Research Article Open Access
Isolation of a Novel Antibiotic Resistance Plasmid DNA from Hospital Isolates of Pseudomonas aeruginosa
Erfaneh Jafari, Mohammad Reza Shakibaie* and Leila PoormasoomiDepartment of Microbiology and Immunology, Kerman University of Medical Sciences, End of 22 Bahman BLVD, 76167-14111, Kerman, Iran
- *Corresponding Author:
- Mohammad Reza Shakibaie
Department of Microbiology and Immunology
Kerman University of Medical Sciences
End of 22 Bahman BLVD, 76167-14111, Kerman, Iran
Tel: +989133408226
Fax: +983413221671
E-mail: mohammadreza.shakibaie@gmail.com
Received Date: May 30, 2013; Accepted Date: June 14, 2013; Published Date: June 16, 2013
Citation: Jafar E, Shakibaie MR, Poormasoomi L (2013) Isolation of a Novel Antibiotic Resistance Plasmid DNA from Hospital Isolates of Pseudomonas aeruginosa. J Clin Exp Pathol 3:140. doi: 10.4172/2161-0681.1000140
Copyright: © 2013 Jafar E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Clinical & Experimental Pathology
Abstract
Emergence and dissemination of antibiotic resistance plasmids are major concern for hospital care system and increases the cost and decreases effectiveness of available antibiotics used in treatment of hospitalized patients. In this study two Pseudomonas aeruginosa, two Escherichia coli, and a Klebsiella pneumoniae were isolated from Intensive Care Unit (ICU) of a university hospital, in Kerman, Iran. K. pneumoniae exhibited resistance to all antibiotics routinely used in our hospital for treatment of patients except meropenem, while, the other isolates were sensitive to carbapenems and ciprofloxacin. Plasmid analysis of the selected isolates showed the presence of a single plasmid with molecular weight 65Kb in the P. aeruginosa isolate1. The plasmid was named as pKUM and belonged to incompatibility group -2 (IncP-2). Conjugation by filter mating revealed that resistance associated with gentamicin, kanamycin, cefotaxime, and ceftazidime phenotypes were transferred to E. coli ATCC25922 (Rifr) recipient cells at the frequencies of 3.13 × 10-5 and 5.3 ×10-7 respectively. The results were further supported by curing and transformation experiments. MIC to cefepime was ≥30 μg/mL both in donor as well as the transconjugants while decreased to 0.5μg/ mL in cured derivative. The plasmid pKUM was quite stable (86%) in both donor cells and the recipient. From above results we concluded that resistances to third generation of cephalosporins, aminoglycosides and cefepime in P. aeruginosa isolate1 were indeed encoded by a conjugative plasmid. Acquisition of cefepime resistance through plasmid complicates the therapy of neutropenic patients in ICU and increases the cost and mortality of these patients.
Keywords
Pseudomonas aeruginosa; Antibiotic resistance; Plasmid; Conjugation; Transformation; Curing
Introduction
Pseudomonas aeruginosa is gram negative short rod belong to family Pseudomonacaeae. It is motile, oxidase positive, non-spore forming bacteria and grows on simple as well as complex medium. P. aeruginosa occupy very important position as nosocomial pathogen and cause serious infections like fulminative septicemia, meningitis or pneumonia in patients hospitalized in the hospitals across the world. This organism is also responsible with high mortality and morbidity in patients with impaired immune system and cystic fibrosis [1].
Antibiotic resistance associated with hospital isolates of P. aeruginosa created serious health care concerns particularly in the ICU where seriously ill patients are hospitalized [2]. Multiple Drug Resistance (MDR) in P. aeruginosa is defined as the resistance to 3 or 4 of the following antibiotic classes; penicillins, cephalosporins, monobactams, carbapenems, aminoglycosides, and quinolones. These strains constantly cumulate several resistance mechanisms as a consequence of multiple genetic events such as chromosomal mutations or horizontal transfers of resistance genes [3,4]. Some of these are widely prevalent in southern Europe, Turkey, and Southeast Asia [1].
