Research Article Open Access
Microbial Degradation of Reactive Red by Pseudomonas spp. MPS-2
| Maulin P Shah1*, Kavita A Patel1, Sunu S Nair1, Darji AM1 and Shaktisinh Maharaul2 | |
| 1Industrial Waste Water Research Laboratory, Applied & Environmental Microbiology Lab, Enviro Technology Limited (CETP), Plot No: 2413/2414, GIDC, Ankleshwar-393 002, Gujarat, India | |
| 2Laboratory of Environmental Bioremediation, Narmada Clean Tech Limited (FETP), Nr. Gujarat Gas, Surti Bhagor, Umarvada Road, Ankleshwar-393001, India | |
| Corresponding Author : | Maulin P Shah Industrial Waste Water Research Laboratory Applied & Environmental Microbiology Lab Enviro Technology Limited (CETP), Plot No: 2413/2414 GIDC, Ankleshwar- 393 002, Gujarat, India Tel: +91-90 999 65504 Fax: +91-2646-250707 E-mail: shahmp@uniphos.com |
| Received June 19, 2013; Accepted August 02, 2013; Published August 04, 2013 | |
| Citation: Shah MP, Patel KA, Nair SS, Darji AM, Maharaul S (2013) Microbial Degradation of Reactive Red by Pseudomonas spp. MPS-2. J Bioremed Biodeg 4:197. doi:10.4172/2155-6199.1000197 | |
| Copyright: © 2013 Shah MP, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Azo dyes are a widespread class of poorly biodegradable industrial pollutants. In anaerobic environments, azo bonds are reductively cleaved yielding carcinogenic aromatic amines, many of which are assumed to resist further metabolism by anaerobes bacteria. The latter compounds generally require aerobic conditions for their degradation. A reactive group of azo dye called C.I: Reactive Red was found to be degraded using pseudomonas spp. MPS- 79 to α-ketoglutaric acid with transient accumulation of 4-aminobenzenesulphonic acid (sulphanilic acid), 4-amino, 3-hydronapthalenesulphonic acid and 4-amino, 5-hydronapthalene 2,7 disulphonic acid as a degradation intermediate in anaerobic facultative batch culture. Colour and Total Organic Carbon (TOC) was successfully removed more than 95% and up to 50% respectively. There is no significant correlation between pH and oxygen depletion since there is slightly change in pH was observed (pH from 7.21 to 7.25) though the anaerobiosis was found developed throughout the experiment (redox potential from 0.7 to 1.6 mV). The anaerobic metabolism of glucose as co-metabolite also shown to provide the electrons required for the initial reductive cleavage of the azo group. This finding suggest that it is possible to mineralize the azo dye in the environment; thereby, avoiding accumulation of toxic intermediates in the water.
| Keywords |
| Reactive red; Pseudomonas; Anaerobiosis; Azo dye |
| Introduction |
| Textile industry is one of the oldest industries in India with over 1000 industries. Taking into account the volume and composition of effluent, the textile wastewater is rated as the most polluting among all in the industrial sectors. In general, the wastewater from a typical textile industry is characterized by high values of BOD, COD, color and pH. It is a complex and highly variable mixture of many polluting substances ranging from inorganic compounds and elements to polymers and organic products [1]. Reactive dyes, including many structurally different dyes, are extensively used in the textile industry because of their wide variety of color shades, high wet fastness profiles, ease of application, brilliant colors, and minimal energy consumption. The three most common groups are azo, anthroquinone and phthalocyanine [2]. The textile finishing generates a large amount of waste water containing dyes and represents one of the largest causes of water pollution [3]. Azo dyes have been used increasingly in industries because of their ease and cost effectiveness in synthesis compared to natural dyes. However, most azo dyes are toxic, carcinogenic and mutagenic [4]. Environmental pollution has been recognized as one of the major hazard of the modern world. Due to rapid industrialization, lot of chemicals including