Research Article Open Access
Molecular Cloning and In Silico Sequence Analysis of Glycine Betaine Biosynthesis Genes in Bacillus subtilis
Lawrance Anbu Rajan*, Krishnakumar Vinodhini, Yeragudipati Rajalakshmi and Vetrivel UmashankarSchool of Biosciences, Department of Bioinformatics, SRM University, Ramapuram, Chennai, Tamil Nadu, India
- Corresponding Author:
- L. Anbu Rajan
School of Biosciences, Department of Bioinformatics
SRM University, Ramapuram, Chennai, Tamil Nadu, India
E-mail:anburajanl@yahoo.co.in
Received date: October 15, 2010; Accepted date: February 24, 2011; Published date: March 21, 2011
Citation: Rajan LA, Vinodhini K, Rajalakshmi Y, Umashankar V (2011) Molecular Cloning and In Silico Sequence Analysis of Glycine Betaine Biosynthesis Genes in Bacillus subtilis. J Biotechnol Biomaterial 1:103. doi:10.4172/2155-952X.1000103
Copyright: © 2011 Rajan LA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
Glycine betaine (N, N, N-trimethylglycine) is a effective compatible solute, which maintains fluidity of membranes and protects the biological structure of the organisms under stress. In this study, betaine aldehyde dehydrogenase (GbsA) and betaine alcohol dehydrogenase (GbsB) genes encoding for glycine betaine biosynthesis were PCR amplified from genomic DNA of Bacillus subtilis isolated from salted anchovies (Thrissina thryssa) collected from retail fish market of Cochin, Kerala, India. The amplified genes were cloned and nucleotide sequences were determined. The sequencing results showed that GbsA and GbsB genes contain ORF of 1473 bp and 1182 bp long, encoding 474 and 266 amino acids respectively (GenBank accession nos. FJ823257 and FJ823258). In silico sequence analysis revealed that the GbsA and GbsB sequences of B. subtilis were conserved in many eubacteria.
Keywords
Betain aldehyde dehydrogenase; Betaine alcohol dehydrogenase; Compatible solutes; Salt stress
Introduction
Soil microorganisms are subjected to frequent fluctuations in the osmotic conditions of their habitat due to drying and wetting of the soil. Bacteria must have active mechanisms to compete successfully for their ecological niche [1,2]. The more effective defense against these conditions is the accumulation of osmoprotectants, which can be amassed to high intracellular levels without disturbing essential functions of the cell [3]. One of the most important osmoprotectants is glycine betaine [4]. Synthesis of glycine betaine from choline is a two-step oxidation process with glycine betaine aldehyde as the intermediate [5]. Characterization of glycine betaine has been most intensively studied at both the molecular and biochemical levels for E. coli [6]. In E. coli, choline dehydrogenase (betA) oxidizes choline to glycine betaine aldehyde and betaine aldehyde dehydrogenase (betB), converts glycine betaine aldehyde to the osmoprotectant glycine betaine. The choline-glycine betaine synthesis pathway is an important facet in Bacillus subtilis to high osmolarity stress [5]. However, the genetic and biochemical details governing choline uptake and glycine betaine synthesis have remained largely unknown. In this study, for the first instant we report the molecular characterization and in silico sequence analysis of GbsA and GbsB genes in B. subtilis isolated from salted fish. Moreover, the sequence analysis of glycine betaine aldehyde dehydrogenase and glycine betaine alcohol dehydrogenase from our isolate shows several base substitutions with that of reported sequences in GenBank, resulting in the altered amino acid sequences of the translated protein structures. implant [13,14]. Since few reports, to our knowledge, analyze the effects of Trabecular TitaniumT on stem cells [15] and none focuses on the genetic effects, the expression of genes related to the osteoblast differentiation was analyzed using cultures of bone marrow derived human mesenchymal stem cells (BM-hMSCs) treated with Trabecular Titanium T for seven days, in order to detect the early effects of the common part of BMPs on stem cells.