Different plasmids have been reported in this bacterium mediate resistance to third generation of cephalosporins and penicillins through β- lactamase enzymes [5]. Cefepime has an extended spectrum of activity against Gram-positive and Gram-negative bacteria than third generation agents. Cefepime is usually reserved to treat moderate to severe nosocomial pneumonia, infections caused by multiple antibiotic resistant P. aeruginosa and empirical treatment of febrile neutropenia [6].
Similarly, most studies have indicated that around 10% of P. aeruginosa isolates are aminoglycosides resistant [7]. Plasmids have been reported to carry genes for enzyme aminoglycosides, acetyltransferase and phosphotransferase [7,8]. In one investigation two clinical isolates of P. aeruginosa, was found to transfer gentamicin resistance to other Pseudomonas by conjugation rate of 1 × 10-3, but not to E. coli or other enterobacteriaceae [9].
Antibiotic resistance phenotypes and plasmid content of 35 multiple drug resistant P. aeruginosa strains were showed that 10 isolates exhibited high level resistance to both gentamicin and tobramycin [10]. Briand et al. [11] isolated a plasmid was transferable to P. aeruginosa with a transfer frequency between 10−5 to 10−7 per recipient strains and also to E. coli K12.
The resistant pattern and antibiotic susceptibility of P. aeruginosa have been changing over the past years particularly, in ICU and surgical wounds [12]. A study was undertaken to characterise P. aeruginosa strains isolated from burned patients in Tehran, Iran indicated 98% isolatedstrains were resistant to cefoxitin, 97% to cefotetan, 93% to ticarcillin, 89% to ticarcillin/clavulanic acid, 76% to gentamicin and imipenem, 63% to piperacillin, 49% to tetracycline, and 20% to meropenem [13].
In previous study we reported emergence of ciprofloxacin resistance phenotype among P. aeruginosa isolated from burn patients [14]. In this investigation we isolated an antibiotic resistant plasmid in P. aeruginosa carried resistance to third generation of cephalosporins; cefepime and aminoglycosides.This organism is a major concern in nosocomial infections and should therefore be monitored in surveillance studies.
Materials and Methods
Bacterial source and identification
Five multiple drug resistance nosocomial pathogens were isolated from ICU patients hospitalized in Afzalipoor hospital, Kerman, Iran. Patients were either admitted directly to the ICU or transferred from other wards, namely surgery, obstetrics and pediatric wards. Postoperative patients requiring ventilation were admitted to the critical care unit, while patients with medical conditions necessitating ventilation were admitted to the ICU. The patients showed sign such as high fever, bacterimia and sever chill. Samples were collected from blood of each patient and inoculated in to 5 mL Stuart Transport medium (ST) and transferred to microbiology laboratory for further analysis. 200 µl of the samples were inoculated onto MacConkey and 5% sheep blood agar medium (Merck, Germany) and identified according to standard biochemical tests [2]. The identified isolates were mixed with 40% glycerol in True North™ Cryogenic Vials (TNC) containing 1mL sterile Trypticase Soy Broth (TSB) and preserved at -70°C for further investigation.
Antibiotic susceptibility
Antibiotic sensitivity of above isolates was determined by disk diffusion method of Bauer et al. [15] on Mueller-Hinton agar [MHA] (Hi-Media, India) using commercially available paper disks (Padtan-Teb, Iran). Antibiotics both anti-pseudomonal and non-antipseudomonal were used in the following concentrations (in µg mL-1): Nalidixic acid (NA) [30 µg], Ceftazidime (CAZ) [30 µg], Imipenem (IPM) [10 µg], sulfametoxazole (SXT) [10 µg], Ciprofloxacin (CP) [5 µg], Tetracycline (Te) [30 µg], Chloramphenicol (C) [30 µg], Amoxicillin (AMX) [25 µg], Cefotaxime (CTX) [30 µg], CAZ + clavulanic acid (CZA) [30µg + 10 µg], Gentamicin (Gm) [10 µg], Amikacin (AN) [30 µg] and Kanamycin (Km) [10 µg] . Zone of inhibition surrounding each disk was measured and labeled as resistance, intermediate, sensitive according to CLSI procedure [16,17]. In case of E-test, inoculums preparation and plating, strip application, and subsequent minimum inhibitory concentration (MIC) determinations were carried out in accordance with the manufacturer’s instructions and CLSI guidelines. E. coli ATCC25922 was included as a control strain for susceptibility testing.