dyes manufactured and used in day to day life [5]. Dyes usually have a synthetic origin and complex aromatic molecular structures which make them more stable and more difficult to biodegrade [6]. Approximately 10,000 different dyes and pigments are used industrially and over 0.7 million tons of synthetic dyes are produced annually, worldwide. The three most common groups of dyes are azo, anthraquinone and phthalocyanine [7], most of which are toxic and carcinogenic. Disposal of these dyes into the environment causes serious damage, since they may significantly affect the photosynthetic activity of hydrophytes by reducing light penetration and also toxic to aquatic organisms due to their break down products [8,9]. One of the most pressing environmental problems related to dye effluents is the improper disposal of waste water from dyeing industry [10]. Traditional methods for the cleanup of azo dyes in the textile waste water usually involve the removal of unwanted materials through sedimentation, filtration and subsequent chemical treatments such as flocculation, neutralization and electro-dialysis before disposal. These processes may not guarantee the treatment of toxic dye in the effluent. Moreover, considering the volume of wastes released during the industrial production process these are often laborious and expensive [11]. Over the past decades, biological decolorization has been investigated as a method to transform, degrade or mineralize azo dyes [12]. A number of microorganisms have been found to be able to decolorize textile dyes including bacteria, fungi, and yeasts [13]. They have developed enzyme systems for the decolorization and mineralization of azo dyes under certain environmental conditions [14]. Several methods are used in the treatment of textile effluents to achieve decolorization but they have many disadvantages and limitations [15]. Biological processes provide alternative technologies that are more cost-effective and environmentally friendly [16]. Many microorganisms belonging to the different taxonomic groups of bacteria have been reported for their ability to decolorize azo dyes [17]. Rapid industrialization has necessitated the manufacture and use of different chemicals in day to day life [18]. Reactive dyes, including many structurally different dyes, are extensively used in the textile industry because of their wide variety of color shades, high wet fastness profiles, ease of application, brilliant colors, and minimal energy consumption. The three most common groups are azo, anthroquinone and phthalocyanine [3]. Results presented in this study suggesting that the biodegradation of Reactive Red 195 under facultative anaerobic conditions was degraded into aromatic amines which can be further degraded into safe end byproducts. |
| Materials and Methods |
| Isolation of dye-degrading bacteria |
| Four bacterial strains (MPS-1, MPS2, MPS-3, MPS-4) isolated in this study were previously isolated from textile wastewaters. Filter Sterilized Textile wastewater (FSTW) was prepared by diluting the filter sterilized raw textile wastewater collected from a textile industry in Ankleshwar, Gujarat, India in distilled water at 1:1 ratio and supplemented with glycerol (1 g/L). Cultures were incubated overnight at 37°C prior to isolation of pure strains by single colony isolation onto the same agar medium. Direct isolation was conducted via serial dilution (100-10- 6) followed by spread plate onto Filter Sterilized Textile Wastewater (FSTW) or Autoclaved Textile Wastewater (AWW) agar medium. Potential bacteria that can grow and produce clear zone on the agar medium was selected for further screening. |
| Screening of bacteria for color removal |
