Research Article Open Access
Isolation of New Hexapeptides?JBIR-39 and JBIR-40?from a Marine Sponge-Derived Streptomyces sp. Sp080513SC-24
Ikuko Kozone1, Miho Izumikawa1, Keiichiro Motohashi1, Aya Nagai1, Masahito Yoshida2, Takayuki Doi2, Motoki Takagi1* and Kazuo Shin-ya3*1Biomedicinal Information Research Center (BIRC), Japan Biological Informatics Consortium (JBIC), 2-4-7 Aomi, Koto-ku, Tokyo 135-0064, Japan
2Graduate School of Pharmaceutical Sciences, Tohoku University, 6-3 Aza-aoba, Aramaki, Aobaku, Sendai 980-8578, Japan
3Biomedicinal Information Research Center (BIRC), National Institute of Advanced Industrial Science and Technology (AIST), 2-4-7 Aomi, Koto-ku, Tokyo 135-0064, Japan
- *Corresponding Authors:
- Dr. K Shin-ya
Biomedicinal Information Research Centre (BIRC)
National Institute of Advanced Industrial Science and Technology
(AIST), 2-4-7 Aomi, Koto-ku
Tokyo 135-0064, Japan
E-mail: k-shinya@aist.go.jp - Dr. M Takagi
Biomedicinal Information Research Centre (BIRC)
Japan Biological Informatics Consortium (JBIC)
2-4-7 Aomi, Koto-ku
Tokyo 135-0064, Japan
E-mail: motoki-takagi@aist.go.jp
Received date December 27, 2010; Accepted date February 15, 2011; Published date February 17, 2011
Citation: Kozone I, Izumikawa M, Motohashi K, Nagai A, Yoshida M, et al. (2011) Isolation of New Hexapeptides-JBIR-39 and JBIR-40 from a Marine Sponge- Derived Streptomyces sp. Sp080513SC-24. J Marine Sci Res Development 1:101. doi: 10.4172/2155-9910.1000101
Copyright: © 2011 Kozone I, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Marine Science: Research & Development
Abstract
Streptomyces sp. Sp080513SC-24 was isolated from a marine sponge, Haliclona sp., which is inhabited by diverse Actinobacteria , and then its culture was comprehensively searched for secondary metabolites. New two hexapeptides, JBIR-39 (1), and -40 (2), were isolated from the fermentation broth of Sp080513SC-24. The structures of 1 and 2 were elucidated on the basis of 1D and 2D NMR spectroscopy and MS analyses.
Keywords
Haliclona; Hexapeptide; Marine sponge; Piperazic acid; Streptomyces
Abbreviations
HPLC: High-performance liquid chromatography; MPLC: Medium-performance liquid chromatography; HR-ESI-MS: High-resolution-electrospray ionization-mass spectrometry; DQFCOSY: Double quantum filtered-correlation spectroscopy; HSQC: Heteronuclear single quantum coherence; HMBC: Heteronuclear multiple bond correlation
Introduction
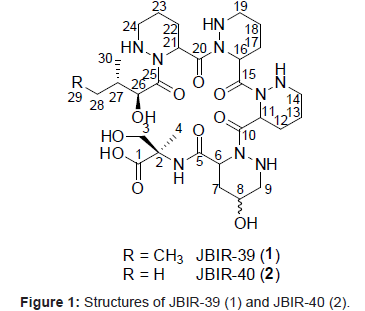
Marine microorganisms, particularly marine Actinobacteria, have attracted considerable attention as one of the most important resources for new biologically active metabolites [1]. For example, new compounds have been isolated from Actinobacteria of sponge origin [2-5]. Our group was recently engaged in the isolation of Actinobacteria from marine sources. Some of the isolated Actinobacteria have been found to produce new compounds, namely, the teleocidin analog JBIR-31, [6] the isoprenoids JBIR-46, JBIR-47, and JBIR-48, [7,8] the salicylamide derivative JBIR-58, [9] and a terpen JBIR-65 [10]. Thus, we isolated a number of phylogenetically diverse Streptomyces including six novel members of the genus Streptomyces from a marine sponge, Haliclona sp., which is inhabited by diverse Actinobacteria, and then comprehensively searched for secondary metabolites in the cultures of isolated strains [11]. We succeeded in isolating 2 new anthracyclines, tetracenoquinocin [12] and 5-iminoaranciamycin, [12] and 2 new tetrapeptides, JBIR-34 and JBIR-35 [13]. In this study, we purified 2 new compounds, termed JBIR-39 (1) and JBIR-40 (2) (Figure 1), from the fermentation broth of a new species (Sp080513SC-24) of Streptomyces isolated from Haliclona sp. This paper describes the fermentation, isolation, and structure elucidation of 1 and 2.