Development of rifampicin resistant mutants
For induction of rifampicin mutation, 1mL of the overnight culture of E. coli ATCC25922 has spread throughout nutrient agar plate containing gradient concentration of rifampicin (Sigma grade) and incubated at 37°C for 2 days. The colonies grown on the highest gradient concentration of the rifampicin were selected and restreaked on Muller-Hinton agar plate supplemented with 100 µg/mL rifampicin. Mutant strain was designated as E. coli ATCC25922 (Rifr). The rifampicin was selected because the resistant gene located on the chromosome and all isolates were susceptible to this antibiotic. The geneunity of the mutants was confirmed by existence of metallic sheen on EMB plate.
Plasmid DNA extraction
Extraction of the plasmid DNA from all isolates was carried out using alkaline lysis method [18] and observed on 0.7% agarose gel (Merck-Germany). Electrophoresis was conducted in horizontal bed apparatus for 3 hours at 60 mA using 500 mL 1mM Tris- Borate- EDTA (TBE × 1) buffer (pH-8.3). Plasmid bands were photographed by a camera attached UV gel documentation system (UV Tech- Cambridge) after stained with 0.5 µg/mL ethidium bromide for 5 minutes. The molecular weight of the plasmid was determined by running λ phage DNA as ladder digested with HindIII restriction enzyme.
Conjugation and plasmid transformation
Conjugation between P. aeruginosa isolate1 as donor and rifampicin-resistant E. coli ATCC 25922 (Rifr) as recipient cells was carried out by membrane filter technique as described previously [19]. The transconjugants were selected on Muller – Hinton agar medium containing CTX (30 µg/mL) + Rif (100 µg/mL) and Gm (30 µg/mL) + Rif (100 µg/mL). Conjugation frequency was calculated as the number of transconjugants divided by the recipient cells multiply dilution factor. Simultaneously, a control of conjugation was carried out along side of the test.
Transformation procedure was adopted as described by Sambrook et al. [18] with some modification. Briefly, an E. coli ATCC25922 cell was made recombinant deficient (recA-) by exposing cells to subinhibitory concentration of ethidium bromide (0.25 µg/mL) and UV irradiation at 260nm as described by Clark [20]. The cells were made competent for transformation by addition of 200µL of 50 mM ice cold calcium chloride (CaCl2) [Merck, Germany] to 500 µL of log phase bacterial suspension for 30 minutes on ice powder. The suspension was transferred to a microfuge tube and centrifuged at 8000 rpm for 10 minutes. The cell pellet was resuspended in 200µL of 50 mM ice cold CaCl2 followed by addition of 400µL extracted of plasmid. Microfuge tubes incubated for additional 5min in laboratory temperature. 100 µL of the transformed cells were spread onto selective medium (MHA containing 100µg/mL CTX and 30µg/mL Gm separately) and on nonselective medium. The petri plates incubated overnight at 37°C and colonies were checked for presence of the plasmid.
