Matching on Race and Ethnicity in Case-Control Studies as a Means of Control for Population Stratification
Received: 24-Aug-2011 / Accepted Date: 20-Sep-2011 / Published Date: 29-Sep-2011 DOI: 10.4172/2161-1165.1000101
Abstract
Some investigators argue that controlling for self-reported race or ethnicity, either in statistical analysis or in study design, is sufficient to mitigate unwanted influence from population stratification. In this report, we evaluated the effectiveness of a study design involving matching on self-reported ethnicity and race in minimizing bias due to population stratification within an ethnically admixed population in California. We estimated individual genetic ancestry using structured association methods and a panel of ancestry informative markers, and observed no statistically significant difference in distribution of genetic ancestry between cases and controls (P=0.46). Stratification by Hispanic ethnicity showed similar results. We evaluated potential confounding by genetic ancestry after adjustment for race and ethnicity for 1260 candidate gene SNPs, and found no major impact (>10%) on risk estimates. In conclusion, we found no evidence of confounding of genetic risk estimates by population substructure using this matched design. Our study provides strong evidence supporting the race- and ethnicity-matched case-control study design as an effective approach to minimize systematic bias due to differences in genetic ancestry between cases and controls.
Keywords: Population stratification; Genetic susceptibility; Case-control; Matching.
158411Introduction
In genetic epidemiology studies, potential confounding by genetic ancestry, known as population stratification, may bias results [1-3]. Although there is no agreement on the magnitude of bias that might be attributable to population stratification, many genetic epidemiology studies have taken measures to guard against it. Large genome-wide association studies often restrict analyses to subjects who conform to a certain degree of ethnic/genetic homogeneity [4]. In studies where such restriction is impossible or even undesirable, estimates of genetic ancestry based on structured association methods are often calculated [4] and adjusted for by regression analyses. However, some investigators argue that controlling for self-reported race or ethnicity, either in statistical analysis or in study design, is sufficient to mitigate unwanted influence from population stratification [2]. Such approaches are desirable under certain circumstances, such as when a study population is being used to type a limited number of genetic variants as part of a replication study, and costs of high-density genotyping are prohibitive. In this report, we examine the effectiveness of a study design involving matching on self-reported ethnicity and race in minimizing bias due to population stratification within an ethnically admixed population.
Materials and Methods
Since 1995, we have been conducting a population-based case control study of childhood leukemia in 35 counties of Northern and Central California. The study population includes 41% Hispanics, a recently admixed population, and 44% non-Hispanic whites, with the remaining 15% comprising smaller numbers of blacks, Asians and other races/ethnicities. The study has been described previously [5]. Briefly, incident cases of childhood leukemia (age under 15 years) were ascertained through a rapid reporting system established with participating hospitals. Controls were selected from the California birth registry and individually matched to cases on date of birth, gender, maternal race and child's Hispanic ethnicity, that is, mother's report of either parent being Hispanic. This study was reviewed and approved by institutional review committees at the authors' academic institutions, the California Department of Public Health (CDPH), and the participating hospitals. Written informed consent was obtained from all parent respondents; participating case and control children over age 7 also provided written assent.
We used a panel of ancestry informative markers (AIMs) developed specifically to estimate individual genetic ancestry of the major ancestral populations comprising Hispanics: Africans, Amerindians, and Europeans [6]. This panel of SNPs was selected based on high allele frequency differences between the ancestral populations and low linkage disequilibrium (r2<0.6) between the SNPs within each population. Using a custom 1536-single nucleotide polymorphism (SNP) Illumina GoldenGate genotyping panel, we attempted to genotype 95 AIMs in our study subjects. The remaining SNPs included in the Illumina panel were variants in 183 candidate genes. Most of these were haplotype-tagging SNPs, selected to capture genetic variation at r20.80, based on data from the 30 Caucasian trios in the Hap Map project (Release 19, Build 34, www.hapmap.org) and the 23 Hispanics in the SNP500Cancer project (www.snp500cancer. nci.nih.gov). After applying an Illumina GenCall threshold of 0.25, and SNP-wise and subject-wise call rate thresholds of =90% and =95%, respectively, we successfully genotyped 80 AIMs and 1260 candidate gene SNPs in whole-genome amplified DNA extracted from buccal cytobrush specimens (73.4%) or archived newborn dried blood spot specimens (26.6%) collected from 376 subjects with childhood acute lymphocytic leukemia (ALL) and 447 controls.