Materials and Methodstable
General experimental procedures
Optical rotations were obtained on an SEPA-300 polarimeter (Horiba, Kyoto, Japan). UV and IR spectra were measured on a DU730 UV/Vis spectrophotometer (Beckman Coulter, CA, USA) and an FT- 720 spectrophotometer (Horiba), respectively. NMR spectra were recorded on a Varian NMR System 600 NB CL (Varian, Palo Alto, CA, USA) in DMSO-d6 (2.50 ppm for 1H, 39.5 ppm for 13C) with the residual solvent peak as the internal standard. HR-ESI-MS data were recorded on an LCT-Premier XE mass spectrometer (Waters, Milford, MA, USA). MPLC was performed using a Purif-pack ODS-100 column (100 µm, Shoko Scientific, Yokohama, Japan). Analytical reversed-phase HPLC was carried out using an L-column2 ODS column (5.0 µm, 4.6 i.d. × 150 mm; Chemicals Evaluation and Research Institute, Tokyo, Japan) equipped with a 2996 photodiode array detector (Waters) and a 3100 Mass Detector (Waters). Preparative reversed-phase HPLC was carried out using an L-column2 ODS column (5.0 µm, 20 i.d. × 150 mm; Chemicals Evaluation and Research Institute, Tokyo, Japan).
Microorganism
Streptomyces sp. strain Sp080513SC-24 has been reported as a new species of genus Streptomyces isolated from Haliclona sp. and was termed Streptomyces spongiae NBRC 106415τ [11].
Fermentation
Streptomyces sp. Sp080513SC-24 was cultivated in 50-ml test tubes, each containing 15 ml of a seed medium consisting of starch (Kosokagaku, Tokyo, Japan) 1.0%, polypeptone (Nihon Pharmaceutical, Tokyo, Japan) 1.0%, molasses (Dai-Nippon Meiji Sugar, Tokyo, Japan) 1.0%, and meat extract (Extract Ehlrich, Wako Pure Chemical Industry, Osaka, Japan) 1.0% before sterilization with pH adjusted to 7.2. The test tubes were shaken on a reciprocal shaker (355 r.p.m.) at 27°C for 2 days. Aliquots (2.5 ml) of the broth were transferred to 500-ml baffled Erlenmeyer flasks containing 100 ml of a production medium, consisting of starch 2.5%, soybean meal (Nisshin Oillio, Tokyo, Japan) 1.5%, dry yeast (Mitsubishi Tanabe Pharma, Osaka, Japan) 0.2%, CaCO3 (Kozaki Pharmaceutical, Tokyo, Japan) 0.4%, and Diaion HP-20 resin (Mitsubishi Chemical, Tokyo, Japan) 1.0% before sterilization with pH adjusted to 7.0. The fermentation was carried out on a rotary shaker (180 r.p.m.) at 27°C for 5 days.
Isolation
The supernatant of whole broth (2 l) collected by centrifugation was successively partitioned with AcOEt (1 l × 3) and n-BuOH (1 l × 2). The n-BuOH layer was evaporated to dryness. The dried residue (413 mg) was fractionated by reversed-phase MPLC (Purif-Pack ODS-100) with a MeOH-water gradient system (0-100% MeOH) and fractions including major metabolites were collected by LC-MS monitoring. The eluate was subjected to preparative reversed-phase HPLC using an L-column2 ODS column (5.0 µm, 20 i.d. × 150 mm; Chemical Evaluation and Research Institute, Tokyo, Japan) developed with 60% aqueous MeOH containing 0.1% formic acid (flow rate: 9.5 ml min-1) to give JBIR-39 (1, 5.3 mg; Retention time (Rt), 19 min) and JBIR-40 (2, 3.1 mg; Rt, 13 min).