Plasmid curing and stability
Curing experiments were performed using ciprofloxacin, SDS, Eucalyptus plant extract and two temperature 42°C and 44°C as curing agents. Briefly, overnight culture of P. aeruginosa isolate 1 was grown in presence of sub-inhibitory concentration of curing agents for 24 hours at 37°C. A loopful of the organism from the highest concentration of curing agents streaked on MacConkey agar to obtain isolated colonies. In case of high temperature, the overnight growth of the organism was streaked on nutrient agar plates and incubated at 42°C and 44°C separately for 48 hours. The individual colonies were inoculated and incubate at the same temperatures. The colonies from all curing agents were then replica plated by sterile toothpick on MHA medium containing antibiotics (CAZ 100 µg/mL & Gm 30 µg/mL) and on nonselective medium. The colonies that failed to grow on selective medium were considered as putative cured derivatives. The physical loss of plasmid in the cured derivatives was confirmed by agarose gel electrophoresis. The percentage curing was estimated as number of colonies with cured phenotype per 100 colonies tested. Similarly, stability of the plasmid was investigated in presence and absence of antibiotic CTX at intervals of 1, 2, 4, 6 and 24 hours for 1000 generation as described by Lee et al. [21].
Plasmid incompatibility
Plasmid incompatibility was carried out by method of Eaton and Rawlings [22]. Plasmid containing E. coli ATCC25922 (Rifr) cells were transformed with a second plasmid belong to IncP, IncW, IncQ and IncC (received from institute pasture, Iran branch) and plated on nutrient agar plates with antibiotic selection for both plasmids. Single colonies were inoculated in to 20 mL sterile nutrient broth incubated at 37°C for 24 hours. Survival of the plasmids was then tested by removing selection for both plasmids and growing the cells in 5mL nutrient broth for 100 generations, with transfer of approximately 1,000 cells to fresh medium at 20 generation intervals. Finally, 70 colonies were replica plated to antibiotic containing nutrient agar plates to score for plasmid retention. Cells containing individual plasmids were similarly grown and plated as a control to account for spontaneous plasmid loss.
Results and Discussion
Over all resistance patterns of the ICU isolates showed that, P. aeruginosa isolate1 had broad spectrum resistance to all antibiotics routinely used in our hospital for treatment of patients except imipenem, meropenem, ciprofloxacin, chloramphenicol and it was intermediate to tetracycline (Table 1). It also exhibited MIC >5 µg/mL to piperacillin/ tazobactam and >30 to cefepime (Table 2). The isolate was also resistant to amikacin and ceftazidime + clavulanic acid. P. aeruginosa isolate2 was susceptible tomeropenem, imipenem, chloramphenicol and tetracycline. The reason behind of chloramphenicol sensitivity of both P. aeruginosa strains might be due to discontinued administration of this antibiotic in our ICU. In case of K. pneumoniae, it was only sensitive to meropenem, tetracycline and resistant to all 14 antibiotics tested. Similarly E. coli isolate1 and E. coli isolate2 were both sensitive to nalidixic acid, imipenem, meropenem, piperacillin/tazobactam, amikacin and ciprofloxacin. In addition, E. coli isolate2 was sensitive to ceftazidime and gentamicin and intermediate to kanamycin (Table 1). Our data indicated that the MDR strains of bacteria are dominant in our ICU and supported the administration of the antibiotics with high antipseudomonal activity, particularly ciprofloxacin and carbapenems. The other antibiotics especially third generation of cephalosporins, cefepime and aminoglycosides were ineffective in routine therapy of infected patients in our hospital.