Individual estimates of genetic admixture, i.e. percent contribution of each of the three ancestral populations, were obtained from maximum likelihood estimation models as described previously [7,8]. We used Hotelling's T test to assess the statistical significance of the association between computed genetic ancestry distribution and casecontrol status.
To assess the degree of potential confounding due to case-control differences in genetic ancestry, we calculated the confounding risk ratio (CRR) for each of remaining successfully genotyped SNPs in the Illumina panel. The CRR is defined as the ratio of the unadjusted odds ratio (OR) to the OR adjusted for a potential confounder (in this case, genetic ancestry) [9]. Because genotyping data were not available for every individually matched case-control set, we report the results of an unmatched analysis, using unconditional logistic regression to calculate the CRR and compare risk estimates adjusted for the matching factors (age, gender, self-reported race and Hispanic ethnicity) to those further adjusted for estimated genetic ancestry. The results reported here were similar to those from a matched analysis using conditional logistic regression of the 278 complete matched case-control sets (201 pairs and 77 triplets). Accordingly, to maximize sample size and statistical power, we present only the results of the unmatched analysis.
Results
The mean percentages of African, European, and Amerindian genetic ancestry by case-control status are presented in (Table 1). European ancestry dominated our total population, followed by Amerindian ancestry, then African ancestry, present at just 7%. Overall, we observed no statistically significant difference in ancestry distribution between cases and controls (P=0.46). Similarly, stratification by Hispanic ethnicity showed no significant case-control differences in ancestry distribution (P=0.66 and 0.60 for Hispanics and non-Hispanics, respectively).
| % European Ancestry a | % African Ancestry a | % Amerindian Ancestry a | |||||||
| N | Mean | (SD) | Mean | (SD) | Mean | (SD) | P b | ||
| All subjects | Cases | 376 | 68.5 | (27.3) | 7.3 | (14.7) | 24.2 | (23.7) | 0.46 |
| Controls | 447 | 70.7 | (26.9) | 7.1 | (14.5) | 22.2 | (23.8) | ||
| Hispanics only | Cases | 156 | 51.8 | (17.4) | 7.2 | (8.3) | 41.0 | (16.8) | 0.66 |
| Controls | 179 | 52.7 | (17.1) | 7.7 | (8.6) | 39.5 | (16.1) | ||
| non-Hispanics only | Cases | 220 | 80.4 | (26.8) | 7.3 | (18.0) | 12.3 | (20.5) | 0.60 |
| Controls | 268 | 82.7 | (25.6) | 6.6 | (17.3) | 10.7 | (19.0) | ||
bHotelling’s T test
Table 1: Percent Estimated Genetic Ancestry Among Childhood ALL Cases and Controls, by Ethnicity. *
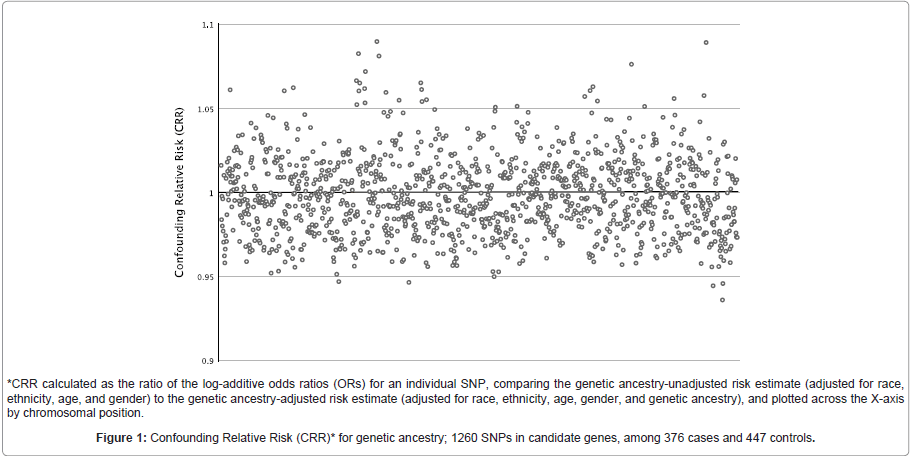
The CRRs for the 1260 candidate gene SNPs assessing potential confounding by genetic ancestry are shown in (Figure 1). Overall, we found no major differences (>10%) in risk estimates due to adjustment for genetic ancestry over and above adjustment for race and ethnicity, though the observation of more CRRs above 1.05 than below 0.95 suggests that risk estimates unadjusted for genetic ancestry tend to be somewhat inflated.
