July 16, 2014
Citation: Gondim DMF, Vasconcelos IM, Moreno FBMB, Monteiro-Moreira ACO, Araújo-Filho JH, et al. (2014) 2D-PAGE of Cashew Stem Coupled to LC ESI Q-TOF MS/MS Reveals Abundance of Antioxidant Enzymes and Heat Shock Proteins, Compatible with the Crop Adaptation to the Semi-Arid Conditions of Tropical Countries. J Anal Bioanal Techniques S6:004. doi: 10.4172/2155-9872.S6-004
Copyright: © 2014 Gondim DMF, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Cashew crop grown in the semi-arid conditions of tropical countries produces cashew nut, an important commodity both for internal consumption and exportation. Cashew is very well adapted to abiotic stresses such as drought, high temperature, high salinity, and solar radiation predominant in the environmental regions where cashew is cultivated. Besides cashew is threatened by a great variety of fungal diseases amongst them gummosis caused by the devastating fungus Lasiodiploidea theobromae that has increased its severity in all northeastern Brazil producing states. Therefore there is a great interest in understanding the biochemical/physiological traits associated with both the climate adaptation of cashew and the resistance/susceptibility to L. theobromae.
This paper reports on the evaluation of a proteomic approach to study the proteins of the cashew stems, a recalcitrant plant tissue, where the L. theobromae infection establishes. After testing different methods for extracting proteins from cashew stems, the precipitation with trichloroacetic acid/acetone combined with the use of an optimized phenol extraction method produced a cashew protein sample free of interfering compounds that showed a highquality 2D-PAGE pattern. The extraction method devised allowed the fractionation of approximately 615 spots from which 130 proteins were identified. Of them 31% are related to plant disease/defense, which is consistent with the excellent fit of cashew to the semi-arid conditions. Therefore, this pioneering map derived from CCP (Premature Cashew Clone) 76, a semiarid-tolerant cashew clone, provides the basis for further investigations of cashew physiology such as detection of genetic reprogramming induced by biotic and abiotic stresses.
Keywords
Anacardium occidentale L.; Cashew; Defense proteins; Heat shock proteins; Proteomic analysis
Abbreviations
CHAPS:3-[(3-Cholamidopropyl dimethylammonio] -1- Propanesulfonate; PMSF: Phenylmethane Sulfonylfluoride; PVPP: Polyvinylpolypyrrolidone; Q-TOF-MS: Quadrupole Time-of- Flight Mass Spectrometer; TCA: Trichloroacetic Acid; TFA: Trifluoroacetic Acid; 2-ME: β-Mercaptoethanol; UPLC: Ultra Performance Liquid Chromatography System.
Introduction
Cashew (Anacardium occidentale L.) is a fruit tree belonging to the Anacardiaceae family that occupies around 3.39 million hectares in the world [1]. The cashew industry has contributed to the social and economical development of some Brazilian, Asian, and African regions that have few economic options to offer to their population. The main economic products of cashew are the nuts and the nut shell liquid. Brazil is the world’s third largest producer of cashew nuts. It is most cultivated at the Northeast semi-arid regions (94% of the national production) of Brazil where it is continually exposed to environmental stresses, such as low rainfall, high temperature daylight, low relative humidity and high soil salinity, besides to biotic stresses caused mainly by fungi [2-4].
Recently, it was reported that cashew orchards planted in Northeastern Brazil have been threatened by a highly severe disease referred to as gummosis that leads to low productivity and eventually death [2,5]. This disease is caused by a necrotrophic pathogenic fungus called Lasiodiplodia theobromae (Pat.) Griffon & Maubl whose primary infection site is the cashew stem [2,6,7]. Plants are equipped with constitutive and inducible defense mechanism to cope with attempted attacks by pathogens and plagues [8]. Most of these mechanisms rely on proteins, some responsible for the recognition of the invading pathogen and others that act directly to kill them or are over- or subexpressed in response to the biotic stress [9-11]. For instance, in some species of the Anacardiaceae family such as in Astronium fraxinifolium (Zebrawood), Pistacia Atlantica (Mt. Atlas mastic tree), Mangifera indica (Mango) and Myracrodruon urundeuva (Aroeira) abiotic and biotic stress-related proteins such as heat shock proteins, 14-3-3-like protein, proteins involved in glutathione metabolism, peroxidase, polyphenol oxidase, cysteine protease, serine protease, chitinase and thionins have been reported [12-15]. The set of all proteins expressed by the genome under specific conditions is defined as the proteome [16,17]. Recently, tremendous progress has occurred in plant proteomics, mostly due to developments in sample preparation, highquality two-dimensional gel electrophoresis (2D-PAGE), and major developments in mass spectrometry dedicated to protein analyses [17-19]. However, all these progress in proteomic could be meaningless without a robust protein sample preparation protocol. For example, high-quality protein preparations from plant materials for 2D-PAGE and mass spectrometry analyses are difficult to obtain because plant tissues are usually rich in interfering substances, such as pigments, polysaccharides, lipids, and secondary metabolites. Additionally, various plant tissues contain relatively low concentrations of proteins that can eventually be hydrolyzed and oxidized by endogenous proteases and oxidative enzymes, respectively, during extraction. The appropriate protocol will therefore depend on the properties of the plant tissue, the proteins of interest, and the downstream analysis that will be performed [18,20,21].
Cashew stems is a very recalcitrant tissue for protein extraction and subsequent proteomic analyses because it has low amount of proteins and besides has several interfering compounds. Thus the establishment of an appropriate protocol to extract proteins from cashew stems to produce reliable 2D maps and the subsequent identification of proteins by mass spectrometry is of utmost importance. Therefore, herein, we report the first proteome study carried out in cashew stem aimed to produce references 2D maps that could help in future studied on the proteins which are differentially expressed upon cashew infection by L. theobromae.