| Antibiotic | P.aeruginosa isolate 1 | P.aeruginosa isolate 2 | K. pneumoniae | E.coli isolate 1 | E.coli isolate 2 | E.coli ATCC 25922 |
|---|---|---|---|---|---|---|
| NA | R | R | R | S | S | S |
| CAZ | R | R | R | R | S | S |
| MEN | S | S | S | S | S | S |
| IPM | S | S | R | S | S | S |
| SXT | R | R | R | R | I | S |
| CP | S | R | R | S | S | S |
| Te | I | S | S | R | R | S |
| AMX | R | R | R | R | R | R |
| CTX | R | R | R | R | R | S |
| Gm | R | R | R | R | S | S |
| Km | R | R | R | R | I | S |
| C | S | S | R | I | S | S |
| CPM | R | ND | ND | ND | ND | S |
| PIT | R | R | R | S | S | S |
Mueller – Hinton agar was used for antimicrobial disk diffusion susceptibility
testing. CFU was adjusted to 106
NA: Nalidixic Acid; CAZ: Ceftazidime; MEN: Meropenem; IPM:
Imipeneme; SXT: Trimethoprim; CP: Ciprofloxacin; Te: Tetracycline;
AMX: Amoxicillin; CTX: Cefotaxime; Gm: Gentamicin; Km: Kanamycin;
C: Chloramphenicol; PIT: Piperacillin/Tazobactam; CPM: Cefepime
S: Sensitive; I: Intermediate; R: Resistant; ND: Not determined
Table 1: Antibiotic resistance pattern of nosocomial bacteria to 14 antibiotics isolated from ICU patients hospitalized in the Afzalipoor hospital, Kerman, Iran.
| Antibiotic | P.aeruginosa isolate1 | MIC(µg/mL) E.coli transconjugants | P.aeruginosa Plasmid Cured |
|---|---|---|---|
| C | 0.5 | 0.5 | 0.5 |
| Gm | >5 | >5 | 0.1 |
| PIT | >5 | >5 | 0.5 |
| CPM | >30 | >30 | 1.0 |
C: Chloramphenicol; Gm: Gentamicin; PIT: Piperacillin / Tazobactam; CPM:
Cefepime;
MIC: Minimum Inhibitory Concentration;
The inoculums size was 108 CFU/mL.
Muller-Hinton agar was used for MIC determination
Table 2: Minimum Inhibitory Concentration (MIC) of four antibiotics against P.aeruginosa isolate1, E.coliATCC25922 (Rifr) transconjugants and plasmid cured derivative.
Many researches were conducted across the globe on emergence of multiple drug resistance P. aeruginosa strains isolated from clinical samples, in one study 94% were susceptible to imipenem, 90.2% to ciprofloxacin, 89% to amikacin and 78% to ceftazidime but 82% were resistant ofloxacin, 58% to pefloxacin and 35% to gentamicin [23].
Conjugation between our ICU isolates as donors and E. coli ATCC 25922 Rifr as a recipient revealed that only P. aeruginosa isolate1 could transferred CTX and Gm resistant phenotype to the recipient cells with frequency of 3.13 × 10-5 and 3.2 × 10-5 respectively (Table 3), however, other nosocomial pathogens could not transfer any resistant gene to the above recipient. Co- transfer study of antibiotic resistant phenotypes in the transconjugantsshowed that CAZ, and Km resistant phenotypes were co-transferred along with CTX and Gm (Table 3). Furthermore, MIC to CPM and PTI increased from 0.5 to 30 and >5 µg/mL respectively E. coli ATCC 25922 Rifr transconjugants exhibited following antibiotic susceptibility pattern by disk diffusion; SXT s, CP s, IPM s, NA s, Te s, Gmr, CAZ r, CPM r CTX rand Rif r. Our data are in agreement with work carried out previously by the other authors [24,25]
| Donor cells | Recipient� � cells | Selective medium | Conjugation frequency | phenotype Co-transferred |
|---|---|---|---|---|
| P. aeruginosa isolate1 PITr, CAZr, CPMr, CTXr, Gmr, Kmr |
E.coli ATCC 25922 (Rifr) |
MHA* +
CTX(30�µg/mL) Rif(100�µg/mL) & Gm(30�µg/mL) Rif(100�µg/mL |
3.13Ã?Â?10-5
3.2 Ã?Â? 10-5 |
CAZ, Km |
*MHA: Muller � Hinton agar.
A control for conjugation was carried out and it was 1 x 10-9.