JBIR-39 (1): colorless oil; [α]25 D -11.0 (c 0.1, MeOH); UV (MeOH) λmax (ε) 233 (3,100); IR (KBr) νmax 3430, 1720, 1640 cm-1; 1H NMR (600 MHz, CDCl3) and 13C NMR (150 MHz, CDCl3), see Table 1; HR-ESIMS m/z 698.3828 [M + H]+ (calcd. for C30H52N9O10, 698.3837).
| 1 | 2 | |||||
| Position | 13C | 1H (J in Hz) | 13C | 1H (J in Hz) | ||
| α-MeSer | ||||||
| 1 | 174.7 | 175.3 | ||||
| 2 | 59.5 | 60.0 | ||||
| 3 | 62.8 | 3.89, d (10.4) | 62.8 | 3.88, d (10.2), 3.38, d (10.2) | ||
| 4 | 20.6 | 1.29, s | 21.0 | 1.28, s | ||
| 2-NH | 7.90, s | 7.90, s | ||||
| γ-OH-Pip | ||||||
| 5 | 170.1 | 170.6 | ||||
| 6 | 48.5 | 4.78, m | 49.0 | 4.77, m | ||
| 7 | 33.4 | 2.25, 1.94, m | 33.9 | 2.24, 1.92, m | ||
| 8 | 61.3 | 3.65, m | 61.8 | 3.67, m | ||
| 9 | 52.7 | 2.79, m | 53.2 | 2.78, m | ||
| 9-NH | 5.28, dd (11.2. 3.1) | 5.28, dd (12.3, 3.2) | ||||
| Pip1 | ||||||
| 10 | 172.9 | 173.4 | ||||
| 11 | 48.4 | 5.60, m | 49.0 | 5.60, m | ||
| 12 | 25.8 | 1.99, 1.78, m | 26.5 | 1.99, 1.74, m | ||
| 13 | 21.3 | 1.52, 1.42, m | 21.8 | 1.51, 1.43, m | ||
| 14 | 46.8 | 2.98, 2.60, m | 47.4 | 2.98, 2.58, m | ||
| 14-NH | 5.15, m | 5.15, m | ||||
| Pip2 | ||||||
| 15 | 172.2 | 172.7 | ||||
| 16 | 48.4 | 5.60, m | 49.0 | 5.60, m | ||
| 17 | 25.8 | 1.99, 1.78, m | 26.5 | 1.99, 1.74, m | ||
| 18 | 21.3 | 1.52, 1.42, m | 21.8 | 1.51, 1.43, m | ||
| 19 | 46.8 | 2.98, 2.60, m | 47.4 | 2.98, 2.58, m | ||
| 19-NH | 5.15, m | 5.15, m | ||||
| Pip3 | ||||||
| 20 | 170.7 | 173.1 | ||||
| 21 | 48.4 | 5.60, m | 49.0 | 5.60, m | ||
| 22 | 25.8 | 1.99, 1.78, m | 26.5 | 1.99, 1.74, m | ||
| 23 | 21.3 | 1.52, 1.42, m | 21.8 | 1.51, 1.43, m | ||
| 24 | 46.8 | 2.98, 2.60, m | 47.4 | 2.98, 2.58, m | ||
| 24-NH | 5.00, dd (12.5, 3.1) | 5.00, dd (12.2, 3.4) | ||||
| a-hydroxylic acid | ||||||
| 25 | 174.9 | 175.3 | ||||
| 26 | 71.8 | 4.43, m | 72.3 | 4.40, m | ||
| 27 | 37.4 | 1.71, m | 31.0 | 1.95, m | ||
| 28 | 22.7 | 1.23, 1.00, m | 20.5 | 0.88, d (6.9) | ||
| 29 | 12.2 | 0.79, t (7.5) | ||||
| 30 | 16.7 | 0.87, d (6.8) | 16.4 | 0.70, d (6.9) | ||
13C (150 MHz) and 1H (600 MHz) NMR spectra were taken on a NMR System 600 NB CL in DMSO-d6, and the solvent peak was used as an internal standard (δC 39.5, δH 2.50).