Materials and Methods
Plant material and reagents
Cashew plant samples came from a commercial orchard of dwarf cashew (Planalto farm, 6°43’30’’ S, 40°35’19’’ W, 730 m asl). Branches of around 40 cm length from four-year-old-cashew (CCP 76 clone) were collected, stored at -20°C for up to 24 h, and taken to the lab where they were stored at -83°C. Before lyofilization, the branches were fragmented to disks of around 3 cm diameter with a saw, lyophilized for 15 days (8 hours/day), carefully powdered (0.5 mm mesh) using a mill (Lee household flour meal, Lee Engineering Co., Milwaukee, Wis., USA) toward avoiding heating , and stored at -83°C until extraction of proteins. Samples were processed from four biological replicates and each sample was assayed three times.
Acetone, acetonitrile, ammonium acetate, bovine serum albumin (BSA), 3-[(3-cholamidopropyl)dimethylammonio]- 1-propanesulfonate (CHAPS), coomassie brilhante blue (CBB) G-250, formic acid, iodoacetamide, β-mercaptoetanol (2-ME), polyvinylpolypyrrolidone (PVPP), trichloroacetic acid (TCA), thiourea, Tris-saturated phenol, sodium dodecyl sulfate (SDS), sucrose, and urea were from Sigma Chemical (St. Louis, MO). Ditiotreitol (DTT), ethylenediaminetetraacetic acid (EDTA), iodoacetamide, IPG buffer 3-10, phenylmethanesulfonyl fluoride (PMSF), immobilized pH gradient (IPG) strips (13 cm, linear pH gradient from pH 3-10 or pH 4-7) were from GE Healthcare Life sciences (Piscataway, NJ). All other chemicals were of analytical grade.
Protein extraction and quantification
Seven different methods of protein extraction for cashew stems were tested to allow posterior proteomic analysis. Six of them, following previously described methodologies [21-26] were not adequate because they did not remove interfering compounds, the low amount of proteins obtained and also the small number of protein bands revealed in one dimensional electrophoresis. The method of choice was one based on phenol as previously described [27,28] with various modifications. The cashew stem fine (0.5 mm mesh) powder (1 g) was resuspended in 25 mL of ice-cold acetone containing 10% TCA) and 2% 2-ME, vortexed for 1 h at 4°C and stored at -83°C overnight. After centrifugation (18,000 × g, 30 min, 4°C), the remaining pellet was rinsed twice with ice-cold acetone containing 2% 2-ME and centrifuged at 18,000 g for 20 min at 4°C. The pellet was dried, and the proteins were extracted in 15 mL of extraction buffer (0.1 M Tris-HCl, pH 8.65, containing 30% sucrose, 2% SDS, 1 mM PMSF, 2% 2-ME, 1% PVPP). The sample was vortexed for 1 h at 4°C, and an equal volume of Tris-saturated phenol (pH 8.0) was added and homogenized for 30 min at 4°C. After centrifugation (10,000 × g, 20 min, 4°C), the upper phenol phase was collected and re-extracted with 15 mL of extraction buffer. The sample was centrifuged (10,000 × g, 20 min, 4°C), and the proteins in the phenol phase were precipitated with five volumes of 0.1 M ammonium acetate in methanol, at -83°C for 2 h, and centrifuged (16,000 × g, 20 min, 4°C). The pellet was washed twice with 0.1 M ammonium acetate in methanol and twice with ice-cold 80% acetone in water. The resuspended pellet obtained between washes was maintained at -20°C for 30 min. After centrifugation (16,000 × g, 20 min, 4°C), the protein pellet was dried in a desiccator containing silica gel at 4°C, resuspended in 300 μL of 7 M urea, 2 M thiourea, 2% CHAPS, and the proteins solubilized by sonication (Ultracleaner 1400A, Unique Ind. Com., Brazil) for 20 min at room temperature (23 ± 2°C), followed by shaking at 200 rpm for 20 min at 4°C. After centrifugation (10,000 × g, 10 min, 4°C) to remove debris, the supernatant (Extract A) was kept at -83°C for subsequent analysis. The protein content was evaluated using bovine serum albumin as a standard [29].
Two-dimensional gel electrophoresis and gel image analysis
The first dimension electrophoresis (isoelectric focusing) was carried out on immobilized pH gradient (IPG) strips (13 cm, linear pH gradient from pH 3-10 or pH 4-7; 4% acrylamide) rehydrated for 16 h with 300 μg of cashew stem protein in 250 μL of the rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS, 2% (v/v) carrier ampholytes, pH 3-10, and traces of bromophenol blue). Isoelectric focusing (IEF) was performed on IPGphor II (Amersham Bioscience) at 20°C, with a current limit of 50 μA/strip using the following schedule: 200 V for 2 h, 500 V for 2 h, 5,000 V for 2 h, and 10,000 V up to 20,000 V/h. After the IEF, proteins in strips were reduced for 15 min in equilibration buffer (50 mM Tris-HCl, pH 8.8, 6 M urea, 30% glycerol, 2% SDS, and 0.1 M DTT), followed by alkylation for 15 min with 0.1 M iodoacetamide in equilibration buffer without DTT. The second dimension electrophoresis was performed in a vertical system with uniform 14% acrylamide separating gel (15 × 15 cm) at 15°C. The runs were carried out at 20 mA/gel for the first 30 min followed by 30 mA/gel for 4.5 h. Proteins were visualized by colloidal Coomassie Brilhante Blue (CBB) staining [30]. At least three replicates were performed for each sample. The gels were scanned using an image scanner (Amersham Bioscience), and the images were analyzed using the ImageMaster 2D Platinum Version 6.0 Analysis Software (Amersham Bioscience) according to the user’s manual.