| Pure bacteria were inoculated (10% v/v) into Chemically Synthetic MgSO4.7H2O (0.1), CaCl2 (0.02), (NH4)2SO4, (1), Glycerol (1), and Reactive Red dye (0.1), adjusted to pH 7 prior to sterilization. Bacteria were grown under facultative anaerobic conditions (1:1 ratio of medium to the total volume of container) for 24 h at 37°C, prior to monitor the decolorization activity. Other parameters were also screened for their removal such as sulfate, phosphate, nitrate and COD. The culture (10 mL) was taken out gradually every 24 hours and reagent was mixed and read by using Shimadzu UV-1800 Spectrophotometer. The decrease of these parameters can increase the quality of the wastewater treated. |
| Identification of bacteria using 16S rRNA |
| The total genomic DNA of MPS-2 was isolated using Promega WIZARD® Genomic DNA Purification kit. The forward primers, FD1 (5′- AGAGTT TGATCC TGGCTCAG -3′) and the reverse primer, RD1 (5′- AAGGAGGTGATCCAGCC -3′) were used to amplify the 16S rRNA sequence of MPS-2. PCR conditions included an initial denaturation for 5 min at 95°C followed by 30 cycles of 1 min at 95°C, 1min at 50°C and 2 min at 72°C. The amplicons were purified and sequenced. The 16S rRNA sequence of MPS-2 was analyzed using Basic Local Alignment Search Tool (www.ncbi.nlm.nih.gov/BLAST). |
| Phylogenetic analyses of MPS-2 16S rRNA gene sequences |
| Genomic DNA was isolated from the pure culture pellet using consensus primers and partial 16s rRNA genes were amplified by PCR using forward primer (5′√ʬ?¬?GAGCGGATAACAATTTCACACAGG-3′), reverse primer (5′- CGCCAGGGTTTTCCCAGTCACGAC-3′) and internal primer (5′- CAGCAGCCGCGGTAATAC- 3′). The amplified 16s rRNA gene was sequenced. The obtained sequence data was aligned and analyzed for identification and finding the closest homology for the isolate. The next closet homo- logy was found with Pseudomonas and it was designated as Pseudomonas spp. MPS-2. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 [19]. The evolutionary history was inferred using the Neighbor- Joining method [20]. The bootstrap consensus tree inferred from 500 replicates is taken to represent the evolutionary history of the taxonomy analyzed [21]. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxonomy clustered together in the bootstrap test (500 replicates) is shown next to the branches [21]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method [22] and are in the units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). There were a total of 1420 positions in the final dataset. This tree was rooted with gram negative bacteria Eschericia coli strain K12 MG1655. |
| Batch culture experiments |
| Culture bacteria of Pseudomonas spp. MPS-2 was incubated at 37°C under facultative anaerobes condition (Schott Bottle filled up with CDM and incubation without shaking). After an optimization processes, the CDM broth have been modified which contains the following chemicals (g/L): K2HPO4 (7), KH2PO4 (2), MgSO4.7H2O (0.1), CaCl2 (0.02), NH4Cl2 (0.5), Glucose (1), and Reactive Red dye (0.1), and was adjusted to pH 7 prior to sterilization. The 1 L Schott bottle bacterial culture was incubated for a total 350 hours period in incubator without any agitation. Samples were withdrawn using syringe without any introduction of air to maintained facultative anaerobic condition. Initially at primary phase, which redox potential tend to decrease from 2.5 to 0.5 mV; samples were taken at 4 hours interval from 0 to 96 h. Meanwhile, after redox potential tend to become steady state at/or less than 0.5 mV (secondary phase), samples were taken daily from 96 h to 350 h. |
| Localization and detection of azoreductase activity |
| Fresh overnight culture of Pseudomonas spp. MPS-2 was harvested after growth for 24 hours in CDM broth without shaking at 37°C. At this time, the cell culture had reached its late exponential phase. The cells were harvested by centrifugation, and washed 2-3 times in cold potassium phosphate buffer (50 mM, pH 7.5). Supernatant (S) was separated from the cell pellets in 1 mL eppendorf tube. Meanwhile, the cells were then re-suspended in the same potassium phosphate buffer, and pellet maintained as resting cells (Rc). The remaining cell suspension was sonicated for a total of 2 minutes, with 2 minutes intervals after each 15 seconds of treatment. The cells were incubated on ice during sonication using an ultrasonic processor of 600 W at 0% amplitude. The homogenate (H) was centrifuged at 10,000 rpm at 4°C for 30 minutes. The supernatant labeled as crude Cell Free Extract (CFE) while pellet contained as Cell Debris (CD). All bacterial fractions were test for their azo reductase activity using following assay condition. |
