Research Article Open Access
Is the Low-Resolution Mass Spectrometer Capable of Detecting Cancer at Early Stages?
Thais MG de Francisco1, João C Gasparetto1, Caroline P Uber1, Luize ZB Carrenho1, Noemi Nagata2, Almeriane W Santos3, Letícia B Cerqueira1, Francinete R Campos1* and Roberto Pontarolo1
1Department of Pharmacy, Universidade Federal do Paraná, Curitiba, Brazil
2Department of Chemistry, Universidade Federal do Paraná, Curitiba, Brazil
3Medical Pathology Department, Universidade Federal do Paraná, Curitiba, Brazil
- *Corresponding Author:
- Campos FR
Department of Pharmacy
Universidade Federal do Paraná
632 Lothário Meissner Avenue
80210-170, Curitiba–PR, Brazil
Tel: +55-41-33604076
Fax: +55-41-3360-4106
E-mail: francampos@ufpr.br
Received date: December 04, 2012; Accepted date: December 21, 2012; Published date: December 31, 2012
Citation: de Francisco TMG, Gasparetto JC, Uber CP, Carrenho LZB, Nagata N, et al. (2013) Is the Low-Resolution Mass Spectrometer Capable of Detecting Cancer at Early Stages? J Anal Bioanal Tech 4:158. doi: 10.4172/2155-9872.1000158
Copyright: © 2013 de Francisco TMG, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
It is known that cancer promotes changes in the metabolites found in biofluids and, therefore, the early detection of these metabolites allows for the best prognosis of the disease. Since ESI-MS fingerprinting has proved to be an excellent tool for ‘‘first pass’’ metabolome analysis of complex biological samples, we have presented, for the first time, the use of direct infusion electrospray ionization low-resolution mass spectrometry associated with chemometric analysis (ESI-DIMS-PCA) for the detection of early stage cancer in mice. Ehrlich tumors were induced in Balb-C and Swiss mice, and blood was collected 1, 2, 3, 5, 7 and 10 days after tumor induction. Data from the mass spectra were analyzed via principal component analysis (PCA). Sample classifications were obtained within 3 days of tumor induction, which reflects a precocious time for cancer detection. ESI-DIMS-PCA was also used to detect cancer in groups of Swiss mice inoculated with sarcoma 180. Sample classifications were obtained 3 days after tumor induction, which indicates ESI-DIMS-PCA can be used to detect distinct neoplasms. Loading plot analysis demonstrated that the ions differentiating between the experimental and control groups are lysophosphatidylcholines and phosphatidylcholines. Partial least-squares analysis demonstrated that ESI-DIMS-PCA is capable of determining unknown samples in different stages of cancer development. ESI-DIMS with chemometric analysis is a promising technique for diagnosing cancer in the early stages without the need for invasive procedures.
Keywords
DIMS; Metabolomic fingerprinting; Early cancer detection; PCA
Introduction
It is known that cancer promotes characteristic changes in one or more of the metabolites found in biofluids. The early detection of these metabolites allows for the best prognosis and treatment of the disease and long-term survival [1].
Many approaches have been developed to detect cancer in its early stages; one such technique is the discovery of biomarkers [1,2]. In biomarkers discovery, metabolomics has been suggested as an important technology because it provides reliable information on biological systems [2].
The most commonly used techniques in metabolomics are nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS). Both techniques are capable of detecting a wide range of metabolites with relatively high specificity and reproducibility [3-7]. However, NMR has limited sensitivity and resolution and can, therefore, only safely detect the most abundant metabolites [8]. Therefore, MS using distinct ionization techniques is the primary candidate for fulfilling the criteria of metabolomic approaches, i.e., a wide range of detected masses with high selectivity and sensitivity [9]. Direct infusion mass spectrometry (DIMS) is considered to be a particularly powerful technique for sample fingerprinting because it provides quick processing, high reproducibility and rapid characterization in complex matrices [3,9-12]. Although DIMS has been explored for metabolomic analysis using low-resolution mass spectrometry, most applications generally involve metabolite profiling focusing on microbial and plant species [12-15]. DIMS has been used in clinical studies to diagnose and screen for inherited metabolic diseases and for biomarker discovery, but high-resolution mass spectrometry with time of flight (TOF-MS) analyzers are often used [16-18].