Mass spectrometry and protein identification
Protein spots were excised from CBB-stained polyacrylamide gels and destained with 100 μL of 25 mM NH4HCO3 in 50% acetonitrile (ACN) until the CBB stain had faded sufficiently. Next, gel fragments were washed twice in 100% ACN for 10 min, until they became opaque, and then dried under vacuum for 30 min. The dried gel fragments were incubated with 10 ng/μL in 50 mM NH4HCO3 trypsin (Promega, Madison, WI, USA), at 37°C in a water bath overnight. The peptides generated were extracted from the trypsin-treated gel fragments, for 30 min (three times), with 50% ACN/5% TFA. Extracts were dried using centrifuge vacuum concentrators. Prior to mass spectrometry (MS) identification, dried peptides were dissolved in 10 μL of 0.1% formic acid. MS/MS analyses were performed in an electrospray ionization (ESI) quadrupole time-of-flight (Q-TOF) Micro™ mass spectrometer coupled to a nanoACQUITY® UltraPerformance liquid chromatography system (Waters, Milford, US). A nanoflow ESI source was used with a Lockspray™ dual electrospray ion source (Waters) for lockmass measurement during all the chromatographic runs. The peptides were separated on a Nanoease C18 (75 μm ID) capillary column equilibrated with 98% solution A (0.1% formic acid/water) and 2% B (ACN/0.1% formic acid). Elution was done with the gradient schedule: 2-60% B for 13 min; 60-95% B for 6 min; 95-2% B for 11 min. Data were acquired in data-dependent mode (DDA) and multiple charged peptide ions (+2 and +3) were automatically mass selected and dissociated in MS/MS experiments. Typical LC and ESI conditions consisted of a flow of 600 nL/min, nanoflow capillary voltage of 3.5 kV, block temperature of 100°C, and cone voltage of 50 V. The acquired mass spectra were processed using the Mascot Distiller software (Matrix Science, London, UK), and the generated MGF files were queried against the NCBI database using the MASCOT software v. 2.2 (Matrix Science, London, UK). Searches were conducted with the consideration of a maximum of one missed cleavage, the carbamidomethylation of cysteine, the possible oxidation of methionine, peptide tolerance of 0.2 Da, and MS/ MS tolerance of 0.2 Da. The significance threshold was set at p<0.05, and identification required that each protein contained at least one peptide with an expected value <0.05. Homology searches were performed against the NCBI protein databases choosing “Viridiplantae taxa” as the taxonomy category. Proteins were classified according to their function in the categories described by Bevan et al. [31]. Localization prediction was analyzed by using the PSORT software program [32].
Results and Discussion
Protein extracts from cashew stem
The main modifications introduced in the methodologies previously described [27,28] are as follow: pulverization of the freeze-dried stem tissue to a fine powder (0.5 mm mesh) to achieve complete tissue disintegration before protein extraction; suppression of the step in which silicon dioxide was used [28]; pre-wash of the fine power with cold TCA/β-mercaptoethanol/acetone solution instead of cold pure acetone [28]; lower storage temperature (-83°C) overnight instead of -20°C [27,28]; use of the extraction buffer consisting of 0.1 M Tris-HCl, pH 8.65, containing 30% sucrose, 2% SDS, 1 mM PMSF, 2% 2-mercaptoethanol and 1% PVPP instead 0.1 M Tris-HCl, pH 8.0, containing 30% sucrose, 2% SDS, 5% 2-mercaptoethanol [27] or 0.05 M Tris-HCl, pH 8.65, containing 2% SDS, 30% sucrose, and 2% 2-mercaptoethanol [28]; a much longer contact time with the extraction buffer (vortexed for 1 h instead of 30 s [27] or 5 min [28]); re-extraction of the phenol phase (high protein content) with Tris-saturated phenol, pH 8.0, for better protein isolation; longer precipitation time with ammonium acetate in methanol (2 h instead of ½ h [27,28]); a final wash with 80% acetone (in water) instead of pure acetone [27]; the use of the protein rehydration solution consisting of 7 M urea, 2 M thiourea, 4% CHAPS, 2% IPG buffer instead of 8 M urea, 4% CHAPS, 2% IPG buffer, 20 mM DTT; and suppression of lyophilization of the final pellet done in [28] to avoid irreversible insolubilization of the extracted proteins. With such alterations, the protein content achieved for the cashew stem extract was 46 ± 10 mg protein/g dry tissue. Moreover, a high number of protein spots (683 ± 42) were obtained after 2D PAGE (Figure 1). Most of them were resolved within the molecular mass (Mr) range of 17 and 50 kDa and pIs of 4.5 and 7.5. The maps were made in triplicate from independent preparations by using different cashew stem powder, and we observed an excellent reproducibility (data not shown).
Figure 1: 2D-PAGE pattern of the cashew stem proteins extracted by the Phenol-Tris based method. Proteins (300 μg) were separated by IEF on a 3-10 linear pH gradient strip (13 cm) followed by SDS-PAGE and visualized after staining with colloidal CBB. Numbered protein spots were excised from the gel and identified using nano-UPLC-ESI-Q-TOF analysis. The identified proteins are listed in Table 1 and Table S1 (supplementary material).
Various plant tissues contain low amounts of proteins and also a variety of secondary compounds that represent a pitfall in the successful extraction of proteins for 2D-PAGE analysis [33]. Cashew stems have low protein content, but it is rich in polysaccharides, and secondary metabolites such as phenols, tannins, lignin, and others [34] that interfere in 2D PAGE quality. Moreover, protein extraction from mechanically resistant tissue such as stems in woody perennials, as cashew, is also challenging.
Amongst the published protocols for analyzing plant tissues, the phenol-based method has been proven to be highly effective with recalcitrant materials containing high levels of interfering compounds [18,35-37]. The results presented herein reinforce the high cleanup capacity of the phenol reagent. This is due to its ability to better solubilize proteins, to reduce molecular interactions between proteins and interfering materials, and the low solubility of polysaccharides and nucleic acids [28,36]. Moreover, pre-washing the plant sample with a cold acetone solution removes most of the interfering compounds (phenolics and lipids), disrupts lipid-protein complexes, and weakens the plant tissue, increasing the contact surface with the extraction buffer and improving the quality and efficiency of the protein extraction protocol for recalcitrant plant tissues, as for the cashew stem [28,38]. To the best of our knowledge, this is the first time that an effective protocol for protein extraction from cashew stem has been described for proteomics analysis. The protocol is expected to accelerate the proteomic study of cashew tissues and other woody plants.