| Assay for azoreductase activity |
| The azoreductase activity was detected and measured using a modified procedure by Robinson et al. [23]. The homogenate or cell extract (150 μL) was added to a sparged anaerobic solution containing Tris-HCI buffer (50 mM, pH 7.5), RR (0.03 mM) and FAD (0.05 mM). The reaction mix was flushed with nitrogen gas to create an anaerobic condition. For aerobic assay, sparging with nitrogen gas to the reaction mix was not required. The reaction (1 mL) was initiated by the addition of NADH (0.8 mM), followed by 10 minutes incubation. The reaction was followed using UV-Visible spectrophotometer (Shimadzu UV 1800, Japan) at the RR dye maximum wavelength absorbance of 517 nm. The azoreductase activity was calculated from the decrease in absorption at 517 nm. One unit (U) of the enzyme activity is defined as the amount of enzyme catalyzing the reduction of 1 ppm of dye per minute. |
| pH and temperature optimum and stability of the azoreductase |
| For determination of the pH dependence of azoreductase, the buffer used for the assay was set to pH values of 5.5 to 8.5. NADH solution and dyestuff solution were prepared with the corresponding buffers. For the determination of the temperature optimum, all solutions were mixed after brought to the corresponding temperature and the spectrophotometer (Shimadzu UV 1800, Japan) was temperature controlled during measurement. Enzyme stability was tested at five different pH values and five different temperatures. For this test, 150 μL of enzyme solution was mixed with 850 μL of buffer and incubated in a thermo-controlled water bath. Samples were taken after 10 minutes incubation, immediately frozen at -20°C, and subsequently analyzed. Buffers used for incubation were as follows: For pH values of 5.5, a 50 mM sodium acetate buffer system, pH of 6.5-7.5, phosphate buffer system and for pH 8.5, Tris-HCL buffer system were used respectively. |
| Analytical methods |
| Decolorization activity was expressed as the percentage of color removed by the bacteria using a modified method described by Yatome et al. [24]. Azo dye of Reactive Red reduction was monitored at an absorbance maximum (λmax) of 517 nm, using UV-Visible spectrophotometer (Shimadzu UV 1800, Japan) in 1 cm quartz cuvette. The pH and ORP value was determined immediately after sampling to avoid any change due to the CO2 production, using a pH meter and ORP meter (Hanna Instrument). All the other analytical determinations such as sulfate, phosphate, nitrate and Chemically Oxygen Demand (COD) were performed as described in Standard Methods for Examination of Water and Wastewater (APHA, 1985). Total Organic Carbon (TOC) was measured using TOC analyzer Multi N/C 2100 provided by Analytic Jena. Meanwhile, samples for High Performance Liquid Chromatography (HPLC) were concentrated using equal volume of ethyl acetate of solvent extraction method [2]. A High Performance Liquid Chromatography (HPLC) Agilent 1100 Series model, equipped with an auto sampler and Water Hypersil C18, 5 μm (4.6 m√?¬?250 mm) reverse phase column was used to separate individual compounds of intermediates that were detected using UVVis Detector. The mobile phase (HPLC grade) consisted of methanol (50%) and nano pure water (50%) in ratio of 1:1. The column was run at flow rate 0.5 mL/min, without controlling the temperature and the eluant was monitored at wavelength 254 nm using isocratic elution. |