Table 1: 1H and 13C NMR spectral data for JBIR-39 (1) and JBIR-40 (2).
JBIR-40 (2): colorless oil; [α]25 D -7.8 (c 0.4, MeOH); UV (MeOH) λmax (ε) 235 (2,800); IR (KBr) νmax 3400, 1730, 1640 cm-1; 1H NMR (600 MHz, CDCl3) and 13C NMR (150 MHz, CDCl3), see Table 1; HR-ESIMS m/z 684.3681 [M + H]+ (calcd for C29H50N9O10, 684.3680).
Results and Discussion
Structure Elucidation of 1
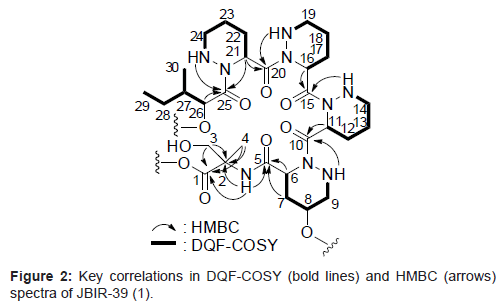
Compound 1 was isolated as a colorless oil and its HR-ESI-MS was consistent with a molecular formula of C30H51N9O10. The 1H and 13C NMR spectral data for 1 are shown in Table 1. From IR absorption (νmax 3430, 1720, and 1640 cm-1) and 13C NMR spectral data, 1 was considered to be a peptide compound. The structural information for 1 was further obtained by a series of 2D NMR analyses, including HSQC, HMBC, and DQF-COSY spectra (Figure 2). The 1H-13C longrange couplings from singlet methyl proton 4-H (δH 1.29) to a carbonyl carbon C-1 (δC 174.7), a quaternary carbon C-2 (δC 59.5), and a methylene carbon C-3 (δC 62.8), from a doublet methylene proton 3-H (δH 3.89) to C-1, C-2, and C-4 (δC 20.6), and from an amide proton 2-NH (δH 7.90) to C-2 revealed an a-methylserine (α-MeSer) moiety. The sequence from a typical a-methine proton 6-H (δH 4.78, δC 48.5) to an amine proton 9-NH (δH 5.28) through methylene protons 7-H (δH 2.25, 1.94), an oxymethine proton 8-H (δH 3.65), and methylene proton 9-H (δH 2.79) was established by the DQF-COSY spectrum. In the HMBC spectrum, 1H-13C long-range couplings from the α-methine proton 6-H and methylene proton 7-H to an amide carbonyl carbon C-5 (δC 170.1), and from the amine proton 9-NH to an amide carbonyl carbon C-10 (δC 172.9) were established. These data revealed that C-6 (δC 48.5) and C-9 (δC 52.7) are connected to the different nitrogen atoms and by taking into consideration the peptidyl structure of 1, the existence of a γ-hydroxyl piperazic acid (γ-OH-Pip) as an amino acid unit, but not a proline residue, was suggested. In the same manner, the 2 identical sequences from α-methine protons 11-/16-H (δH 5.60, δC 48.4) to amine protons 14-NH/19-NH (δH 5.15) through methylene protons 12-/17-H (δH 1.99, 1.78), 13-/18-H (δH 1.52, 1.42), and 14-/19-H (δH 2.98, 2.60), and another resembled sequence from an α-methine proton 21-H (δH 5.60, δC 48.4) to an amine proton 24-NH (δH 5.00) through methylene protons 22-H (δH 1.99, 1.78), 23-H (δH 1.52, 1.42), and 24-H (δH 2.98, 2.60) were also established by the analyses of the DQF-COSY spectrum of 1. The 1H-13C long-range couplings from 21-H to an amide carbonyl carbon C-20 (δC 170.7) and C-25 (δC 174.9), and from 24-NH to C-25 proved a piperazic acid (Pip) moiety. From the molecular formula and 1H and 13C NMR chemical shift values, other 2 units were also determined as Pip moieties. The sequence from an oxymethine proton 26-H (δH 4.43, δC 71.8) to a triplet methyl proton 29-H (δH 0.79) through a methine proton 27-H (δH 1.71), which was in turn coupled to a doublet methyl proton 30-H (δH 0.87), and a methylene proton 28-H (δH 1.23, 1.00) was established by the DQF-COSY spectrum. The 1H-13C long-range couplings from 26-H and 27-H to a carbonyl carbon C-25 (δC 174.9) established an isoleucic acid moiety. The connectivity of these partial structures was determined by 1H-13C long-range couplings from 6-H, 7-H, and 2-NH to an amide carbonyl carbon C-5, from 9-NH and 11-H to an amide carbonyl carbon C-10, from 14-NH and 16-H to an amide carbonyl carbon C-15 (δC 172.2), from 19-NH and 21-H to an amide carbonyl carbon C-20, and from 24-NH and 26-H to C-25, as shown in Figure 2. From the molecular formula of 1, the structure of the compound was established, as shown in Figure 1. The amino acid sequence of 1 was further confirmed by HR-ESI-MS/MS (Waters, SYNAPT G2 HDMS) data, as shown in Figure 3.