Proteomic analysis of cashew stem
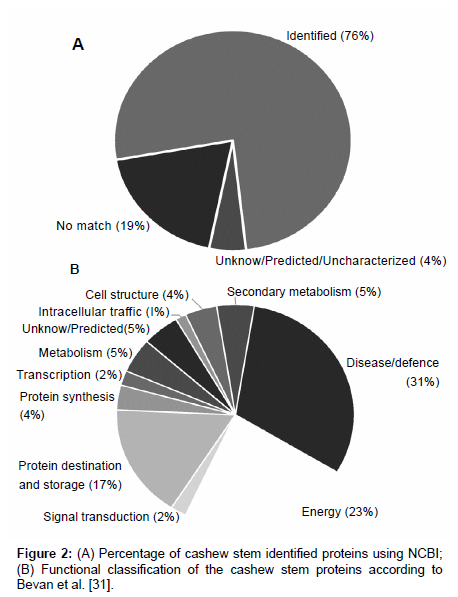
One hundred and seventy one different spots (Figure 1) were excised from the gel, fragmented, trypsin digested, and analyzed by mass spectrometry. One hundred and thirty proteins (~76%) were indentified and 8 (~5%) were unknown/predicted/uncharacterized proteins (Figure 2A) using the NCBI bank (Table S1 - supplementary material). Thirty four spots (19%) do not match with proteins in the database (Figure 2A), suggesting that they might have not been previously described or were not yet deposited in the databases. These indentified proteins were all categorized (Figure 2B) according to their function as described by Bevan et al. [31]. Thirty one percent of them (43 spots) were grouped at “Disease/Defense” (spots 1, 2, 3, 10, 32, 33, 34, 80, 81, 89, 104, 106, 109, 116,117,118, 119, 122, 123, 124, 127, 129, 130, 131, 132, 133, 134, 135, 136, 137, 139, 140, 141, 143, 156, 157, 158, 159, 160, 167, 168, 169, 173); 23% (32 spots) “Energy” (spots 8, 14, 18, 19, 20, 22, 24, 25, 26, 27, 28, 30, 31, 47, 49, 51, 52, 54, 55, 57, 59, 60, 61, 63, 64, 67, 68, 69, 84, 87, 105, 121); 17% (23 spots) “Protein destination and Storage” (spots 13, 15, 16, 17, 29, 36, 37, 38, 71, 78, 92, 93, 100, 101, 102, 110, 111, 112, 120, 145, 163, 164, 172). Other categories were less represented: “Metabolism” (~5%; 7 spots: 23; 40; 50; 58; 115; 142; 170); “Cell structure” (~4%; 6 spots: 35; 44; 45; 46; 150; 151); “Protein biosynthesis” (~4%; 5 spots: 75; 82; 83; 126; 149); ”Signal transduction” (~2%; 3 spots: 114; 147; 148); “Secondary metabolism” (~5%; 6 spots: 70; 72; 73; 76; 77; 90); “Intracellular traffic” (~1%; 2 spots: 39; 108); “Transcription” (~2%; 3 spots: 75; 82; 83); “Unknown/predicted/ uncharacterized” (~5%; 8 spots: 7; 74; 79; 94; 113; 146; 152; 155).
In the environment, cashew plants are subjected to different stresses such as high temperature, drought and salinity, and the attack of pathogens and herbivores. Moreover, the majority of the cashew orchards are managed using technologically unsophisticated practices [3]. To overcome these challenges, cashew plants have efficient protection mechanisms, including those depending on differential protein expression.
In this present work, a relative large number (30 spots) of heatshock proteins (HSPs), particularly sHSPs (small HSPs), was detected in cashew stems (Table 1), which is consistent with the stress-tolerance trait exhibited by this culture cultivated in tropical regions of the world where high temperature, high solar radiation and drought are prevalent. Various studies indicate that sHSPs have very important roles in thermotolerance and plant adaptation to the environment [39]. Indeed, high temperature, drought and salinity are known to induce HSPs in plants [40-42]. HSPs expression is normally limited in normal conditions but it is strongly expressed under stress situations. In general, HSPs act as molecular chaperones and participate in the correct folding of proteins [41,43-45]. Plants can express up to 40 types of sHSPs that are induced under various stress conditions. They are apparently not essential for basal cell functions but very important for plant survival under heat stress. sHSPs are responsible for capturing unfolding proteins to form stable complexes and prevent their irreversible aggregation. It is also described their role in maintaining the integrity of cell membranes under stress conditions [45,46].
| Spot No. | Accession No. |
Protein identification | Organism | Subcellular localization (PSORT) |
Mr (KDa)/pI | Sequence covered (%) |
Score | ||
|---|---|---|---|---|---|---|---|---|---|
| Exp | Theor | ||||||||
| 1 | gi|110623251 | heat shock protein | Triticum durum | Unclear | 87.46/6.23 | 100.62/6.07 | 2 | 121 | |
| 2 | gi|4558484 | heat shock protein | Triticum aestivum | Unclear | 87.46/6.30 | 101.29/5.95 | 4 | 126 | |
| 3 | gi|37718900 | heat shock protein | Oryza sativa | Cytosol | 87.82/6.39 | 82.64/5.43 | 3 | 91 | |
| 10 | gi|108707472 | heat shock cognate protein | Oryza sativa | Cytosol | 60.21/5.20 | 71.93/5.30 | 12 | 276 | |
| 117 | gi|255560519 | heat-shock protein | Ricinus communis | Unclear | 20.81/6.48 | 21.71/6.45 | 10 | 71 | |
| 118 | gi|255558876 | heat-shock protein | Ricinus communis | Cytosol | 20.10/6.95 | 17.50/5.34 | 29 | 208 | |
| 119 | gi|255557799 | heat-shock protein | Ricinus communis | Unclear | 19.63/7.37 | 22.17/6.21 | 12 | 137 | |
