Research Article Open Access
Isolation and Identification of A Novel Aporphine Alkaloid SSV, An Antitumor Antibiotic from Fermented Broth of Marine Associated Streptomyces sp. KS1908
Sunanda Kadiri1*, Nagendra Sastry Yarla2 and Siddaiah Vidavalur11Department of Organic Chemistry and FDW, Andhra University, Visakhapatnam, 530 003, India
2Department of Biochemistry and Bioinformatics, School of Life Sciences, Institute of Science, GITAM University, Visakhapatnam, 530045, India
- *Corresponding Author:
- Sunanda Kadiri
Department of Organic Chemistry and FDW
Andhra University, Visakhapatnam
530 003, India
Tel: +91-0891-2844683
Fax: +91-0891-2525611
E-mail: sunandakadiri65@gmail.com
Received date: September 26, 2013; Accepted date: October 24, 2013; Published date: October 29, 2013
Citation: Kadiri S, Yarla NS, Vidavalur S, Duddukuri GR (2013) Isolation and Identification of A Novel Aporphine Alkaloid SSV, An Novel Antitumor Antibiotic from Fermented Broth of Marine Associated Streptomyces sp. KS1908. J Marine Sci Res Dev 3:137. doi: 10.4172/2155-9910.1000137
Copyright: © 2013 Kadiri S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Marine Science: Research & Development
Abstract
A marine actinomycete, Streptomyces sp. KS1908 was isolated from a marine sediment sample collected in
the Bay of Bengal, India and identified based on morphological, cultural, physiological, biochemical characteristics,along with the cell wall analysis and 16S rRNA gene sequence analysis. Strain KS1908 has more similarities with Streptomyces albus but some variations were observed. A novel aporphine alkaloid SSV was isolated according to bioactivity guided fractionation of fermented broth through solvent extraction and chromatography. Chemical structure of the aporphine alkaloid SSV was elucidated on the basis of spectroscopic analysis, including two-dimensional (2D) NMR and HR-ESI-MS data. Aporphine alkaloid SSV showed significant antibacterial, antifungal activity and also possesses considerable anticancer activity against human larynx (HEp-2), cervical (HeLa), leukemia HL-60 and MCF-7 breast cancer cell lines.
Keywords
Antitumor antibiotic; Streptomyces sp. KS1908; Aporphine alkaloid SSV; Fermented broth
Introduction
Streptomycetes have been shown to possess the ability to synthesize antibacterial, antifungal, insecticidal, antitumor [1-5], antiinflammatory, anti-parasitic, antiviral, anti-infective, antioxidant and herbicidal compounds [1,2,6-9]. Hence, these are widely recognized as industrially important microorganisms [10]. Moreover, approximately 60% of the antibiotics discovered in the year 1990 and most of the antibiotics are from the genus Streptomyces [11]. These characteristics make this genus an important research area. Earlier literature suggests that many antimicrobial molecules have been isolated from Streptomyces albus. Salinomycin, a new polyether antibiotic was produced by strain of Streptomyces albus ATCC 21838 [12]. A new macromolecular peptide antibiotic, named AN-1 was isolated from the culture broth of Streptomyces albus AJ 9003 [13]. An antibiotic complex identical to Paulomycins A and B active against multiple resistant strains of staphylococci and other gram-positive bacteria was isolated from cultures of Streptomyces albus G [14]. In present study, bioactive actinomycete was collected from marine sediment and identified as Streptomyces sp. KS 1908 further responsible antibiotic was isolated and spectroscopic assignment of structure was demonstrated here.
Isolation and Taxonomy
Marine sediment samples were collected at Bay of Bengal near Gangavaram Coast, Visakhapatnam, India. Sample serially diluted to isolate as pure culture on a starch casein agar (SCA-soluble starch, 10.0 g; vitamin free casein, 0.3 g; KNO3, 2.0 g; NaCl, 2.0 g; K2HPO4, 2.0 g; MgSO4•7H2O, 0.05 g; CaCO3, 0.02 g; FeSO4•7H2O, 0.01 g; agar, 20.0 g; sterilized natural aged sea water, 1.0 L; pH, 7.2; supplemented with rifampicin 25 μg/ml and cycloheximide 75 μg/ml to inhibit bacterial and fungal contamination, respectively) plate, which had been seeded with a sediment sample suspension and incubated at 28°C for 14 days [10]. The isolated actinomycete colonies being filamentous, compact, often leathery giving a conical appearance, and maintained on YEME (Yeast Extract Malt Extract) slants at 4°C and as a glycerol suspension (20%, v/v) at −20°C [15]. This pure culture was later used for taxonomic and bioactivity studies.