Structure elucidation of 2
Compound 2 was isolated as a colorless oil and its HR-ESI-MS was consistent with a molecular formula of C29H49N9O10. In the NMR spectral data for 2, the triplet methyl protons 29-H had disappeared and instead, 2 doublet methyl protons (δH 0.88 and 0.70) were observed. The 13C NMR spectrum of 2 also showed the disappearance of the methyl signal at C-29 in 1, which indicated the existence of a valinic acid residue. In the same way as in 1, the amino acid sequence of 2 was supported by HR-ESI-MS/MS data (Figure 3). These collective spectroscopic data proved that the isoleucic acid moiety in 1 was displaced by a valinic acid residue in 2.
Absolute configuration of 1
The absolute configuration of 1 was defined by Marfey's method [14] applied for the acid hydrolysate of 1 in comparison with standard amino acids. Compound 1 (1.0 mg) was hydrolyzed in 6N HCl at 110°C for 12h. After the compound was concentrated to dryness, the residue was dissolved in 10 ml of EtOAc-H2O (1: 1). The amino acid mixture recovered in the aqueous layer was dried in vacuo and was added to 5% NaHCO3 (500 µl) and 0.2 mg Nα-(5-fluoro-2,4-dinitrophenyl)-lalaninamide (FDAA) in acetone (500 µl). The solution was heated in an oil bath at 80°C for 3h. The reaction products were analyzed by the ultra performance liquid chromatography (UPLC) system (Waters) as follows: column, Acquity UPLC BEH C18 column (2.1 i.d. × 50 mm, Waters); flow rate, 0.3 ml min-1; solvent, 25% aqueous MeCN containing 0.1% formic acid to detect α-MeSer. The retention times of FDAA derivatives were analyzed by LC-MS monitoring using negative mode (m/z = 370). Retention times of the standard FDAA derivatives were as follows: (R)-α-MeSer, 1.30 min and (S)-α-MeSer, 1.53 min. The chromatogram of the hydrolysate derivatives showed a peak corresponding to (R)-α-MeSer. To determine the absolute configuration of three piperazic acids, the reaction products were analyzed by the UPLC system using 10-100% (7 min) aqueous MeCN containing 0.1% formic acid. Retention times of the standard FDAA derivatives of (R) - (S)-piperazic acid were 2.36 min and 2.55 min, respectively. The chromatogram of the hydrolysate derivatives showed that the ratio of peaks corresponding to (R)-piperazic acid and (S)-piperazic acid are 2: 1. Although two (R)-piperazic acids and a (S)-piperazic acid were determined in 1, the sequence of three piperazic acids are unknown. The analysis of the order of each piperazic acid unit is presently underway.