| 124 | gi|156711718 | chloroplast small heat shock protein | Rhododendron breviperulatum | Cytosol | 23.04/4.84 | 12.84/4.84 | 7 | 46 | |
| 127 | gi|189014946 | small heat shock protein | Mangifera indica | Cytosol | 20.12/4.91 | 19.85/5.23 | 8 | 49 | |
| 129 | gi|349591294 | class I small heat shock protein | Solanum lycopersicum | Cytosol | 21.13/5.42 | 17.62/5.82 | 14 | 131 | |
| 130 | gi|123539 | class I heat shock protein | Glycine max | Cytosol | 21.25/5.58 | 17.52/5.98 | 14 | 98 | |
| 131 | gi|349591294 | class I small heat shock protein | Solanum lycopersicum | Cytosol | 20.43/5.68 | 17.62/5.82 | 20 | 149 | |
| 132 | gi|349591294 | class I small heat shock protein | Solanum lycopersicum | Cytosol | 19.93/5.69 | 17.62/5.82 | 20 | 162 | |
| 133 | gi|255558876 | heat-shock protein | Ricinus communis | Cytosol | 19.16/5.72 | 17.50/5.34 | 13 | 122 | |
| 134 | gi|283482292 | class I small heat shock protein | Rhododendron mariesii | Cytosol | 18.51/5.74 | 16.32/5.36 | 27 | 160 | |
| 135 | gi|255585824 | Heat-shock protein | Ricinus communis | Cytosol | 18.17/5.86 | 17.77/5.85 | 14 | 101 | |
| 136 | gi|75279027 | class I heat shock protein | Solanum peruvianum | Cytosol | 17.67/5.73 | 17.56/5.22 | 16 | 160 | |
| 137 | gi|357465797 | Class II small heat shock protein | Medicago truncatula | Cytosol | 17.23/5.66 | 17.72/6.17 | 20 | 134 | |
| 156 | gi|283482308 | cytosolic class I small heat shock protein type 1 | Rhododendron ovatum | Cytosol | 17.57/6.06 | 16.36/5.55 | 26 | 128 | |
| 157 | gi|284433776 | heat-shock protein | Jatropha curcas | Cytosol | 17.75/6.26 | 18.07/6.85 | 28 | 170 | |
| 158 | gi|349591294 | class I small heat shock protein | Solanum lycopersicum | Cytosol | 17.32/6.29 | 17.62/5.82 | 20 | 141 | |
| 159 | gi|123555 | class I heat shock protein | Pisum sativum | Cytosol | 19.23/6.00 | 18.07/5.83 | 8 | 67 | |
| 160 | gi|123555 | lass I heat shock protein | Pisum sativum | Cytosol | 19.74/6.29 | 18.07/5.83 | 14 | 95 | |
| 139 | gi|37704391 | class I small heat shock protein | Nicotiana tabacum | Cytosol | 18.41/5.42 | 15.62/5.39 | 6 | 52 | |
| 140 | gi|255558876 | heat-shock protein | Ricinus communis | Cytosol | 18.05/5.42 | 17.50/5.34 | 17 | 124 | |
| 141 | gi|1350520 | class II cytoplasmic small molecular weight heat shock protein | Picea glauca | Cytosol | 17.36/5.40 | 17.08/5.54 | 15 | 120 | |
| 143 | gi|349591294 | class I small heat shock protein | Solanum lycopersicum | Cytosol | 17.45/5.17 | 17.62/5.82 | 14 | 70 | |
| 168 | gi|284433776 | heat-shock protein | Jatropha curcas | Cytosol | 17.52/7.07 | 18.07/6.85 | 26 | 267 | |
| 169 | gi|255558876 | heat-shock protein | Ricinus communis | Cytosol | 17.02/7.07 | 17.50/5.34 | 27 | 149 | |
| 173 | gi|21592809 | heat shock protein | Arabidopsis thaliana | Cytosol | 17.62/7.24 | 17.62;6.85 | 9 | 89 | |
| 32 | gi|262192812 | Catalase | Citrus maxima | Cytosol | 45.47/7.13 | 38.71/6.00 | 10 | 188 | |
| 33 | gi|262192812 | Catalase | Citrus maxima | Cytosol | 45.59/7.26 | 38.71/6.00 | 12 | 197 | |
| 34 | gi|262192812 | Catalase | Citrus maxima | Cytosol | 45.35/7.38 | 38.71/6.00 | 12 | 251 | |
| 104 | gi|15222163 | glutathione S-transferase DHAR2 | Arabidopsis thaliana | Cytosol | 26.57/6.66 | 23.50/5.79 | 8 | 130 | |
| 106 | gi|297824877 | glutathione S-transferase | Arabidopsis lyrata | chloroplast | 23.53/6.64 | 24.32/6.31 | 3 | 60 | |
| 122 | gi|161778782 | manganese superoxide dismutase | Vitis vinifera | Mitochondria | 22.61/5.93 | 25.35/6.79 | 16 | 141 | |
| 123 | gi|161778782 | manganese superoxide dismutase | Vitis vinifera | Mitochondria | 22.75/5.71 | 25.35/6.79 | 16 | 178 | |
| 80 | gi|87042321 | beta-1,3-glucanase | Mangifera indica | chloroplast | 31.01/4.74 | 19.55/5.77 | 27 | 159 | |
| 81 | gi|87042321 | beta-1,3-glucanase | Mangifera indica | Chloroplast | 31.16/4.59 | 19.55/5.77 | 13 | 64 | |
| 89 | gi|16660407 | abscisic acid-responsive protein | Cucumis melo | Unclear | 28.46/6.01 | 12.89/6.22 | 18 | 97 | |
| 109 | gi|21068664 | quinone oxidoreductase | Cicer arietinum | Nuclear | 23.04/7.24 | 21.70/6.51 | 17 | 185 | |
| 116 | gi|255541754 | hevamine-A precursor | Ricinus communis | Chloroplast | 27.93/9.81 | 26.42/9.21 | 5 | 62 | |
| 167 | gi|118162023 | CBS domain-containing protein | Solenostemon scutellarioides | Cytosol | 18.31/7.22 | 22.22/9.24 | 13 | 124 | |
Table 1: Identification of the cashew stem proteins associated to the functional category of “Disease/defense”.