Conjugation was carried out by membrane filter technique with ratio of 2:1 for donor
and recipient cells
Table 3: Conjugation between multiple drugresistance P.aeruginosa isolate 1 as donor and E.coli ATCC 25922(Rifr) as recipient by membrane filter method.
To confirm presence of the plasmid in both P. aeruginosa isolate1 and the E. coli ATCC 25922 Rifr transconjugants, we performed plasmid isolation and subsequent agarose gel electrophoresis. It was found that both organisms carried65 Kb plasmid band (Figure 1). The isolated plasmid was designated as pKUM.
Figure 1: Agarose gel electrophoresis of the plasmid pKUM isolated from P.aeruginosa isolate1. The electrophoresis was conducted in 0.7% agarose gel and stained with 0.5 μg/mL ethidium bromide. The gel was then observed on the UV transilluminatior. The plasmid band was marked by arrow. Lane 1: P.aeruginosa isolate1 wild type, Lane 2: Plasmid pKUM in E.coli ATCC 25922(Rif r) transconjugants, Lane 3: Plasmid pKUM in E.coli transformants, Lane 4: Plasmid pKUM in kmr transformants, Lane 5: P. aeruginosa isolate1 cured derivative.
Plasmid pKUM was further transformed to recA-E. coli ATCC 25922 with a frequency of 5.3×10-7 as shown in (Table 4) Transformation resulted in simultaneous transfer of CTX, Gm, Km and CAZ resistant phenotypes to the transformants. This data were further supported by existence of a plasmid band in the transformants (Figure 1). Transfer of these resistant genes creates serious challenge for physician in treating ICU patients. Similarly, Our data showed that conjugative plasmid in nosocomial bacteria especially in ICU can easily transferred to unrelated species and may resulted in emergence of pan-resistant strains, where no longer any antibiotics could be used. This may create huge problem for low income countries in Africa or Latin America [26].
Curing of plasmid pKUM showed that, SDS (400 µg/mL) had highest percentage of the curing activity (1.38%), while exposing cells to temperature 42°C did not cure the plasmid at all. Eucalyptus extract and ciprofloxacin could cure plasmid with 1.25 and 1.19% respectively. Similarly, loss of antibiotic resistance at 44°C was 1.13%. The curing of the antibiotic resistance phenotypes was indeed associated with physical loss of plasmid on agarose gel as shown in Figure 1.
In one study in India, ethidium bromide was used for plasmid curing [27]. Curing and transformation experiments showed that resistance to amikacin was plasmid mediated. It was noted that the isolates that lost plasmids became susceptible to amikacin, chloramphenicol and tetracycline, while remained resistant to carbenicillin, clindamycin, ampicillin and co-trimoxazole. Isolation of P. aeruginosa strain from pulmonary brush of a patient hospitalized in a suburb of Paris, France [28] revealed that plasmid was transferred by conjugation to rifampin resistant P. aeruginosa pU21 at a frequency of 2 ×10-7 but not to rifampin resistant E. coli K-12 C600.
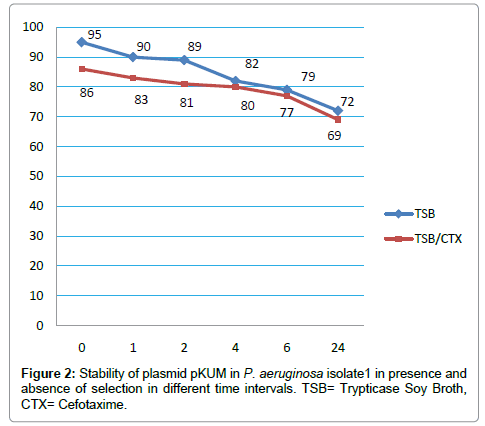
Plasmid incompatibility and stability revealed that pKUM plasmid was belong to IncP-2 group and was quite stable in both donor cells as well as transconjugants. The stability of the plasmid in P. aeruginosa isolate1 was significantly high in absence of CTX in the beginning of the experiment (99%) however; it decreased to 72% after 24 hours of incubation. In presence of CTX, the stability of pKUM plasmid decreased steadily from 86% to 69% (Figure 2).