Taxonomic identification was done by physiological conditions, biochemical tests, chemo taxonomic investigations and molecular characterization. Morphological observation through macroscopic based on growth pattern on different media like yeast malt extract agar (ISP-2), Oatmeal agar (ISP-3), Inorganic salts starch agar (ISP- 4), Glycerol asparagines agar (ISP-5), tryptone yeast glucose agar, peptone agar, and nutrient agar. The color of aerial mycelium, substrate mycelium and soluble pigment were observed by naked eye. Optical and Scanning electron microscopies ((JSM-6610LV, JEOL Ltd.) were used for microscopic observations. Organism growth conditions were studied on SCA at various pH, Temperature and NaCl levels for physiological characterization [16]. Organism was biochemically characterized using the tests viz., Enzymes, H2S production tests, Carbon and nitrogen utilization tests using different substrates12. Cell wall chemical composition was demonstrated according to the procedures of Lechevalier [17] for chemotaxonomic investigation17. Molecular characterization was done by 16s rRNA gene sequencing [16,18,19].
Fermentation
Sporulated isolate prepared with sterile water according to 0.5Mc Farland standard. The resulted spore suspension at 10% level was transferred aseptically into a 250 ml Erlenmeyer flask containing 45 ml of the inoculation medium [8]. The flasks were inoculated and incubated at 28°C for 48 hrs at 120 rpm. Thoroughly washed pellet containing cell mass was suspended in sterile 0.9% sodium chloride solution and used as inoculum was then transferred to the modified production medium [8] and incubated at 30°C for 96 hrs at 160 rpm on a rotary shaker. The fermented broth was used for extraction of the active principle.
Bioactivity guided fractionation and purification
The fermented broth was aseptically collected in a sterile centrifuge tube and centrifuged at 4000 rpm for 15 min at 4°C. The culture filtrate (supernatant) and mycelial pellet obtained were extracted separately for identification of the active principle source. Antibiotics from the cell mass were isolated usually by extraction with polar and nonpolar solvents while that from the fermented medium were extracted by solvent extraction only when the antibiotic has a reasonably high degree of solubility in non-polar organic solvents. Bioautography was performed to identify bioactive fraction. The other alternative technique for the separation of bioactive principle from the culture filtrate is the adsorption of the compound on some inert material like silica [20].
Compound identification and Structure elucidation
Thin Layer Chromatography (TLC) was analyzed on the glass percolated silica gel plates GF254, and spots were checked by UV light, Iodine and spraying with 10% sulfuric acid in methanol followed with heating. The melting point was determined on Fisher-Johns melting point apparatus. FT-IR spectra were recorded on a Perkin- Elmer spectrophotometer with KBr pellet. The sample was scanned between 400 and 4000 cm-1 wave number. High Resolution Mass Spectrum (HRMS) was recorded on QSTAR XL, HYBRID MS System and EI MS was recorded on VG 7070H (70 eV). The NMR data of purified compound was acquired using an AMX-400 spectrometer (Bruker, Rheinstetten, Germany). 1H NMR spectra were obtained at 400.13 MHz and 13C NMR spectra were obtained at 100.6 MHz. All NMR spectra were recorded in DMSO –d6. The chemical shifts were expressed in δ (ppm) using DMSO-d6 as solvent and TMS as internal reference. Dragendorff’s reagent test was used for the identification of alkaloid [21].