Discussion
In this study, we found no significant differences in estimated genetic ancestry between race- and ethnicity-matched cases and controls overall. Furthermore, we found no evidence of confounding of genetic risk estimates by population substructure using this matched design. The results of this study demonstrate that careful study design can overcome potential differences in genetic ancestry between cases and controls that can lead to population stratification.
In order for confounding of a gene-disease association by any factor to exist, evidence of an association between that factor and the disease must be observed. In our matched case-control study, among all races/ethnicities together as well as after stratification by Hispanic ethnicity, we found no evidence of association between disease and genetic ancestry. However, it should be noted that the AIMs we used were optimized for discerning major continental ancestry origins [6]. As such, they are more informative in discerning ancestry among Hispanics than among non-Hispanics. Indeed, the overwhelmingly dominant ancestry among our non-Hispanic subjects was European (80-83%). Thus, potential confounding of results by subtle, intracontinental differences such as Northern vs. Southern European ancestry would have to be investigated by more sophisticated matching (such as matching on parental or grandparental country of origin) and/ or a more extensive set of AIMs than was done in our study.
It should also be noted that the individual matching of our original study design requires adjustment for matching factors when performing an unmatched analysis. These factors included race, ethnicity, gender, and age. Accordingly, assessment of confounding by ancestry via the CRR necessitated inclusion of these matching variables in regression analyses. This was done even though the distributions of the matching factors were balanced between cases and controls by the very design of the study.
Our intention with this investigation was not to gauge the general magnitude of population stratification on results, but to comment on the absence of material changes in risk estimates in race- and ethnicity matched case control studies after adjustment for estimated genetic ancestry. This is particularly relevant given the large proportion of subjects reporting Hispanic ethnicity in our study population. Hispanics are a recently admixed group [10] and are reported to have the highest incidence of childhood leukemia in California [11]. It is therefore imperative to include this group in the search for genetic susceptibility loci for childhood ALL. Accordingly, the absence of a significant association between genetic ancestry and disease within this ethnic group is especially reassuring.
Similar to the results of our investigation, a recent study in New York found little improvement in model fit for specific genotype-phenotype associations due to adjustment for genetic ancestry over self-reported race/ethnicity among ethnically heterogeneous populations and specifically among Hispanics [12]. Another study in a multi-ethnic population compared multiple methods for adjusting for genetic structure, and found that the potential impact of population stratification was effectively mitigated by adjustment for self-reported race/ethnicity or AIMs-based ancestry estimates [13]. The authors attribute the modest extent of bias due to population stratification observed to both the frequency matching of cases and controls on race/ ethnicity as well as the sampling of subjects of same ethnicity from the same geographic area. These findings provide support our conclusion that adjustment for race/ethnicity in admixed populations - by design, analysis, or both - paired with careful attention to repidemioluitment of subjects from comparable geographic regions, can effectively mitigate effects of population stratification.
Matching reduces heterogeneity between cases and controls that may be due to the matching factors, or more specifically, broad unmeasured risk factors that are associated with the matching factors. In the case of matching on self-reported race and ethnicity, these include both genetic and non-genetic components. The matched study design's reduction in heterogeneity improves efficiency to study effects that do not involve these unmeasured components. However, individual matching such as performed for this study is logistically challenging and costly, and matching on race/ethnicity precludes assessment of these factors as potential risk factors. Furthermore, there exists the risk of overmatching with respect to environmental and lifestyle factors particular to certain racial/ethnic groups. Nevertheless, for a rare disease such as childhood ALL (31.9 cases per million/ year in the US [14]) the number of cases available to participate in epidemiologic studies tends to be small, and thus the improvement in statistical efficiency afforded by individual matching of controls is useful.