Regardless of the equipment used, DIMS analysis generates a large amount of data. Identifying patterns to differentiate between healthy subjects and patients with a complex disease using DIMS in this way is difficult. For this reason, chemometric analysis has been used to extract information from multivariate chemical data and principal component analysis (PCA) to reduce the data dimensionality without losing relevant information, which enables exploratory data analysis and visualization [7,19,20]. In summary, direct infusion mass spectrometry combined with chemometric analysis (ESI-DIMS-PCA) is a promising technique for biomarker discovery and early-stage cancer detection.
The aim of this work was to evaluate the use of ESI-DIMS-PCA for early cancer diagnosis in mice receiving an i.p. transplantation of Ehrlich and Sarcoma 180 tumor cells followed by the identification of possible biomarkers using HPLC-MS/MS. Compared to the methods for cancer detection described in the literature, the use of a low-resolution mass spectrometer with chemometric analysis may be a simple, low cost and robust alternative for detecting cancer in the early stages.
Experimental
Chemicals and reagents
Acetonitrile (HPLC grade) and formic acid (88.0%) were purchased from J. T. Baker Chemicals B. V. (Deventer, The Netherlands). Diethyl ether (99.0%), trifluoroacetic acid (99.0%), trichloroacetic acid (99.0%), penicillin G, streptomycin sulfate, dimethylsulphoxide and trypan blue solution (0.4%) was obtained from Sigma-Aldrich (St. Louis, USA). RPMI 1640 medium was purchased from Hy-media, (Tokio, Japan), and fetal calf serum from Cultilab (Campinas, Brazil). Ultrapure water was obtained using a Milli-Q purification system from Millipore (Bedford, USA).
Animals
Experiments were conducted using groups of 7-week-old Swiss and isogenic Balb-C male and female mice (22-30 g). The animals were housed under controlled temperature (22-24°C), humidity (45-65%) and light (12 h light/12 h dark, with lights on at 6:00 a.m.) and received both food and water ad libitum. All of the experimental protocols in this study involving animals were approved by the Ethics Committee of the Universidade Federal do Paraná.
Tumors cells
Erlich and Sarcoma 180 (S180) tumor cells were kept frozen at -80°C in RPMI 1640 medium supplemented with 10% fetal calf serum, 0.3 g mL-1 penicillin G, 50 I.U. mL-1 streptomycin sulfate and 10% dimethylsulphoxide. For the experiments, cells were quickly thawed and seeded in RPMI 1640 full medium for at least two passages (105 cells mL-1) during which their viability always exceeded 90% based on the trypan blue exclusion test. Tumor cells were maintained in the ascitic form by a weekly i.p. transplantation of 106 tumor cells into Swiss mice.
Tumor induction and serum sample collection
The ascitic fluid was removed by opening the belly and carefully collecting it with a sterile syringe. The cells were found to be more than 90.0% viable using the trypan blue dye exclusion method. The final concentration of the tumor cell suspensions were adjusted with 0.9% saline to 2×106 viable cells mL-1 [21].
The animals were deprived of food for 12 hours before blood collection, which occurred 1, 2, 3, 5, 7 and 10 days post-tumor injection. Each animal was anesthetized by the inhalation of diethyl ether before cardiac puncture. Approximately 0.5 to 1.0 mL of blood was collected from each animal before the animal was sacrificed by cervical dislocation. The blood samples were centrifuged for 6 min at 12,000 rpm at room temperature, and the serum was transferred to a new plastic tube. The samples were stored by freezing at -80°C.
A quality control (QC) sample was prepared by pooling an equal volume from each sample.
Serum sample preparation
The serum samples were thawed at room temperature, and a 50 μL aliquot was transferred to a 2 mL plastic centrifuge tube. A total of 100 μL of acetonitrile containing 0.1% trifluoroacetic acid (TFA) was added to this tube, and the samples were then incubated in an ice bath for 5 min. After cooling, the samples were vortexed for 10 min and centrifuged at 14,000 rpm for 5 min at 4°C using an Eppendorf centrifuge model 5810R (Hamburg, Germany).
The supernatant was transferred to new plastic centrifuge tube and evaporated at 40°C in a sample concentrator (Labconco CentriVap, Kansas City, USA). The samples were resuspended in 150 μL of acetonitrile/water/formic acid (70:30:0.1, v/v/v) before injecting into the mass spectrometer. A blank run was inserted between consecutive samples to identify any sample carryover and avoid cross contamination. A quality control (QC) sample was analyzed after every 5 samples to check the sample stability.