Materials and Methods
Bacterial Strain, growth conditions and DNA isolation
B. subtilis was isolated from salted anchovies procured from local fish markets in Cochin, Kerala, India. Microbial identification and biochemical characterization of B. subtilis was carried out as per [U.S. Food and Drug Administration (USFDA)] methods. B. subtilis was grown aerobically in nutrient broth medium containing 0.5% peptone, 0.1% beef extract, 0.1% yeast extract, 0.5% NaCl and incubated at 37°C. Genomic DNA of B. subtilis was prepared as described by [10].
Polymerase chain reaction
The glycine betaine biosynthesis genes, GbsA and GbsB were amplified by using gene specific primers. The PCR reaction was performed with the final volume of 50 µL that contained; 0.5 µM each of forward and reverse primers, 1.0µL of crude genomic DNA, 200µM of dNTPs, 1X Taq buffer, 2.5 mM MgCl2, 1U Taq DNA polymerase (MBI Fermentas, Hanover, Maryland, USA) and autoclaved Millipore water. The PCR was performed using a Master cycler (Eppendorf, Germany) with the following conditions; Initial denaturation at 94°C for 3 min, followed by 30 repeated cycles of 94°C for 30 sec, 50°C for 1 min and 72°C for 2 min and final extension at 72°C for 5 min. The PCR amplified product was analyzed on 1.5% agarose gel along with DNA ladder (MBI Fermentas) and documented using a gel documentation system (Alpha Imager 1220, Alpha Innotech Corporation, San Leandro, CA, USA).
Cloning of PCR product
The amplified DNA fragments of GbsA and GbsB were purified by the use of Perfectprep Gel Cleanup Kit (Eppendorf) and cloned into the cloning vector, pTZ57R/T (MBI Fermentas). The cloned inserts were transformed into E. coli JM109 and plated on LB agar containing ampicillin (100 µg mL-1), IPTG (50 µM) and X-gal (80 µg mL-1). The plates were incubated at 37°C and the transformants were selected and inoculated in 5 mL LB broth containing ampicillin. The recombinant plasmids were isolated from theovernight culture by alkaline lysis method (Sambrook and Russell, 2001) [11].
Characterization of the Recombinant Plasmid
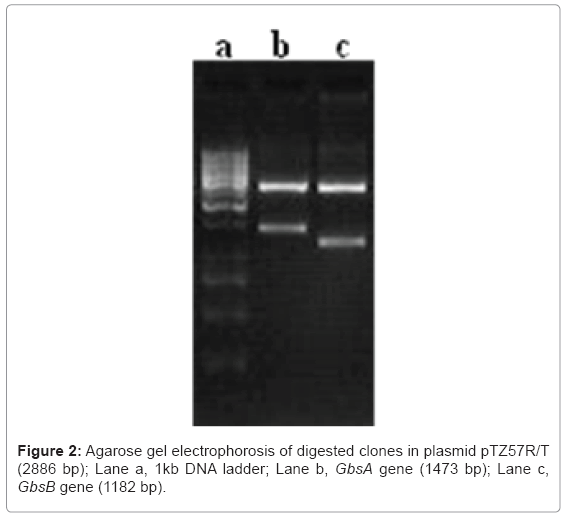
The recombinant plasmid was double digested with restriction enzymes, BamI & XbaI for GbsA and XbaI & HindIII for GbsB. The reaction mixture contained recombinant plasmid 2µL, Enzyme buffer (10X) 2µL, each restriction enzyme (10UµL-1) 0.5µL and volume up to 20mL with autoclaved Millipore water. The reaction mixture was incubated overnight at 37°C in a water bath. The digested products were analyzed on 1.5% agarose gel. The clone with the correct insert as judged by size was sequenced on an ABI PRISM 377 genetic analyzer (Applied Biosystems, Perkin Elmer Co., Foster City, CA, USA).