The isoleucic acid moiety dissolved in the organic layer was dried in vacuo and was added to 0.2 mg p-bromophenacyl bromide and trace amounts of potassium fluoride in DMF (100 µl). The mixture was heated in an oil bath at 50°C for 12h. The reaction product was analyzed by HPLC as follows: column, Daicel CHIRAL PAK IC column (4.6 i.d. × 250 mm, Daicel, Osaka, Japan); flow rate, 1.0 ml min-1; solvent, hexane- isopropanol (9: 1). The retention times of p-bromophenacyl adducts were analyzed by UV (254nm) monitoring. The retention times of the standard p-bromophenacyl derivatives were as follows: l-isoleucic acid, 25 min; d-isoleucic acid, 40 min; allo-l-isoleucic acid, 28 min; and allod- isoleucic acid, 49 min. The retention time of the p-bromophenacyl adduct of the hydrolysate in the chromatogram showed a corresponding peak to l-isoleucic acid.
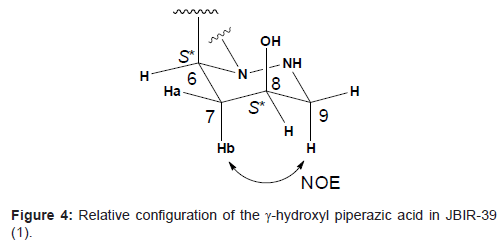
The relative configuration of the γ-hydroxyl piperazic acid ring was established from 3JH-H coupling constants. Since the both spin coupling constants between the α-methine proton 6-H and methylene protons 7-H were relatively small (3J6H-7Ha = 2.3 Hz and 3J6H-7Ha = 5.5 Hz, respectively), 6-H was deduced to be in the equatrial location. The higher-fielded 1H chemical shift of 7-Hb (δH 1.94) comparing with that of lower-fielded 1H chemical shift of 7-Ha (δH 2.25), together with an NOE between 7-Hb and 9-H, supported that 7-Hb is in the axial location and the conformation of this six-membered ring structure is chair form. In addition, both relatively small coupling constants between 7-Ha and 8-H (3J7Ha-8H = 2.6 Hz) and between 7-Hb and 8-H (3J7Hb-8H = 6.5 Hz) suggested that 8-H is in the equatrial location. Thus, the relative configurations were established to be 6S* and 8S*, respectively as shown in Figure 4.
Conclusion
We isolated two novel hexapeptide compounds, 1 and 2, from the culture broth of sponge-derived Streptomyces sp. Sp080513SC-24. The structures of 1 and 2 possessed an Α-methylserine moiety, a γ-hydroxyl piperazic acid moiety, three piperazic acid moieties, and an isoleucic acid moiety or a valinic acid moiety. The structures of 1 and 2 were found to be related to piperidamycin F and D isolated from the artificially streptomycin or rifampicin resistant strains of a soilisolated Streptomyces species [15]. However, Sp080513SC-24 showed sensitivity to streptomycin (MIC = 1 µg/ml) or rifampicin (MIC = 30 µg/ ml). Furthermore, piperazimycins possessing the Pip and γ-OH-Pip moieties have been isolated from a marine-originated Streptomyces [16]. These compounds have been reported to show antimicrobial and cytotoxic activities [15,16]. Therefore, we attempted to investigate the cytotoxic and antimicrobial activities of 1 and 2. The results showed that 1 and 2 did not exhibit cytotoxic activity against several cancer cell lines nor did they show antibacterial activities against Micrococcus luteus and Escherichia coli (data not shown). The results of this study confirm that this sponge contains undiscovered microorganisms that possess the ability to produce new substances. We anticipate that this study will convince chemists that new species of Streptomyces can produce compounds containing unique skeletal structures and also encourage them to investigate such species.
Acknowledgements
This work was supported by a grant from the New Energy and Industrial Technology Department Organization (NEDO) of Japan, a Grant-in-Aid for Scientific Research (20380070 to K.S.) from the Japan Society for the Promotion of Science (JSPS), and the Ministry of Education, Culture, Sports, Science and Technology (MEXT).
References
- Fenical W, Jensen PR (2006) Developing a new resource for drug discovery: marine actinomycete bacteria. Nat Chem Biol 2: 666-673.
- Hughes CC, MacMillan JB, Gaudêncio SP, Jensen PR, Fenical W (2009) The ammosamides: structures of cell cycle modulators from a marine-derived Streptomyces species. Angew Chem Int Ed Engl 48: 725-727.