Overproduction of the reactive oxygen species (ROS) – superoxide anion, hydrogen peroxide, singlet oxygen, and hydroxyl radical – is as common response of plants to environmental adverse factors, including drought, low/high temperature, high light intensities, mechanical stress, and pathogen attacks. Excessive accumulation of ROS due to environmental stresses is a major cause of loss of crop productivity worldwide. In plant systems, whether ROS will act protecting or damaging depends on the equilibrium between antioxidants and ROS [47,48]. ROS are highly reactive and can alter the normal cellular metabolism, oxidize lipids, proteins, and nucleic acids. Under oxidative stress, the ROS scavenging enzymes superoxide dismutase (SOD), ascorbate peroxidase (APX), catalase (CAT), and the enzymes of the ascorbate-glutathione cycle are synthesized de novo toward decreasing the intracellular toxic levels of ROS [49-51]. Furthermore, quinone oxidoreductases scavenge superoxide (O2•) using NADH or NADPH as the electron donor [52] and the cystathionine β-synthase (CBS) domain proteins also help in maintaining the intracellular redox balance [53]. In this context, nine different antioxidant proteins [3 CATs (spots 32, 33, 34) 2 Gluthatione-S-transferases (spots 104, 106), 2 SODs (spots 122, 123), 1 oxidoreductase (spot 109) and 1 CBS domain-containing protein (spot 167)] were detected in cashew stem (Table 1). A close correlation between the antioxidant capacity of these enzymes and the stress tolerance of several crops has been previously demonstrated. For instance, there are similarities of the responses to heat stress and to oxidative stress because both stresses induce the overexpression of antioxidant enzymes and accumulation of HSPs [54,55]. Transgenic plants overexpressing SOD, CAT, APX, glutathione reductase (GR), glutathione transferases (GST), glutathione peroxidase (GPX) and other enzymes are best suited to different stress [27,48]. Additionally, in field occurs cross-tolerance phenomenon by which defense reactions induced by a stress lead to protection against other stress. Mustard seedling primed with heat-shock positively modulates the activities of APX, DHAR (monodehydroascorbate reductase), GR, GST, GPX and glyoxalase I/II as compared to the control [56]. H2O2 may be involved in the regulation of heat-shock- and cadmium-increased APX and GR activities in leaves of rice seedlings [57]. With respect to the cashew, high temperature positively modulates oxidative protection in saltstressed plant by the activation of antioxidant enzymes such as SOD, APX, CAT [58].
Other defense-related proteins expressed and identified in the cashew stems were isoflavone reductases (spots 72, 73, 90) and alcohol dehydrogenases (spots 52, 60), which are involved, respectively, with the synthesis of phytoalexin [59] and the lignin biosynthesis [60]; the pathogenesis-related protein (PR-protein) β-1,3-glucanase (spots 80,81), which hydrolysis β-glucans present in the cell wall of diverse phytopathogens [61]; and the hevamine-A precursor (spot 116) similar to that of Ricinus communis that is a class III chitinase [62]. Chitinases are PR-proteins that hydrolysis chitin and are induced in plants by phytopathogen challenge or elicitors [61].
The responses of plants to salinity, drought, heat or pathogens as well as development of stress tolerance are extremely complex events and several mechanisms appear to be involved, but the exact physiological and biochemical mechanism(s) is poorly understood to most plants. This work demonstrated that ‘“Disease/Defense” related proteins account for 31% of the identified proteins in cashew stem. The high expression of these groups of proteins is consistent with the excellent adaptation of cashew to the semi-arid climate as for the CCP 76 clone that has good productivity in such adverse conditions. Salinity and drought stresses are known to affect seedling germination and the establishment of cashew [63-66]. Both stresses also interfere with the nutrient acquisition in young and adult cashew plants besides to affect photosynthesis and nitrogen metabolism [26,67]. Despite this, studies have shown an efficient antioxidant mechanism in cashew leaves under salinity and high temperature by enzymatic and non-enzymatic systems [58,68]. It was reported that, in response to salt stress, proline accumulated and glutamine synthetase activity increased in cashew leaves [67], and guaiacol peroxidase and ascorbate peroxidase activities increased dramatically in cashew roots [26]. Additionally, high temperature positively modulated activities of CAT, SOD and APX in salt-stressed plants, promoting a favorable change in the ascorbate redox state in order to protect cashew plant [58]. To advance the knowledge of biochemical mechanisms of stress tolerance in cashew, this study sought to determine the overall protein profile of well-adapted cashew plant to field in order to know and understand the contribution of the various proteins in cashew tolerance to semiarid regions.
Conclusion
Our findings suggest that the cashew plants use active mechanisms probably dependant on the relative abundance of antioxidant enzymes and HSPs to withstand the stress conditions imposed by the environmental conditions of the semi-arid regions where they are planted. Nevertheless, this pioneering proteomic investigation of a cashew clone (CCP 76) tolerant to semi-arid environment, but susceptible to the fungus L. theobromae, which cause gummosis, currently the most important disease affecting the cashew culture in the semi-arid conditions of Northeastern Brazil [2,69], can be taken as an useful starting point for investigation of cashew physiology and genetic reprogramming induced by both biotic and abiotic stresses. This approach could provide relevant data that will contribute to the understanding of both the climate adaptability and resistance/ susceptibility of cashew clones to the devastating fungus L. theobromae and other fungi that cause diseases in cashew such as Colletotrichum gloeosporioides, Pilgeriella anacardii, Septoria anacardii, Oidium anacardii, Pythium splendens, Phytophthora heveae and P. nicotiana, Cylindrocladium scoparium and Sclerotium rolfsii [2].
Appendices
Table S1 Identification of the cashew stem protein spots selected from 2D gels by ESI-Q-TOF-MS/MS analysis.
Acknowledgements
This work was supported by the Council for Advanced Professional Training (CAPES), The National Council for Scientific and Technological Development (CNPq), and Research Council of the State of Ceará (FUNCAP). Darcy M. F. Gondim gratefully acknowledges a doctoral scholarship from CAPES and a postdoctoral fellowship from CNPq.
References
- Castro ACR, Bordallo PN, Cavacanti JJV, Barros LM (2011) Brazilian cashew germplasm bank. Acta Hortic 918: 857-861.
- Freire FCO, Cardoso JE, Santos AA, Viana FM (2002) Diseases of cashew nut plants (Anacardium occidentale L.) in Brazil. Crop Prot 21: 489-494.