Antimicrobial assay
Anti-microbial studies were carried out on clinical isolates of human pathogenic bacteria and dermatophytic fungi, Salmonella typhi, Vibrio cholerae, Shigella dysenteriae, Enterococcus faecalis are gastrointestinal pathogens, which were collected at King Gerorge Hospital, Visakhapatnam, India. Proteus vulgaris NCIM 2813, Pseudomonas aeruginosa NCIM 5031 cultures were collected from NCL, Pune, India. Staphylococcus aureus MTCC 7443, Bacillus subtilis MTCC 8141, Aspergillus niger MTCC 6484, Aspergillus awamori MTCC 7711, Candida albicans MTCC 1346, Trichophyton rubrum. MTCC 3272 cultures were collected from Indian microbial technology, India. Clinical isolate of Candida albicans was collected from skin lesions. Zone of inhibitions were determined using agar well diffusion method ad Minimum Inhibitory Concentration (MIC) was done by broth dilution assay. Microbial broth cultures (Mueller Hinton broth for bacteria, Sabouraud Dextrose broth for fungi) were adjusted the absorbance to 0.6 (Optical Density at 620 nm) in Spectrophotometer according to CLSI guidelines. These cultures were used as Inoculums. The agar plates were prepared by pour plate method using 20 ml of sterilized agar medium (MH agar for bacteria, SD agar for fungi). The sterile agar medium was cooled to 45°C and mixed thoroughly with 1ml of growth culture of concerned test organism (inoculum) and then poured into the sterile petri dishes and allowed to solidify. Wells of 6 mm size were made with sterile cork borer and test compounds were added. The agar plates were incubated at for 4 days at 28°C for fungi wile 24 hours at 37°C for bacteria. Zone of inhibitions were measured by Himedia milli meter zone reader. MIC was performed on broth media (10 ml) containing 1000-1 μg/ml of test compound prepared by 10 fold dilution. 0.1 ml of culture inoculums was added. The MIC was determined at which concentration of compound causes nil absorbance (no growth) in the spectrophotometer at 620 nm. All the experiments were conducted according to Clinical Laboratory Standard Institute. Ciprofloxacin (for bacteria) and fluconazole (for fungi) were antibiotics used as positive control [22-26].
MTT assay for anticancer activity
The cytotoxic activities of the compound (SSV) isolated from Streptomyces sp. KS1908, were tested against human larynx carcinoma cells HEp-2, cervical cancer (HeLa), human leukemia HL-60 and MCFbreast carcinoma cell lines. This cell line was obtained from TRIMS and National Centre for Cell Sciences (NCCS), India further cultured at 37°C with 5% CO2, using MEM(minimum essential medium) medium. MTT assay [8] was used to study the cytotoxic properties of the sample. 200 μl of cell culture (2×104 cells/ml) was added in each well containing 100 μl MEM medium in a 96 well plate. After 24 hrs of incubation 20 μl of different test concentrations like 1000 μg /ml, 500 μg /ml, 250 μg /ml, 125 μg /ml, 62.5 μg /ml were added to the respective wells. After incubation of 4 days at 37°C temperature and 5% CO2 in a CO2 incubator. 20 μl of MTT (3-(4, 5- dimethyl-2-thiozolyl)-2,5- diphenyl-2H-tetrazolium bromide) reagent 5mg/ml concentration, was added to each well and incubated for 4hrs. Then the medium was carefully discarded and the Formazan complexes formed in the cells were dissolved in 200 μl of DMSO (Dimethyl sulphoxide). The rate of color appearance was measured at 570 nm in a spectrophotometer. The percent of cell inhibitions were calculated based on control and test absorbance values. Results were expressed as IC50 means concentration of compound where 50% of cancer cell growth inhibition occurred. The experimental measurements were made in five replicates. Camptothecin was used as the standard [27,28].
Molecular docking
Topoisomerase I (PDB ID 1T8I) and IIA (PDB ID 2XCT) crystal structures of proteins were obtained from Protein Data Bank. Cocrystallized ligands and water molecules are removed from target protein using Argus lab. Ligands are prepared using Chemoffice (Cambridge). Energy minimization was done using molecular mechanics. The minimized was executed until root mean square value reached smaller than 0.001 Kcal/mol. Such energy minimized ligands and receptor used for docking studies using Molegro Virtual Docker [29].