In silico Sequence analysis
The nucleotide sequences obtained were compared to the available database sequences by BLAST analysis using the NCBI (http://www. ncbi.nlm.nih.gov) database. The sequences were aligned and clustered using CLUSTAL-X version 1.81 [7]. The output alignments were imported into the GeneDoc program (http://www.psc.edu/biomed/genedoc/) and BioEdit version 7.05 program (www.mbio.ncsu.edu/BioEdit/) to calculate the percent identities among the nucleotide and amino acid sequences. The molecular masses and the theoretical pI values of the polypeptides were predicted using the ProtParam tool (http:// www.expasy.org/tools/protparam.html).
Results
The GbsA and GbsB genes encode the betaine aldehyde dehydrogenase and betaine alcohol dehydrogenase respectively. Together these proteins constitute the glycine betaine biosynthetic pathway. The GbsA and GbsB genes were PCR amplified and is encoded by polynucleotides of 1473 bp and 1182 bp respectively (Figure 1). The GbsA and GbsB encodes proteins of 490 and 393 amino acids with the calculated molecular masses of 53665 and 42386Da., based on the in silico estimates. After PCR amplification, the products were purified from the agarose gel and cloned into pTZ57R/T cloning vector. The recombinant transformants with GbsA and GbsB genes were also confirmed by double digestion with restriction enzymes (Figure 2). The nucleotide sequence of inserts GbsA and GbsB genes were submitted to GenBank and have been given accession nos. FJ823257 and FJ823258.
The search for homologous genes and deduced amino acid sequence were performed using BLAST. The nucleotide sequence of GbsA and GbsB genes matches significantly with the glycine betaine biosynthesis genes from other organisms. The GbsA and GbsB sequences from the B. subtilis isolate were compared with the reported nucleotide and amino acid sequences of other eubacteria viz. Bacillus pumilus (GenBank accession no. CP000813), Bacillus licheniformis (AE017333), Bacillu licheniformis (CP0000023), Bacillus amyloliquefaciens (CP0005601), using Clustal W software. In silico nucleotide sequences analysis of GbsA and GbsB genes revealed a high degree of similarity with other eubacteria. The amino acids analysis revealed that the GbsA gene encoded a protein belongs to the betaine aldehyde dehydrogenase family. The protein showed partial homology with betaine aldehyde dehydrogenase family from other bacteria as follows: B. amyloliquefaciens, 80% identity; B. licheniformis, 76% identity and B. pumilus with 74% identity. In B. amyloliquefaciens various amino acid substitutions were observed in N-terminal, middle and C-terminal regions, in total 53 amino acid substitutions were observed. In B. licheniformis, 64 amino acid substitutions were observed. In B. pumilus, 105 amino acid substitutions were found. The amino acid analysis revealed that GbsB gene encoded a protein belongs to the type III alcohol dehydrogenases family. The protein showed partial homology with betaine aldehyde dehydrogenase family from other bacteria as follows: Bacillus amyloliquefaciens, 80% identity; Bacillus licheniformis with 79% identity and Bacillus pumilus with 78% identity. The sequence analysis of GbsB also revealed various amino acid substitutions in C-terminal than in the N-terminal and middle region of other eubacteria. In B.amyloliquefaciens, various amino acid substitutions were observed in C-terminal regions as predicted in GbsB.