- Bezerra MA, Lacerda CF, Gomes Filho E, Abreu CB, Prisco JT (2007) Physiology of cashew plants grown under adverse conditions. Braz J Plant Physiol 19: 449-461.
- Oliveira VH (2008) Cashew crop. Rev Bras Frutic 30: 1-3.
- Cardoso JE, Paiva JR, Cavalcanti JJV, Santos AA, Vidal JC (2006) Evaluation of resistance in dwarf cashew to gummosis in north-eastern Brazil. Crop Prot 25: 855-859.
- Cysne AQ, Cardoso JE, Maia AHN, Farias FC (2010) Spatial-temporal analysis of gummosis in three cashew clones at northeastern Brazil. J Phytopathol 158: 676-682.
- Muniz CR, Freire FC, Viana FM, Cardoso JE, Cooke P, et al. (2011) Colonization of cashew plants by Lasiodiplodia theobromae: microscopical features. Micron 42: 419-428.
- Anderson JP, Gleason CA, Foley RC, Thrall PH, Burdon JB, et al. (2010) Plants versus pathogens: an evolutionary arms race. Funct Plant Biol 37: 499-512.
- Thurston G, Regan S, Rampitsch C, Xing T (2005) Proteomic and phosphoproteomic approaches to understand plant–pathogen interactions. Physiol Mol Plant Pathol 66: 3-11.
- Mehta A, Brasileiro AC, Souza DS, Romano E, Campos MA, et al. (2008) Plant-pathogen interactions: what is proteomics telling us? FEBS J 275: 3731-3746.
- Afroz A, Ali GM, Mir A, Komatsu S (2011) Application of proteomics to investigate stress-induced proteins for improvement in crop protection. Plant Cell Rep 30: 745-763.
- Agrawal AA, Konno K (2009) Latex: A model for understanding mechanisms, ecology, and evolution of plant defense against herbivory. Annu Rev Ecol Evol Syst 40: 311-331.
- Inbar M, Mayer RT, Doostdar H (2003) Induced activity of pathogenesis related (PR) proteins in aphid galls. Symbiosis 34: 293-300.
- Inglis PW, Ciampi AY, Salomão AN, Costa TSA, Azevedo VCR (2014) Expression of stress-related genes in zebrawood (Astronium fraxinifolium, Anacardiaceae) seedlings following germination in microgravity. Genet Mol Biol 37: 81-92.
- Konno K (2011) Plant latex and other exudates as plant defense systems: roles of various defense chemicals and proteins contained therein. Phytochemistry 72: 1510-1530.
- Chen S, Harmon AC (2006) Advances in plant proteomics. Proteomics 6: 5504-5516.
- Oeljeklaus S, Meyer HE, Warscheid B (2009) Advancements in plant proteomics using quantitative mass spectrometry. J Proteomics 72: 545-554.
- Wang W, Vignani R, Scali M, Cresti M (2006) A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 27: 2782-2786.
- Quirino BF, Candido ES, Campos PF, Franco OL, Krüger RH (2010) Proteomic approaches to study plant-pathogen interactions. Phytochemistry 71: 351-362.
- Gómez-Vidal S, Tena M, Lopez-Llorca LV, Salinas J (2008) Protein extraction from Phoenix dactylifera L. leaves, a recalcitrant material, for two-dimensional electrophoresis. Electrophoresis 29: 448-456.
- Sheoran IS, Ross ARS, Olson DJH, Sawhney VK (2009) Compatibility of plant protein extraction methods with mass spectrometry for proteome analysis. Plant Sci 176: 99-104.
- Hurkman WJ, Tanaka CK (1986) Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol 81: 802-806.
- Gallardo K, Job C, Groot SP, Puype M, Demol H, et al. (2001) Proteomic analysis of arabidopsis seed germination and priming. Plant Physiol 126: 835-848.
- Noir S, Bräutigam A, Colby T, Schmidt J, Panstruga R (2005) A reference map of the Arabidopsis thaliana mature pollen proteome. Biochem Biophys Res Commun 337: 1257-1266.
- Vasconcelos EAR, Nogueira FCS, Abreu EFM, Gonçalves EF, Souza PAS, et al. (2005) Protein extraction from cowpea tissues for 2-D gel electrophoresis and MS analysis. Chromatographia 62: 447-450.
- Abreu CEB, Prisco JT, Nogueira ARC, Bezerra MA, Lacerda CF, et al. (2008) Physiological and biochemical changes occurring in dwarf-cashew seedlings subjected to salt stress. Braz J Plant Physiol 20: 105-118.
- Wang W, Scali M, Vignani R, Spadafora A, Sensi E, et al. (2003) Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis 24: 2369-2375.
- Yao Y, Yang YW, Liu JY (2006) An efficient protein preparation for proteomic analysis of developing cotton fibers by 2-DE. Electrophoresis 27: 4559-4569.
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254.
- Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, et al. (2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25: 1327-1333.
- Bevan M, Bancroft I, Bent E, Love K, Goodman H, et al. (1998) Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391: 485-488.
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, et al. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585-587.
- Görg A, Weiss W, Dunn MJ (2004) Current two-dimensional electrophoresis technology for proteomics. Proteomics 4: 3665-3685.
- Menezes JB, Alves RE (1995) Physiology and technology postharvest of Cashew apple. Fortaleza: EMBRAPA CNPAT 20.
- Saravanan RS, Rose JK (2004) A critical evaluation of sample extraction techniques for enhanced proteomic analysis of recalcitrant plant tissues. Proteomics 4: 2522-2532.
- Carpentier SC, Witters E, Laukens K, Deckers P, Swennen R, et al. (2005) Preparation of protein extracts from recalcitrant plant tissues: an evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics 5: 2497-2507.
- Sarma AD, Oehrle NW, Emerich DW (2008) Plant protein isolation and stabilization for enhanced resolution of two-dimensional polyacrylamide gel electrophoresis. Anal Biochem 379: 192-195.
- Shaw MM, Riederer BM (2003) Sample preparation for two-dimensional gel electrophoresis. Proteomics 3: 1408-1417.