Results
Isolation and Taxonomy of isolated marine actinomycete
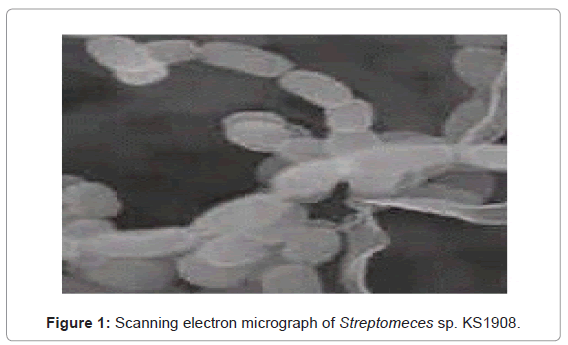
The isolated bioactive actinomycete colonies being filamentous, compact, often leathery giving a conical appearance, dry surface on SCA, which can easily be distinguished from fungi and non filamentous bacteria. Morphological and cultural observations of the isolate grown on different ISP media given in Table 1, revealed that vegetative mycelium showed yellow-brown color, aerial hyphae were abundant, well-developed with white color on different test media and substrate mycelium with pale yellow color. It didn’t produce any pigments but faint yellow color pigmentation on Yeast-malt extract agar (ISP-2). The scanning electron micrograph of the strain KS1908 revealed that aerial mycelia were monopodially branched with compact spirals of sporophore terminating in long open coils. Each spore chain consisted of 8-20 white, oblong to cylindrical shaped spores, 0.6 ∼ 0.7 x 0.8 ∼ 0.9 μm in size, having smooth surface (Figure 1). The chemotaxonomic investigations revealed that the cell wall peptidoglycan of isolate contained L-diaminopimelic acid and glycine. This indicates that isolate belongs to cell wall type I which is characteristic of the genus Streptomyces.
| Medium | Characteristics | ||||
|---|---|---|---|---|---|
| Growth | Vegetative mycelia | Aerial mycelia | Spore mass | Soluble pigment | |
| Nutrient agar | Good | Moderate, pale yellow | Moderate, white | Poor, white | Nil |
| Yeast – malt extract agar (ISP-2) | Abundant | Moderate, yellow | Abundant, white | Moderate, white | Faint yellow |
| Oatmeal agar (ISP-3) | Abundant | Good, pale-yellow | Abundant, white | Moderate, white | Nil |
| norganic salts starch agar (ISP-4) | Good | Moderate, yellow | Good, white | Poor, white | Nil |
| Glycerol asparagines agar (ISP-5) | Good | Moderate, yellow | Good, white | Moderate, white | Nil |
| Tyrosine agar (ISP-7) | Moderate | Moderate, pale-yellow | Good, white | Poor, white | Nil |
| Yeast – starch aga | Moderate to good | Good, pale yellow | Good, white | Moderate, white | Nil |
| Sucrose – nitrate agar | Moderate | Moderate, pale yellow | Moderate, white | Poor, white | Nil |
Table 1: Cultural characteristics of the isolated Streptomyces sp. KS1908.
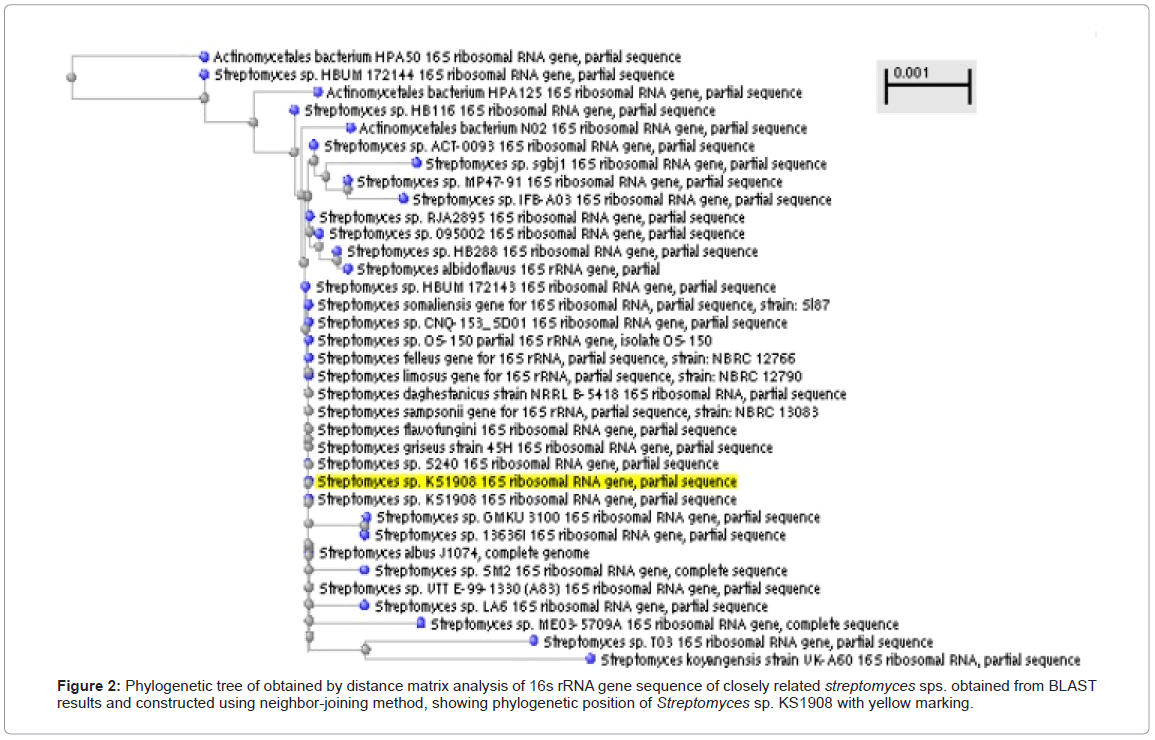
16S rRNA gene sequence analysis (Genbank accession no. KC556777) and other cultural, biochemical physiological, chemotaxonomic characteristics revealed that Strain KS1908 has close similarities with Streptomyces albus [12] (Figure 2 and Tables 1 and 2) but some variations were observed so named as Streptomyces SP. KS 1908.