- Lee HS, Shin HJ, Jang KH, Kim TS, Oh KB, et al. (2005) Cyclic peptides of the nocardamine class from a marine-derived bacterium of the genus Streptomyces. J Nat Prod 68: 623-625.
- Mitova MI, Lang G, Wiese J, Imhoff JF (2008) Subinhibitory concentrations of antibiotics induce phenazine production in a marine Streptomyces sp. J Nat Prod 71: 824-827.
- Pimentel-Elardo SM, Gulder TAM, Hentschel U, Bringmann G (2008) Cebulactams A1 and A2, new macrolactams isolated from Saccharopolyspora cebuensis, the first obligate marine strain of the genus Saccharopolyspora. Tetrahedron Lett 49: 6889-6892.
- Izumikawa M, Khan ST, Komaki H, Takagi M, Shin-ya K (2010) JBIR-31, a new teleocidin analog, produced by salt-requiring Streptomyces sp. NBRC 105896 isolated from a marine sponge. J Antibiot 63: 33-36.
- Khan ST, Izumikawa M, Motohasi K, Mukai A, Takagi, M, et al. (2010) Distribution of 3-hydroxyl-3-methylglutaryl coenzyme A reductase gene and isoprenoid production in marine-derived Actinobacteria. FEMS Microbiol Lett 304: 89-96.
- Khan ST, Tamura T, Takagi M, Shin-Ya K (2010) Streptomyces tateyamensis sp. nov., Streptomyces marinus sp. nov. and Streptomyces haliclonae sp. nov., three novel species of Streptomyces isolated from marine sponge Haliclona sp. Int J Syst Evol Microbiol 12:2775-2779
- Ueda JY, Khan ST, Takagi M, Shin-ya K (2010) JBIR-58, a new salicylamide derivative, isolated from a marine sponge-derived Streptomyces sp. SpD081030ME-02. J Antibiot 63: 267-269.
- Takagi M, Motohashi K, Khan ST, Hahimoto J, Shin-ya K (2010) JBIR-65, a new diterpene, isolated from a sponge-derived Actinomadura sp. SpB081030SC-15. J Antibiot 63: 401-403.
- Khan ST Higher incidence of discovery of novel compounds from novel members of Streptomyces associated with a marine sponge Haliclona sp. Environ Microbiol in press.
- Motohashi K, Takagi M, Shin-ya K (2010) Tetracenoquinocin and 5-iminoaranciamycin from a sponge-derived Streptomyces sp. Sp080513GE-26. J Nat Prod 73: 755-758.
- Motohashi K, Takagi M, Shin-ya K (2010) Tetrapeptides possessing a unique skeleton, JBIR-34 and JBIR-35, isolated from a sponge-derived actinomycete, Streptomyces sp. Sp080513GE-23. J Nat Prod 73: 226-228.
- Bhushan R, Bruckner H (2004) Marfey's reagent for chiral amino acid analysis: a review. Amino Acids 27: 231-247.
- Hosaka T, Ohnishi-Kameyama M, Muramatsu H, Murakami K, Tsurumi Y, et al. (2009) Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat Biotechnol 27: 462-464.
- Miller ED, Kauffman CA, Jensen PR, Fenical W (2007) Piperazimycins: cytotoxic hexadepsipeptides from a marine-derived bacterium of the genus Streptomyces. J Org Chem 72: 323-330.
Relevant Topics
- Algal Blooms
- Blue Carbon Sequestration
- Brackish Water
- Catfish
- Coral Bleaching
- Coral Reefs
- Deep Sea Fish
- Deep Sea Mining
- Ichthyoplankton
- Mangrove Ecosystem
- Marine Engineering
- Marine Fisheries
- Marine Mammal Research
- Marine Microbiome Analysis
- Marine Pollution
- Marine Reptiles
- Marine Science
- Ocean Currents
- Photoendosymbiosis
- Reef Biology
- Sea Food
- Sea Grass
- Sea Transportation
- Seaweed
Recommended Journals
Article Tools
Article Usage
- Total views: 15293
- [From(publication date):
May-2011 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 10678
- PDF downloads : 4615