| Results |
| Isolation and screening study |
| Four isolates designated MPS-1, MPS-2, MPS-3 and MPS-4, showed their ability to remove colour. All of these isolates were further grown on Filter Sterilized Textile Wastewater agar (FSTW agar), without the addition of any carbon and nitrogen sources. They showed the ability to grow on the FSTW agar after 48 h of incubation at 37°C. Screening results for color removal was done under two different conditions, facultative anaerobic and aerobic condition. Bacteria MPS-2 gives the highest color removal under facultative anaerobic condition, which is more than 63%. Meanwhile, MPS-4 showed the reverse trends that is, color removal occurred under aerobic condition, but only 28.5%. Subsequently, the forthcoming decolorization studies were carried out under facultative-anaerobic condition using pure culture of MPS-2. |
| Sequence analyses of gene encoding for the 16S rRNA from bacterium MPS-2 |
| The 16s rRNA gene sequences were compared by using BLAST similarity searches and the closely related sequences were obtained from GenBank. On the basis of morphological and biochemical analysis in combination with phylogenetic analysis, the strain ETL- 2013 was identified as Pseudomonas. The phylogenetic tree (Figure 1) constructed by the MEGA4 [18] displays MPS-2′s position in relation to other members of the Pseudomonas spp., and 98% most closely related to Pseudomonas otitidis strain 81f and Pseudomonas spp. GD6. |
| Degradation of RR by Pseudomonas spp. MPS-2 |
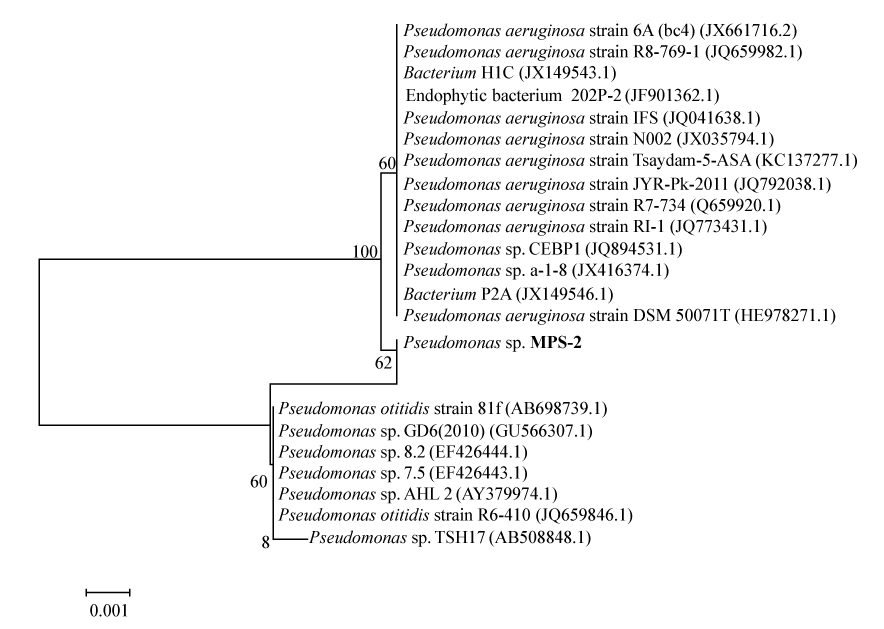
| Further decolorization studies under optimized conditions in chemically defined medium supplemented with (gL-1) of glucose (1), NH4Cl (0.5), Reactive red (0.1), adjusted to pH 7 with inoculum (10% v/v) and grown under partial anaerobic condition at 37°C, showed that bacterium MPS-2 successfully removed more than 95% colour and up to 50% of Total Organic Carbon (TOC). While the colour was removed more than 70%, the ORP values were decreased from 2.5 mV to 0.47 mV and the pH maintained from 7.21-7.25 in the phosphate buffered system (Table 1). In addition, data in Figure 2 show that the decolorization activity of Pseudomonas spp. MPS-2 decreased slightly from early-log phase (3.29 mg g cell-1 h-1) to early-stationary phase (1.24 mg g cell-1 h-1). Meanwhile, biomass concentration or cell dry weight of the bacteria increased from 0.48 to 1.36 g L-1. First-order rate constants (k), resulting from fitting equation (1) [25] to the whole curve (monoazo dyes) is, k=4 0.0091 h-1. Based on the k-values, RR which contains triazine as a reactive group had a slow rate of decolourisation as compared to other reactive dye [25]. |
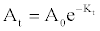 (1) (1) |
| ln (At) = ln(A0)-Kt |
| with: At=light absorbance at λmax at a given time (t) |
| A0=light absorbance at λmax at time 0. |
| Localization and detection of enzymatic activities |
| The enzyme activity of whole cells/resting cells (0.006 U/mL) and supernatant (0.003 U/mL) were significantly lower than the enzyme activity of the same dyes determined with other cellular component (0.020-0.030 U/mL) under anaerobic assay condition (Table 2). But, the highest enzyme activity was found in cell free extract which is 0.033 U/mL as well as homogenate (0.030 U/mL). Otherwise, under aerobic assayed condition (Table 2), the azo reductase activities were significantly high to whole cell supernatant and resting cells of Pseudomonas spp. MPS-2, which are 0.026 U/mL and 0.015 U/mL, respectively. While the azo reductase activities present in membrane fraction/cell debris and homogenate were slightly low (0.001-0.002 U/ mL). No enzyme activity was detected (<0.001 U/mL) in the cytoplasmic fraction/cell free extract of strain MPS-2. This azoreductase enzyme was found to be stable at pH 7 and temperature of 37°C. The enzyme activity tend to decrease more than 20% when the pH was varies from pH slightly acidic and alkaline. While, same trends was observed when it was exposed at lower and higher temperature. |