Bioactivity guided fractionation and purification
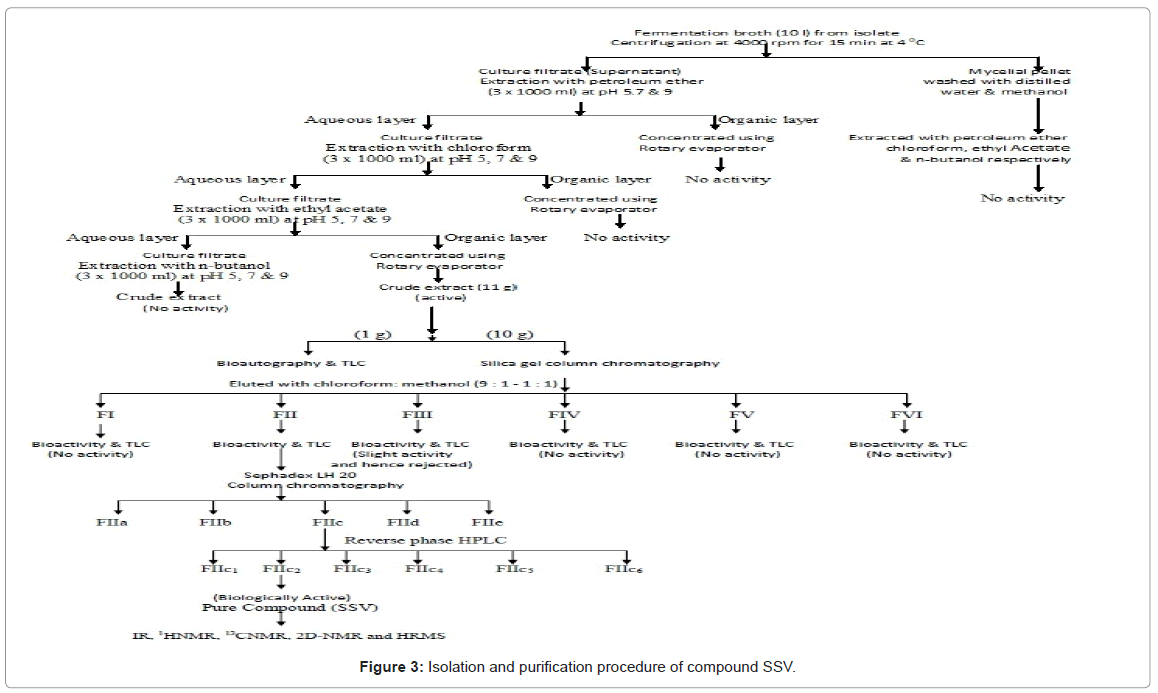
Fermented broth of sterptomyces sp. KS1908 was extracted using various solvents but only ethyl acetate extract showed bioactivity. Then ethyl acetate extract was run by chromatography using silica gel to obtain bioactive fraction II (255 mg), which was further fractionated with Sephadex LH-20 column and separated into five major fractions that included with fraction IIc. Preparative reverse phase HPLC was used to get light brown colored pure bioactive compound SSV in FIIc2 fraction. Schematic representation of detailed fractionation and purification was given in Figure 3.