Previous studies of antibiotic susceptility among hospital isolates in Kerman indicated sensitivity of isolated bacteria to third generation of cephalosporins, carbapenems and ciprofloxacin [29], however, emergence and dissemination of resistance to imipenem, cefepime and ciprofloxacin in our hospital through horizontal gene transfer must be prevented by routine antibiotic surveillance so as to maximize the possibility of administering an effective therapeutic regime whenever there is a need.
Conclusion
As the above results indicated, it can be concluded that among ICU isolates, only P. aeruginosa isolate1 was carried a MDR plasmid. The unique features of the pKUM plasmid were different resistances genes encoded, high stability, low curing efficiency and easy transferred through conjugation to the other nosocomial pathogen. The acquisition of cefepime, third generation of cephalosporins and aminoglycosides resistances through plasmid may complicate the therapy of the patients in ICU and increase the cost, mortality and prolonging hospitalization in the hospital.
Acknowledgements
Authors would like to thank the staff of Department of Microbiology, Kerman University of Medical Sciences for help and providing facilities for this research.
References
- Livermore DM (2002) Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis 34: 634-640.
- Arruda EA, Marinho IS, Boulos M, Sinto SI, Caiaffa HH, et al. (1999) Nosocomial infections caused by multiresistant Pseudomonas aeruginosa. Infect Control Hosp Epidemiol 20: 620-623.
- Lambert PA (2002) Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med 95 Suppl 41: 22-26.
- Gastmeier P, Schwab F, Bärwolff S, Rüden H, Grundmann H (2006) Correlation between the genetic diversity of nosocomial pathogens and their survival time in intensive care units. J Hosp Infect 62: 181-186.
- E Paramythiotou, C Lucet, J Franc, O Timsit, D Vanjak, et al. (2004) Acquisition of Multidrug-Resistant Pseudomonas aeruginosa in Patients in Intensive Care Units: Role of Antibiotics with Antipseudomonal Activity. Clinic Infection Diseases 38: 670–677.
- Chapman TM, Perry CM (2003) Cefepime: a review of its use in the management of hospitalized patients with pneumonia. Am J Respir Med 2: 75-107.
- Hancock RE, Speert DP (2000) Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist Updat 3: 247-255.
- Shahid M, Malik A (2005) Resistance due to aminoglycoside modifying enzymes in Pseudomonas aeruginosa isolates from burns patients. Indian J Med Res 122: 324-329.
- Jacoby GA (1974) Properties of R plasmids determining gentamicin resistance by acetylation in Pseudomonas aeruginosa. Antimicrob Agents Chemother 6: 239-252.
- Tsakris A, Vatopoulos AC, Tzouvelekis LS, Legakis NJ (1992) Diversity of resistance phenotypes and plasmid analysis in multi-resistant 0:12 Pseudomonas aeruginosa. Eur J Epidemiol 8: 865-870.
- Michel-Briand Y, Dupont MJ, Chardon-Loriaux I, Jouvenot M (1981) Isolation of an antibiotic multiresistance plasmid from Pseudomonas aeruginosa. J Antimicrob Chemother 7: 371-378.
- L Poirel, T Naas, N Nicolas, L Collet, S Bellais,, et al. (2000) Nordmann. Characterization of VIM-2, a Carbapenem hydrolyzing metallo-β–Lactamase and its plasmid- and integron-borne gene froma Pseudomonas aeruginosa clinical isolate in France. Antimicrobial Agents and Chemotherapy 44:891–897.
- Smith S, Ganiyu O, John R, Fowora M, Akinsinde K, et al. (2012) Antimicrobial resistance and molecular typing of pseudomonas aeruginosa isolated from surgical wounds in Lagos, Nigeria. Acta Med Iran 50: 433-438.