| Detection and analysis of HPLC products |
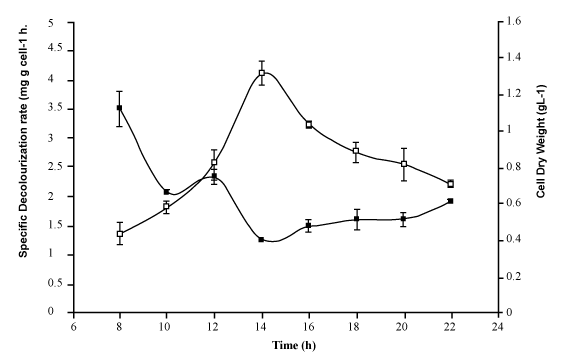
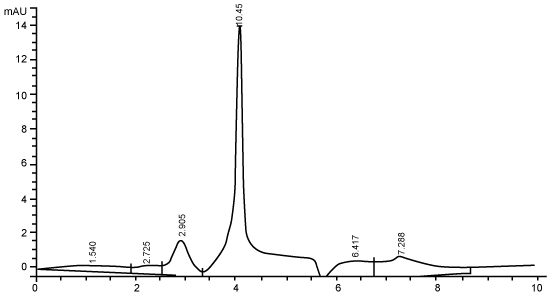
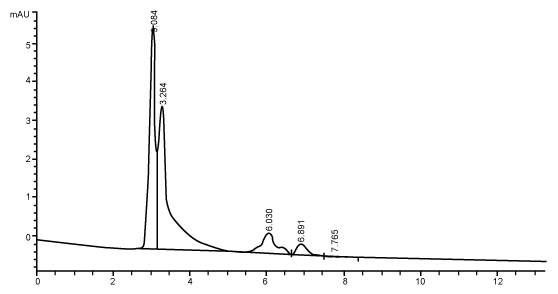
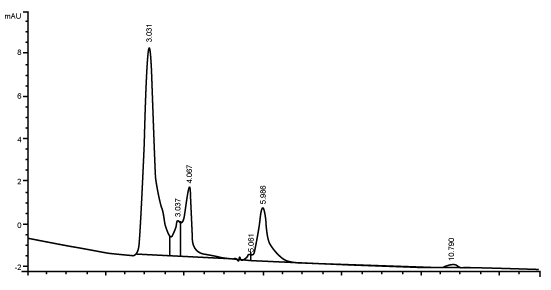
| Figure 3 shows the HPLC chromatograms of RR decolorization before the treatment processes. It was shown that the compound which is unknown metabolite had retention time, Rt=4.045. At the beginning of the anaerobic incubation, the parent compound, Reactive Red (RR) was not detected under these chromatographic conditions. While (Figure 4) shows that the retention time of the parent compound was disappeared (Rt=4.045) and produced lower retention time of 2.984 (A) and 3.254 (B) suggested were 4-amino, 3-hydronapthalenesulphonic acid (± 0.44%) and 4-aminobenzenesulphonate (sulpanilic acid) (± 0.6%) respectively. The sulphanilic acid was suggested can further be degraded by ring opening mechanism of aromatic ring into α-ketoglutaric acid, C with retention time of 3.031(± 0.36%) as shown in Figure 5. The α-ketoglutaric acid is one of TCA cycles intermediates which is a common metabolic in organism [26]. Furthermore, the fact that the UV-detectable area in Figure 4 shifted towards lower retention times (Rt=2.984 and 3.254) which means that these by-products of RR decolorization were more polar than the parent compounds (Rt=4.045) shown in Figure 3. While Figure 4 also showed that when such byproducts were degraded aerobically they formed less aromatic, morepolar compounds, since the UV-detectable area decreased in Figure 4 and was shifted towards more lower retention times (Rt=3.031) than in Figure 4 (Rt=3.254). |
| Discussion |
| Factors affecting color removal |
| Previously, it was observed that physiological factors like pH, oxygen level and temperature had significant effects on the rate of decolorization. This finding on color removal seems to indicate that glucose is probably an essential co metabolites for cell growth, since the was a decrease in specific reduction activity from early log phase to early stationary phase, during which cell growth is the dominant physiological activity [3]. The growth promotes certain metabolite pathways or accumulation of certain coenzyme or reducing equivalents (e.g. NADH and/or flavin) which lead to the decolourization by the reduction of azo bond(s) [27]. |
| Identification of bacteria using 16S rRNA |
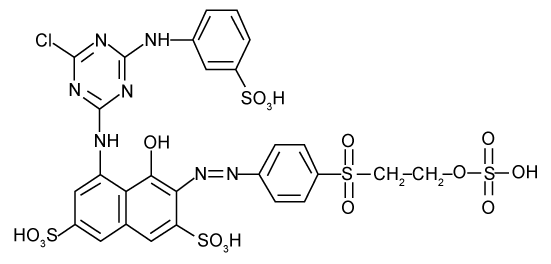
| Since the BLASTn result revealed that the sequence from isolated MPS-2 has 98% identity with Pseudomonas sp., it can possibly suggest that bacteria MPS-2 belong to the genus Pseudomonas. Thus, it can be suggested that Pseudomonas sp. MPS-2 has the ablity to decolourize textile wastewater by reductive cleavage of azo linkages in the azo dyes compounds. There is more than 95% decrease in absorbance (λmax=517 nm) of azo dyes decolorization, RR by Pseudomonas sp. MPS-2 caused the reaction products were usually colourless. But, in this experiment, a new absorption peak was found at 336 nm, close to λmax, in which a shift from red to yellow was observed. In the cases of monoazo dyes, the reaction followed first-order kinetics as shown for the example of RR. In contrast, dyes with more than one azo linkage displayed multiphase kinetics [3]. Based on the k-values, RR which contains triazine as a reactive group (Figure 6) had a slow rate of decolorization as compared to other reactive dye [25]. |
| Localization of azoreductase enzyme |