Structural elucidation of aporphine alkaloid SSV
Compound SSV was obtained as light brown needles having melting point 68-72°C; [α] [25 ]D -28.0 (c = 1.5, MeOH). Dragendorff’s reagent test showed positive [16]. The molecular formula of SSV was assigned as C19H19NO4 from its elemental and mass spectral analyses (HRMS: m/z 326.1386 [M+H]+, 348.1206 [M+Na]+). This was corroborated by the decoupled [13] C-NMR spectrum which showed signals for all the nineteen carbons of the molecule (Table 3). The mass fragmentation pattern of compound SSV showed typical of aprophine alkaloid [17]. The IR absorption bands at 3430 (OH), 945 (-O-CH2-O-) cm-1 indicated the presence of hydroxyl and methylenedioxy groups.
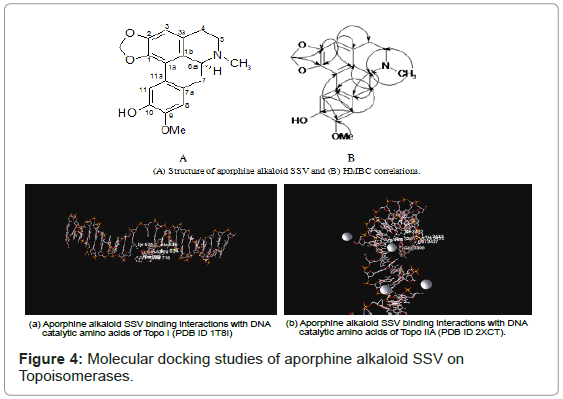
The proton NMR spectrum of compound SSV showed three, one proton singlet peaks at δ 7.55, 6.75 and 6.57 corresponding to H-11, H-8 and H-3 respectively of an aporphine alkaloid (Table 3). The two, one proton singlet peaks at δ 6.11 and 5.97 indicated the presence of one methylenedioxy group on C1-C2 and two, three proton singlet peaks at δ 3.77 and 2.42 attributed to one methoxyl group and one N-methyl group, respectively. The proton NMR spectrum also showed a one proton singlet peak at δ 9.19 attributed to hydroxyl group. Further the correlations observed in HMQC, HMBC and HSQC confirmed that methylenedioxy group was present at C-1 and C-2 carbons, methoxyl group was present on C-9 and free hydroxyl group was present on C-10 (Table 3). Thus from the foregoing spectral studies, the structure of compound SSV was established as 10-hydroxy-9-methoxy-1,2-methylenedioxy-6-methyl- 4,5,6,6a-tetrahydro-7H,6-azabenzanthrene (Figure 4).
Antimicrobial and anticancer activities of aporphine alkaloid SSV
Aporphine alkaloid SSV showed good antimicrobial activity especially on multi drug resistant clinical isolates including bacteria and fungi. Aporphine alkaloid SSV was more effective against bacteria than fungi.
As shown in Table 4, zone of inhibition found to be between 9-14 mm at 30 μg of compound. MIC range found to be between 1-100 μg/ ml. 14 mm was the inhibitory zone showed by SSV on S. typhi and with lowest MIC of 1 μg/ml. Aporphine alkaloid SSV showed potent antibacterial activity against both gram positive and gram negative bacteria. Compound SSV showed very effective activity against gastrointestinal pathogenic bacteria (S. typhi, V. cholerae, E. faecalis and E. coli). Dermatophytic fungi (T. rubrum and C. albicans) showed slight resistance. Aporphine alkaloid SSV showed comparable antimicrobial potency with ciprofloxacin and flucanozole (antibiotics).
Anticancer activity of aporphine alkaloid SSV
Anticancer activity of compound SSV performed by MTT assay and found significant cytotoxic activity on human larynx (HEp-2), cervical (HeLa), human leukemia HL-60 and MCF-breast cancer cell lines with the IC50 values of 1.10, 1.13, 2.85 and 4.44 μg/ml, respectively. Aporphine alkaloid SSV showed comparable potency with camptothecin (Table 5).