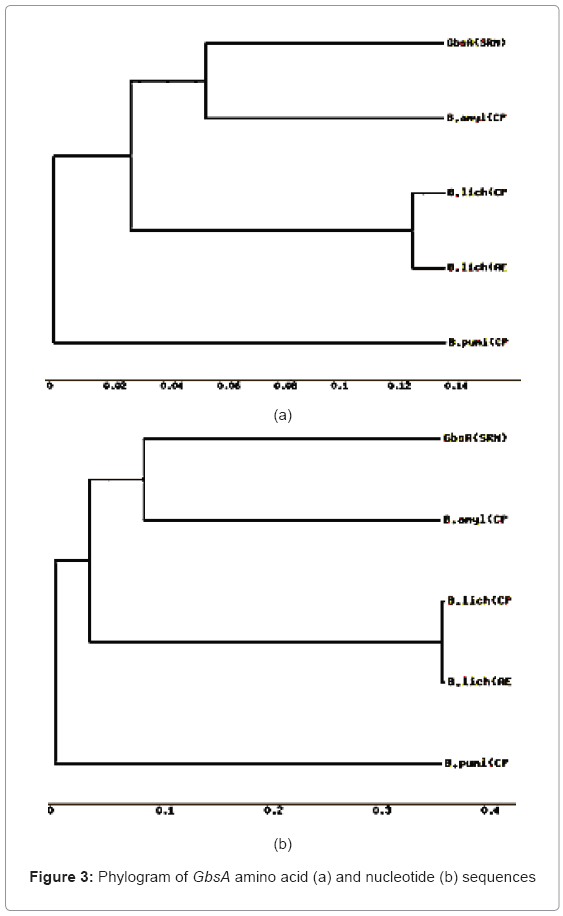
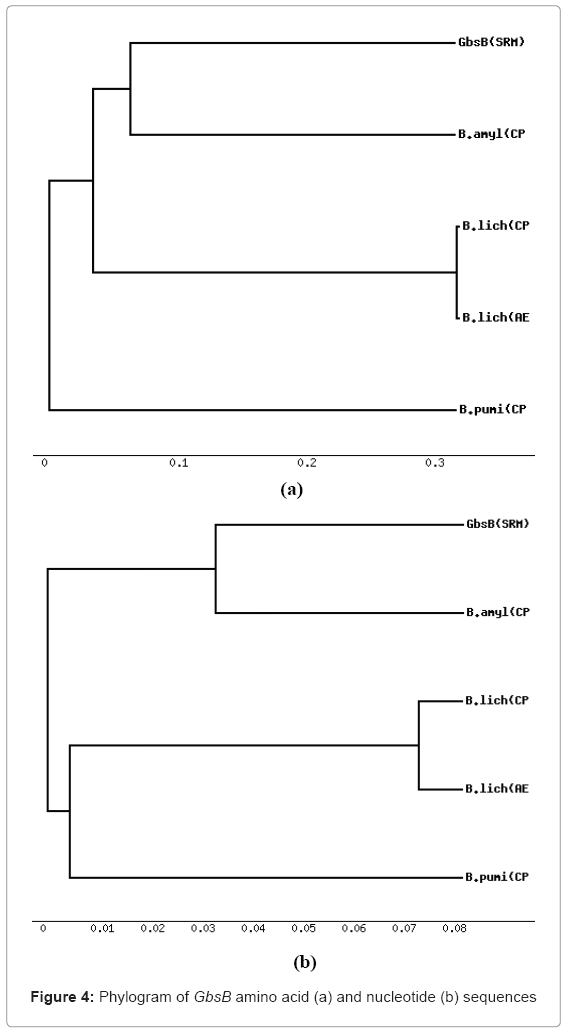
Phylogenetic tree based on evolutionary distances was constructed using nucleotide and amino acid sequences of GbsA and GbsB using MEGA software (Molecular Evolutionary Genetics Analysis, version 3.1) [13] using the neighbour-joining method. The tree at nucleotide and amino acid sequence of GbsA (Figure 3) and GbsB (Figure 4) reveals that B. subtilis and B. amyloliquefaciens forms a single cluster with that of other eubacteria. Many bacterial species switched to different clusters for GbsA and GbsB genes at nucleotide and amino acid level indicating the divergence among the organisms and the degree of divergence in the sequences.
Discussion
Glycine betaine (N, N, N-trimethylglycine) is a very efficient osmolyte [3] found in a wide range of bacteria and plants [12], where it is accumulated at high cytoplasmic concentrations in response to osmotic stress. In addition to its osmoprotectant activity, glycine betaine is also an effective protectant against mutagenic compounds and radiation-induced damage. Glycine betaine can either be taken up directly from the environment, or can be synthesized. A common biosynthetic pathway for glycine betaine is from choline, utilizing a two - step pathway with betaine aldehyde as intermediate.