In summary, our findings support the race- and ethnicity-matched case-control study design as an effective approach to mitigating systematic bias due to differences in genetic ancestry between cases and controls.
Acknowledgements
We thank our clinical collaborators and Northern California Childhood Leukemia Study (NCCLS) participating hospitals: University of California Davis, University of California San Francisco, Children's Hospital of Central California, Lucile Packard Children's Hospital, Children's Hospital Oakland, Kaiser Permanente Roseville, Kaiser Permanente Santa Clara, Kaiser Permanente San Francisco, and Kaiser Permanente Oakland. We thank the staff and students of the NCCLS, the UC Berkeley Survey Research Center, and lab personnel at both UC Berkeley and UC San Francisco for their effort and dedication. Finally, we thank the families who participated in the NCCLS for their strong support and selflessness, without which this research could not have been conducted.
We acknowledge funding support from the National Institute of Environmental Health Sciences (PS42ES04705 and R01ES09137) and the Children with Cancer UK Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIEHS, NIH or the Children with Cancer UK Foundation.
References
- Thomas DC, Witte JS (2002) Point: population stratification: a problem for case-control studies of candidate-gene associations? Cancer Epidemiol Biomarkers Prev 11: 505-512.
- Wacholder S, Rothman N, Caporaso N(2002) Counterpoint: bias from population stratification is not a major threat to the validity of conclusions from epidemiological studies of common polymorphisms and cancer. Cancer Epidemiol Biomarkers Prev 11: 513-520.
- Yu K, Wang Z, Li Q, Wacholder S, Hunter DJ, et al. (2008) Population substructure and control selection in genome-wide association studies. PLoS One 3: e2551.
- Tian C, Gregersen PK, Seldin MF (2008) Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet 17: R143- 150.
- Ma X, Buffler PA, Layefsky M, Does MB, Reynolds P (2004) Control selection strategies in case-control studies of childhood diseases. Am J Epidemiol 159: 915-921.
- Yang N, Li H, Criswell LA, Gregersen PK, Alarcon-Riquelme ME, et al. (2005) Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum Genet 118:382-392.
- Hanis CL, Chakraborty R, Ferrell RE, Schull WJ (1986) Individual admixture estimates: disease associations and individual risk of diabetes and gallbladder disease among Mexican-Americans in Starr County, Texas. Am J Phys Anthropol 70: 433-441.
- Aldrich MC, Selvin S, Hansen HM, Barcellos LF, Wrensch MR, et al. (2009) CYP1A1/2 haplotypes and lung cancer and assessment of confounding by population stratification. Cancer Res 69: 2340-2348.
- Wacholder S, Rothman N, Caporaso N (2000) Population stratification in epidemiologic studies of common genetic variants and cancer: quantification of bias. J Natl Cancer Inst 92: 1151-1158.
- Baye TM, Wilke RA (2010) Mapping genes that predict treatment outcome in admixed populations. Pharmacogenomics J 10: 465-477.
- Campleman SL, Wright WE. Childhood cancer in California 1988 to 1999 Volume I: birth to age 14. Sacramento, CA: California Department of Health Services, Cancer Surveillance Section; 2004 July, 2004.
- Lee YL, Teitelbaum S, Wolff MS, Wetmur JG, Chen J (2010) Comparing genetic ancestry and self-reported race/ethnicity in a multiethnic population in New York City. J Genet 89: 417-423.
- Wang H, Haiman CA, Kolonel LN, Henderson BE, Wilkens LR, et al. (2010) Self-reported ethnicity, genetic structure and the impact of population stratification in a multiethnic study. Hum Genet 128: 165-177.
- Linabery AM, Ross JA (2008) Trends in childhood cancer incidence in the U.S. (1992-2004). Cancer 112: 416-432.
Citation: Chokkalingam AP, Aldrich MC, Bartley K, Hsu LI, Metayer C, et al. (2011) Matching on Race and Ethnicity in Case-Control Studies as a Means of Control for Population Stratification. Epidemiol 1:101. DOI: 10.4172/2161-1165.1000101
Copyright: © 2011 Chokkalingam AP, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 16167
- [From(publication date): 9-2011 - Nov 28, 2025]
- Breakdown by view type
- HTML page views: 11240
- PDF downloads: 4927