Docking of aporphine alkaloid SSV
Molecular docking studies of aporphine alkaloid SSV were on topoisomerases using Molegro Virtual Docker. Docked energy or binding energy was inversely proportional to affinity of compounds towards enzyme. Lower binding energy indicated higher binding affinity. -98.4 kcal was the binding energy of aporphine alkaloid SSV on topo II, which was less than docked score of ciprofloxacin (-79.7 Kcal/ mol). Aporphine alkaloid SSV showed comparable binding energy (-78.6 Kcal/mol) with camptothecin (-77.1 Kcal/mol) on topoisomerase I. Figure 4, demonstrated that Compound SSV binding interactions with DNA catalytic residues of topoisomerase I and II enzymes.
Discussion
Multidrug resistance is the one of major problem in treatment of various microbial infectious diseases and cancer. There is a necessity to develop novel drugs with high potency and less toxic. Most of the antitumor antibiotics have been isolated from marine microbes especially Streptomyces sp. These antitumor antibiotics are commonly interacting with DNA to cause cell death [5]. Anthracyclines are a special class of antitumor antibiotics which act through topoisomerase II inhibition [30]. Pimprinine, an extracellular alkaloid has been isolated from the culture filtrate of Streptomyces CDRIL-312 [31]. Alkaloid group of aporphine antibiotics are topoisomerase I inhibitors from Streptomyces sp [32].
In conclusion, bioactive streptomyces sp. KS1908 was isolated and characterized from marine associated actinomycetes further novel aporphine alkaloid SSV is an antitumor antibiotic which was isolated from bioactive fraction of fermented broth and chemically characterized through advanced spectroscopic data. Therefore, the isolated aporphine alkaloid SSV can be promising agent for treatment of cancer and microbial infections.
Acknowledgements
Sunanda Kadiri thankful to Department of Science and Technology (DST) for the financial assistance by sanctioning a project under the women scientist scheme with Grant No. WOS-A/LS-74/2009. We acknowledged Bioserve, India for help in gene sequencing and Phylogenetic tree construction.
References
- Berdy J (2005) Bioactive microbial metabolites. J Antibiot 58:1-26.
- Sacramento DR, Coelho RRR, Wigg MD, Linhares LFTL, Santos MGM, et al. (2004) Antimicrobial and antiviral activities of an actinomycete (Streptomyces sp.) isolated from a Brazilian tropical forest soil. World J Microbiol Biotechnol20: 225-229.
- Prabavathy VR, Mathivanan N, Murugesan K (2006) Control of blast and sheath blight diseases of rice using antifungal metabolites produced by Streptomyces sp. PM5. Biol Control 39: 313-319.
- Ebrahimi ZM, Shahidi BGH, Padasht DF, Ayatollahi MSA, Rashid FP, et al. (2009) Biological control of rice blast (Magnaporthe oryzae) by use of Streptomyces sindeneus is isolate 263 in greenhouse. Am J Appl Sci6: 194-199.
- Galm U, Martin HH, Steven GVL, Jianhua J, Jon ST, et al. (2005) Antitumor Antibiotics: Bleomycin, Enediynes, and Mitomycin. Chem Rev 105: 739-758.
- Pimentel-Elardo SM, Kozytska S, Bugni TS, Ireland CM, Moll H, et al. (2010) Antiparasitic compounds from Streptomyces sp. strains isolated from Mediterranean sponges. Mar Drugs 8: 373-380.
- Rahman H, Austin B, Mitchell WJ, Morris PC, Jamieson DJ, et al. (2010) Novel Anti-Infective Compounds from Marine Bacteria. Mar Drugs8: 498-518.
- Teppei K, Miho I, Misa O, Hideki Y, Masayuki H, et al. (2012) JBIR-94 and JBIR-125, Antioxidative Phenolic Compounds from Streptomyces sp. R56-07. J Nat Prod 75: 107-110.
- Sousa CS, Soares ACF, Garrido MS (2008) Characterization of Streptomyceteswith potential to promote plant growth and biocontrol. Sci Agric (Piracicaba, Braz) 65: 50-55.
- Ramesh S, Mathivanan N (2009) Screening of marine actinomycetes isolated from the Bay of Bengal, India for antimicrobial activity and industrial enzymes. World J Microbiol Biotechnol 25: 2103-2111.
- Ramesh S, William A (2012) Marine actinomycetes: An ongoing source of novel bioactive metabolitesMicrobiol Res 167: 571-580.
- Yohko K, Shojiroh M, Yukiko O, Akio S (1980) Antibiotic no. 6016, a polyether antibiotic. J Antibiot 33: 1437-1442.