| Location of the enzyme system(s) responsible for the reduction of sulfonated azo dyes by Pseudomonas sp. strain MPS-2 was determined. Majority of the azoreductase enzyme produced by Pseudomonas sp. MPS-2 was probably located inside the cell (intracellular enzyme) which is oxygen sensitive enzyme. Recently, the most generally accepted hypothesis for bacterial reduction of azo dyes is that many bacterial cells possess a rather unspecific cytoplasmic azo reductases which transfers electrons via soluble or bound-flavins to the azo dyes. The oxygen inhibition on enzymatic reduction of azo dyes for bacterial species containing azoreductase system has also been observed [28], and may be attributed to a predominant competition for NADH utilization by aerobic respiration, which triggers an electron transfer from NADH to oxygen to form ATP [29]. Since NADH acts as the electron donor for the reduction of azo bonds that leads to bacterial decolorization of azo dyes, the consumption of NADH by oxidative phosphorylation would become a negative effect on the azoreductasedriven decolorization. Azoreductase was found to be stable at pH 7. This is due to azoreductase configuration and its active site was preserved and stable at pH 7 which promoted optimum decolourization processes. The decolourization was optimum when pH of the system is between pH 6-8. The optimum pH for colour removal is often at a neutral pH value or a slightly alkaline pH value and the rate of colour removal tends to decrease rapidly at strongly acid or strongly alkaline pH values [30]. Meanwhile, the enzyme shows high activity within the range of 30-40°C. The assay was carried out under anaerobic condition at pH 7. The temperature required to produce the maximum rate of colour removal tends to correspond with the optimum cell culture growth temperature of 35-45°C. The decline in colour removal activity at higher temperatures can be attributed to the loss of cell viability or to the denaturation of the azo reductase enzyme [31]. |
| Analysis of reactive red biodegradation |
| The HPLC analysis performed to supernatants taken during the different duration of the anaerobic phases showed the correspondence of the evolution of decolorization metabolite to the color removal. The chromatographic peak areas corresponding to the dye-degradation metabolite increased as the incubation time of anaerobic phase increased. The UV-absorbing area of the metabolite formed in anaerobic phase significantly decreased in the subsequent phase. It should be noted that due to the unavailability of authentic standards, the chromatographic peaks appearing in this sample taken during the anaerobic phase could not be identified or quantified [2]. Another factor is precision which means that all measurements of an analyte should be very close together with the standard. All quantitative results should be of high precision and there should be no more than a ± 2% variation in the assay system [32]. The chromatograms demonstrate that anaerobic treatment generated UV-absorbing compounds which were more polar than the parent compounds. The anaerobic degradation of reactive azo dye has been shown to produce several aromatic and ionic by-products [33]. In addition, the chromatograms also demonstrate that aerobic treatment reduced the levels of these UV-absorbing compounds and increased their polarity. Degradation in the aerobic stage may result in the formation of oxidised and very polar derivatives (e.g., aldehydes, carboxylic acids) having a lower aromaticity, as suggested by Nortemann et al. [34] in a study of 6-aminonaphthalene- 2-sulphonic acid degradation. The fate of aromatic amines in the aerobic stage cannot be conclusively determined. Partial or complete removal of many aromatic amines can be suspected from the decrease or disappearance of the sometimes unidentified- peaks in HPLCchromatograms [3]. |
| Mechanism of colour removal |