- Waters ER (2013) The evolution, function, structure, and expression of the plant sHSPs. J Exp Bot 64: 391-403.
- Swindell WR, Huebner M, Weber AP (2007) Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8: 125.
- Zhang JH, Wang LJ, Pan QH, Wang YZ, Zhan JC, et al. (2008) Accumulation and subcellular localization of heat shock proteins in young grape leaves during cross-adaptation to temperature stresses. Sci Hort 117: 231-240.
- Han F, Chen H, Li XJ, Yang MF, Liu GS, et al. (2009) A comparative proteomic analysis of rice seedlings under various high-temperature stresses. Biochim Biophys Acta 1794: 1625-1634.
- Hamilton EW, Heckathorn SA, Downs CA, Schwarz TE, Coleman JS, et al. (1996) Heat shock proteins are produced by field-grown naturally occurring plants in the summer in the temperate northeast. US Bull Ecol Soc Am 77: 180.
- Sun W, Van Montagu M, Verbruggen N (2002) Small heat shock proteins and stress tolerance in plants. Biochim Biophys Acta 1577: 1-9.
- Al-Whaibi MH (2011) Plant heat-shock proteins: A mini review. J King Saud Univ Sci 23: 139-150.
- Waters ER, Lee GJ, Vierling E (1996) Evolution, structure and function of the small heat shock. J Exp Bot 47: 325-338.
- Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405-410.
- Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48: 909-930.
- Gozzo F (2003) Systemic acquired resistance in crop protection: from nature to a chemical approach. J Agric Food Chem 51: 4487-4503.
- Møller IM, Jensen PE, Hansson A (2007) Oxidative modifications to cellular components in plants. Annu Rev Plant Biol 58: 459-481.
- Saleh L, Plieth C (2009) Fingerprinting antioxidative activities in plants. Plant Methods 5: 2.
- Siegel D, Gustafson DL, Dehn DL, Han JY, Boonchoong P, et al. (2004) NAD(P)H:quinone oxidoreductase 1: role as a superoxide scavenger. Mol Pharmacol 65: 1238-1247.
- Kushwaha HR, Singh AK, Sopory SK, Singla-Pareek SL, Pareek A (2009) Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genomics 10: 200-221.
- Lee HS, Kim KY, You SH, Kwon SY, Kwak SS (1999) Molecular characterization and expression of a cDNA encoding copper/zinc superoxide dismutase from cultured cells of cassava (Manihot esculenta Crantz). Mol Gen Genet 262: 807-814.
- Lee BH, Won SH, Lee HS, Miyao M, Chung WI, et al. (2000) Expression of the chloroplast-localized small heat shock protein by oxidative stress in rice. Gene 245: 283-290.
- Hossain MA, Mostofa MG, Fujita M (2013) Heat-shock positively modulates oxidative protection of salt and drought-stressed mustard (Brassica campestris L.) seedlings. J Plant Sci Mol Breed 2: 1-13.
- Chou TS, Chao YY, Kao CH (2012) Involvement of hydrogen peroxide in heat shock- and cadmium-induced expression of ascorbate peroxidase and glutathione reductase in leaves of rice seedlings. J Plant Physiol 169: 478-486.
- Ferreira-Silva SL, Voigt EL, Silva EN, Maia JM, Fontenele AV, et al. (2011) High temperature positvely modulates oxidatve protecton in salt-stresses cashew plants. Environ Exp Bot 74: 162-170.
- Paiva NL, Edwards R, Sun Y, Hrazdina G, Dixon RA (1991) Stress responses in alfalfa (Medicago sativa L.) 11. Molecular cloning and expression of alfalfa isoflavone reductase, a key enzyme of isoflavonoid phytoalexin biosynthesis. Plant Mol Biol 17: 653-667.
- Lapierre C, Pollet B, MacKay JJ, Sederoff RR (2000) Lignin structure in a mutant pine deficient in cinnamyl alcohol dehydrogenase. J Agric Food Chem 48: 2326-2331.
- van Loon LC, Rep M, Pieterse CM (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44: 135-162.
- Bokma E, Spiering M, Chow KS, Mulder PPMFA, Subroto T, et al. (2001) Determination of cDNA and genomic DNA sequences of hevamine, a chitinase from the rubber tree Hevea brasiliensis. Plant Physiol Bioch 39: 367-376.
- Bezerra IL, Gheyi HR, Fernandes PD, Santos FJS, Gurgel MT, et al. (2002) Germination, formation of rootstocks and grafting of precocious dwarf cashew under salinity stress. Rev Bras Eng Agric Amb 6: 420-424.
- Carneiro PT, Fernandes PD, Gheyi HR, Soares FAL (2002) Germination and initial growth of precocious dwarf cashew genotypes under saline conditions. Rev Bras Eng Agric Amb 6: 199-206.
- Ferreira-Silva SL, Silveira JAG, Voigt EL, Soares LSP, Viégas RA (2008) Changes in physiological indicators associated with salt tolerance in two contrasting cashew rootstocks. Braz J Plant Physiol 20: 51-59.
- Voigt EL, Almeida TD, Chagas RM, Ponte LF, Viégas RA, et al. (2009) Source-sink regulation of cotyledonary reserve mobilization during cashew (Anacardium occidentale) seedling establishment under NaCl salinity. J Plant Physiol 166: 80-89.
- Silveira JA, Viégas Rde A, da Rocha IM, Moreira AC, Moreira Rde A, et al. (2003) Proline accumulation and glutamine synthetase activity are increased by salt-induced proteolysis in cashew leaves. J Plant Physiol 160: 115-123.
- Ferreira-Silva SL, Voigt EL, Silva EN, Maia JM, Aragão TCR, et al. (2012) Partial oxidative protection by enzymatic and non-enzymatic components in cashew leaves under high salinity. Biol Plantarum 56: 172-176.
- Cardoso JE, Wilkinson MJ (2008) Development and characterization of microsatellite markers for the fungus Lasiodiplodia theobromae. Summa Phytopathol 34: 55-57.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 15718
- [From(publication date):
specialissue-2014 - Apr 05, 2025] - Breakdown by view type
- HTML page views : 11049
- PDF downloads : 4669