- Shigeyoshi M, Takao K, Hiroshiro S, Tsuyoshi S (1983) The fermentation, Isolation and Characterization of a Macromolecular peptide antibiotic: AN-1. J Antibiot37: 27-32.
- Majer J, Chater KF (1987) Streptomyces albus G produces an antibiotic complex identical to paulomycins A & B. J Gen Microbiol 133: 2503-2507.
- Williams ST, Cross T (1971) Methods in Microbiology. New York: Academic press.
- Lu J, Ma Y, Liang J, Xing Y, Xi T, et al. (2012) Aureolic acids from a marine-derived Streptomyces sp. WBF16. Microbiol Res67: 590-595.
- Lechevalier HA, Lechevalier MP (1970) A critical evaluation of the genera of Aerobic actinomycetes. 393- 405.
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406-425.
- Sharma AD, Singh J (2005) A non enzymztic method to isolate genomic DNA from bacteria and actinomycetes. Anal Biochem 337: 354-356.
- Venkata RRKD, Murali KKM, Murali YN, Sri RRD (2012) Novel Pyridinium compound from marine actinomycete, Amycolatopsis alba var. nov. DVR D4 showing antimicrobial and cytotoxic activities in vitro. Microbiol Res 67: 346- 351.
- Coe FG, Parikh DM, Johnson CA (2010) Alkaloid presence and brine shrimp (Artemia salina) bioassay of medicinal species of eastern Nicaragua. Pharm Biol 48: 439-445.
- Mackie, Mac cartney (1969) Practical Medical Microbiology, Churchill Livingstone Publisher (Elsevier);14: 141.
- Taplin D, Zaias N, Rebell G, Blank H (1969) Isolation and recognition of dermatophytes on a new medium (DTM). Arch Dermatol99: 203-209.
- Perez C, Pauli M, Bazevque P (1990) An antibiotic assay by the agar well diffusion method. Acta Biologiae et MedicineExperimentalis 15: 113-115.
- Clinical Laboratory Standards Institute Methods for Dilution Antimicrobial Susceptibility Test for Bacteria That Grow Aerobically; Approved Standard-Ninth Edition. M07-A9, 32: 12-20.
- Pfaller MA, Castanheira M, Diekema DJ, Messer SA, Moet GJ, et al. (2010) Comparison of European Committee on Antimicrobial Susceptibility Testing (EUCAST) and E test Methods with the CLSI Broth Microdilution Method for Echinocandin Susceptibility Testing of Candida Species. J Clin Microbiol 48: 1592-1599.
- Mosmann T (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods65: 55-63.
- Govinda Rao Duddukuri, Nagendra Sastry Y, Kaladhar DSVGK, Ajay Babu P, Kamalakara Rao K, et al. (2011) preliminary studies on in vitroanti-tumor activity of tender seed coat extract of borassus flabellifer l. On HeLa cell line. International Journal of Current Research3: 075-077.
- Rene T, Mikael HC (2006) MolDock: A New Technique for High-Accuracy Molecular Docking. J Med Chem 49: 3315 -3321.
- Fujiwara A, Hoshino T, Westley JW (1985) Anthracycline Antibiotics. Critical Reviews in Biotechnology 3: 133-157.
- Naik SR, Harindran J, Varde AB (2001) Pimprinine, an extracellular alkaloid produced by Streptomyces CDRIL-312: fermentation, isolation and pharmacological activity. J Biotechnol 88: 1-10.
- García MT, Blazquez MA (2011) New alkaloid antibiotics that target the DNA topoisomerase I of Streptococcus pneumoniae. J Biol Chem 286: 6402-6413.
Relevant Topics
- Algal Blooms
- Blue Carbon Sequestration
- Brackish Water
- Catfish
- Coral Bleaching
- Coral Reefs
- Deep Sea Fish
- Deep Sea Mining
- Ichthyoplankton
- Mangrove Ecosystem
- Marine Engineering
- Marine Fisheries
- Marine Mammal Research
- Marine Microbiome Analysis
- Marine Pollution
- Marine Reptiles
- Marine Science
- Ocean Currents
- Photoendosymbiosis
- Reef Biology
- Sea Food
- Sea Grass
- Sea Transportation
- Seaweed
Recommended Journals
Article Tools
Article Usage
- Total views: 16408
- [From(publication date):
December-2013 - Nov 20, 2025] - Breakdown by view type
- HTML page views : 11488
- PDF downloads : 4920