- R Ranjbar, P Owlia, H Saderi, S Mansouri, N Jonaidi-Jafari ( 2011) Arjomandzadegan. Characterization of Pseudomonas aeruginosa strains isolated from burned patients hospitalized in a major burn center in Tehran, Iran. Acta MedicaIranica 49: 675-679.
- M.R Shakibaie, S. Adeli, Y. Nikian (2001) Emergence of Ciprofloxacin resistance among Pseudomonas aeruginosa isolated from burn patients, Iranian.Journal of Medical Sciences 26: 155-159.
- Bauer AW, Kirby WM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45: 493-496.
- CLSI (2009) Methods for dilution antimicrobial susceptibility testing of bacteria grow aerobically. Approved standard M7-A7. Clinical and Laboratory Standards Institute Wayne PA, USA.
- MR Shakibaie, KA Jalilzadeh, SM Yamakanamardi (2009) Horizontal transfer of antibiotic resistance genes among gram negative bacteria in sewage and lake water and influence of some physico-chemical parameters of water on conjugation process, Environmental Biology 30: 45-49.
- Sambrook, Fritsch, Maniatis (1988) "Lysis by alkali" Molecular Cloning: A Lab Manual. (2ndedn), Cold Spring Harbor Lab Press. New York, USA.
- Shakibaie MR, Kapadnis BP, Dhakephalker P, Chopade BA (1999) Removal of silver from photographic wastewater effluent using Acinetobacter baumannii BL54. Can J Microbiol 45: 995-1000.
- Lee SY, Yim KS, Chang HN, Chang YK (1994) Construction of plasmids, estimation of plasmid stability, and use of stable plasmids for the production of poly(3-hydroxybutyric acid) by recombinant Escherichia coli. J Biotechnol 32: 203-211.
- Loftie-Eaton W, Rawlings DE (2009) Comparative biology of two natural variants of the IncQ-2 family plasmids, pRAS3.1 and pRAS3.2. J Bacteriol 191: 6436-6446.
- A Olayinka, B Olayinka, B Onile (2009) Antibiotic susceptibility a plasmid pattern of Pseudomonas aeruginosa from the surgical unit of university teaching hospital in north central Nigeria. Journal of Medical Sciences 1: 79-83.
- N Gaouar-Borsali, M Gaouar-Yadi, Z. Babaahmed,, M.Drissi (2012) Antibiotic resistance study of some clinical strains of Pseudomonas aeruginosa characterization by conjugation and cleaning out of plasmid. Der Pharma Chemica 4: 1160-1163.
- Strateva T, Yordanov D (2009) Pseudomonas aeruginosa - a phenomenon of bacterial resistance. J Med Microbiol 58: 1133-1148.
- Blahova J, Hupkova–Lesicka M, Kralikova K, Krcmery V Sr, Kubonova K, et al. (1998) Further studies of transferable antibiotic resistance in strains of Pseudomonas aeruginosa from four clinical settings in three countries. J Chemother 10: 215-220.
- E Raja, G Selvam (2009) Plasmid profile and curing analysis of Pseudomonas aeruginosa as metal resistant. International Journal of Environmental Sciences and Technology 6: 259-266.
- Poirel L, Girlich D, Naas T, Nordmann P (2001) OXA-28, an extended-spectrum variant of OXA-10 beta-lactamase from Pseudomonas aeruginosa and its plasmid- and integron-located gene. Antimicrob Agents Chemother 45: 447-453.
- H Shikh-Bardsiri, MR Shakibaie (2013) Antibiotic Resistance Pattern among Biofilm Producing and Non Producing Proteus Strains Isolated from Hospitalized Patients; Matter of Hospital Hygiene and Antimicrobial Stewardship. Pakistan Journal of Biological Science 16: 1496-1502.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 16056
- [From(publication date):
June-2013 - Oct 19, 2025] - Breakdown by view type
- HTML page views : 11197
- PDF downloads : 4859