| Decolourization usually occurs favorably under facultative anaerobic condition indicated by lower redox potential of the system. This phenomenon may be due to either the electron-withdrawing nature of the azo bond and their resistance to oxygenases attack, or because oxygen is a more effective electron acceptor, therefore having more preference for reducing equivalents than the azo dye [16,31,35]. In the present study under anaerobic condition, a rather high decolorization of Reactive Red azo dyes was observed when incubated them with Cytoplasmic Fraction Extract (CFE) of Pseudomonas spp. strain MPS-2 in anaerobic buffer with NADH as a source of reduction equivalents (Table 2) with the presence of Flavin Adenine Dinucleotide (FAD). A possible explanation for this phenomenon is that FAD is reduced enzymatically to FADH2 by NADH and then spontaneously reduces the RR sulfonated azo dyes to the corresponding amines [15]. In contrast, it was shown that the addition of NADH and FAD did not lead to enhancement of the reduction rates of sulfonated azo dyes by whole cells, resting cells (Rc) and supernatant (S) of Pseudomonas sp. strain MPS-2 (Table 2). Thus, this has generally been explained by the low permeability of the cell membranes for the highly polar sulfonated azo compounds [11]. It was clearly demonstrated in this study that the total activity of flavin reductase, which hypothesized to function under adequate conditions as flavin-dependent azo reductase, was present in the cytoplasmic fraction. Therefore, it appears reasonable that, with intact cells (Rc), intracellular enzymes like flavin reductases are of little importance for reduction of extracellular sulfonated azo compounds by strain MPS-2. These results supported the hypothesis of Russ and coworkers that the reduction of sulfonated azo dyes by reduced flavins formed by cytosolic flavin-dependent azo reductases is mainly observed in vitro and in vivo is of insignificant importance [15]. Thus, in the intact cells, other enzyme systems, which does not require transport of the azo dyes though the cell membrane, are presumably responsible for the unspecific reduction of various sulfonated azo dyes by Pseudomonas spp. strain MPS-2. In addition, it could be demonstrated in the cell-fractioning experiments that the azoreductase activity was almost restrictively present in the membrane fraction, cell debris (CD), (Table 2) These results suggest the existence of a nonspecific azoreductase in Pseudomonas spp. strain MPS-2 membranes that catalyze the reduction of endogenous reducing equivalent, maybe responsible for the reduction of exogenous reducing equivalents which then transfer reduction equivalents to sulfonated azo dyes outside the cells. Meanwhile, the rather high reduction rate of RR sulfonated azo dyes in the present of oxygen (aerobic condition) found in whole cells (Rc) and supernatant (S) experiment (Table 2) Thus, we cannot involved in the aerobic reduction of sulfonated azo dyes by whole cells of Pseudomonas spp. strain MPS-2. We are currently attempting to explore the other extracellular electron transferring mechanisms of strain MPS-2 which are involved for the reduction of the azo dye to clarify this mechanism. |
| Conclusion |
| In summary, we successfully isolated a decolorizing bacterium; Pseudomonas spp. strain MPS-2 that can degrade azo dyes to yellowish colored intermediates of aromatic amines and/or even to partially mineralize them into simpler compound that is a common metabolic in organism, which are safe and less toxic to the environment. We also have shown that an azoreductase is responsible for azo dye reduction by Pseudomonas spp. strain MPS-2. Future investigations should focus on the application of this highly potential azo dye-degrading bacterium for the treatment of textile dyeing effluents. |
| Acknowledgement |
| Authors are highly grateful to the management of Enviro Technology Limited., Ankleshwar, Gujarat, India for allowing us to carry out such a noble work for the sustainable environment. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 |
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 |
 |
 |
 |
| Figure 4 | Figure 5 | Figure 6 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 14128
- [From(publication date):
July-2013 - Apr 04, 2025] - Breakdown by view type
- HTML page views : 9533
- PDF downloads : 4595