Based on the sequence analysis, it was previously reported that the GbsA gene of B. subtilis codes for betaine aldehyde dehydrogenase and GbsB gene of B. subtilis codes for alcohol dehydrogenase [8,9]. To date, only least information on the characterization of GbsA and GbsB genes have been reported [9]. As a first step towards the molecular characterization of betaine aldehyde dehydrogenase and alcohol dehydrogenase, in this study we cloned and analyzed the GbsA and GbsB genes from B. subtilis cells isolated from salted fishes. Evaluation of the deduced amino acid sequence of GbsA and GbsB genes with reported sequences in the database revealed a maximum similarity. However, the sequence analysis of GbsA and GbsB of our isolate showed several base substitutions with that of reported sequences, resulting in the altered amino acid sequences of the translated proteins.
In conclusion, to our knowledge, this study represents the first instance in which GbsA and GbsB genes from B. subtilis isolated from salted fish has been cloned and characterized in detail. Moreover, the determination of protein structure modification due to the nucleotide substitutions will certainly provide the basis for performing site-directed mutagenesis to improve the production and configuration of the osmolytes of biotechnological interest.
Acknowledgements
Authors are grateful to the Pro-Vice Chancellor, SRM University, Ramapuram, Chennai for providing the necessary facilities to carry out this research work. The research concept was developed by L.A.R and research experiments were performed by K.V. and Y.R; V.U. organized for and provided the necessary facilities.
References
- Csonka LN, Hanson AD (1991) Prokaryotic osmoregulation: genetics and physiology. Annu Rev Microbiol 45: 569-606.
- Lucht JM, Bremer JN (1994) Adaptation of Escherichia coli to high osmolarity environments: osmoregulation of the high-affinity glycine betaine transport system ProU. FEMS Microbiol Rev 14: 3-20.
- BochJ, Kempf B, Bremer E (1994) Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J Bacteriol 176: 5364-5371.
- Kempf B, Bremer E (1995) OpuA: an osmotically regulated binding proteindependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. J Biol Chem 270: 16701-16713.
- Lamark T, Kaasen I, Eshoo MW, Falkenberg P, McDougall J, et al. (1991) DNA sequence and analysis of the bet genes encoding the osmoregulatory cholineglycine betaine pathway of Escherichia coli. Mol Microbiol 5: 1049-1064.
- Lamark T, Rokenes TP, McDougall J, Strom AR (1996) The complex bet promoters of Escherichia coli: regulation by oxygen (ArcA), choline (BetI), and osmotic stress. J Bacteriol 178: 1655-1662.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876-4882.
- Boch J, Kempf B, Schmid R, Bremer E (1996) Synthesis of the osmoprotectant glycine betaine in Bacillus subtilis: Characterization of the gbsAB Genes. J Bacteriol 178: 5121-5129.
- Caldas T, Demont Caulet N, Ghazi A, Richarme G (1999) Thermoprotection by glycine betaine and choline. Microbiol 145: 2543-2548.
- Ausubel FM, Brent R, Kingston RE, Moore DD, Scidman JG, et al. (1994) Current protocols in molecular biology Johnwiley and Sons NewYork: 2.0.1-2.14.8
- Sambrook J, Russell DW (2001) Molecular cloning: A laboratory manual, 3rd ed. New York: Cold Spring Harbor Laboratory Press.
- Jagendorf AT, Takabe T (2001) Inducers of glycinebetaine synthesis in barley. Plant Physiol 127: 1827-1835.
- Kumar S, Tamura K, Nei M (2004) MEGA3:integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5:150-163.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 14937
- [From(publication date):
March-2011 - Nov 21, 2025] - Breakdown by view type
- HTML page views : 10164
- PDF downloads : 4773