DIMS analysis
Mass spectrometry experiments were performed on an Applied Biosystems MDS Sciex API 3200 Triple Quadrupole Mass Spectrometer (Toronto, Canada) equipped with an ESI source. A Harvard 22 Dual Model syringe pump (Harvard Apparatus, South Natick, USA) with a flow rate of 10 μL min-1 was used to directly infuse the samples into the mass spectrometer. The ESI source was operated in the positive ion mode because more compounds can be ionized in this mode and because it is more widely used in serum metabolomics [22,23].
The following ion-source parameters were used: ion spray voltage (IS): 5500 V, curtain gas (CUR): 10 psi, nebulizer gas (GS1): 15 psi; declustering potential (DP): 40 eV; and entrance potential (EP): 10 eV. High-purity nitrogen was used as both the CUR and GS1 and was produced using a nitrogen generator from PEAK Scientific Instruments (Chicago, USA). The mass spectra were acquired over a scan range from 50 to 1800 Daltons by accumulating 100 scans of 3 seconds each. Data acquisition was performed using an MS Workstation with Analyst 1.4 software (ABI/Sciex).
HPLC-MS/MS analysis
Biomarker identification was accomplished via HPLC/MS-MS experimentation. The analyses were conducted using an Agilent 1200 HPLC System (Wilmington, USA) consisting of a G1312B binary pump, G1379B degasser and G1316B column oven. The HPLC was connected to a CTC Sample Manager (Model 2777, Waters Corporation, Milford, USA). The HPLC system was coupled to an Applied Biosystems MDS Sciex API 3200 Triple Quadrupole Mass Spectrometer (Toronto, Canada) equipped with an electrospray ionization (ESI) source operating in the positive ion mode. The analytical separations were achieved on a Poroshell 300SB-C18 2.1×75 mm (5 μm particle size) column coupled with a Poroshell 300SB-C18 2.1×12.5 mm (5 μm particle size) guard column. The injection volume was 10 μL, and the column temperature was maintained at 25°C. The mobile phase consisted of acetonitrile (A) and water containing 0.1% formic acid (B). The flow rate was maintained at 200 μL/min, and the gradient profile was as follows: t0-5 min:A=35%; t5.1-7 min:A=40%; t7.1-9 min:A=40%; t9.1-11 min:A=45%; t11.1-60 min:A=90%. The following mass spectrometer parameters were used: ion spray voltage (IS):5500 V; entrance potential (EP):10 eV; declustering potential (DP):40 eV; curtain gas (CUR):10 psi; nebulizer and auxiliary gases (GS1 and GS2): 40 psi. Nitrogen was used as the collision gas in the MS/MS experiments, and the collision energy (CE) was adjustable over the range from 20 eV to 40 eV.
Data analysis
All mass spectra data obtained from both the control and experimental groups were subtracted from those obtained from the blank injections. The primary DIMS parameters were set as follows: a mass range from 50-1800, mass window of 0.5, and noise elimination level of 5. Origin Pro 8 software (OriginLab) was used to convert the mass spectra into a variable table displaying the mass and associated intensities as columns for all of the samples. Principal components analysis (PCA) and partial least squares (PLS) were performed using PLS-Toolbox 3.0 (Eigenvector Research, Inc.) operated in Matlab® 7.0.1 (MathWorks, Inc.). The samples were classified using the mean-centered pre-processing data. Score plots of the principal components (PCs) were constructed to distinguish between the experimental and control groups and loadings plot allowed the visualization of the ions most relevant to group discrimination. PLS analyses were performed to model the data and predict the unknown samples.
Results and Discussion
In this work, groups of Balb-C and Swiss mice received separately i.p. injections of Sarcoma 180 and Ehrlich tumor cell suspensions. The possibility of early cancer detection in different stages of the disease was assessed using ESI-DIMS with chemometric analysis.
Mice have made several contributions to cancer diagnoses because there is scientific evidence that they have similar genetic, genomic, physiological, biochemical and metabolic profiles to humans [24]. Therefore, mouse models are the most commonly used animal model to elucidate carcinogenesis, detect cancer and translate this information to humans [25].
Ehrlich and Sarcoma 180 (S180) tumor cells were selected because they grow rapidly and progressively in all strains of mice, and short-term studies can thus be achieved. In addition, Ehrlich and S180 tumor cells are homogeneous and easily transplantable, which allows for reproducible models [26,27]. Therefore, these types of tumors were the first choice for evaluating the possibility of early cancer detection using ESI-DIMS-PCA.
After inoculating the animals with the tumor cells, serum samples from the both the experimental and control groups were obtained by intracardiac puncture. The mass spectra of serum samples of experimental and control groups were obtained by ESI-DIMS and compared via PCA to differentiate between the groups. Another experimental group was also maintained for survival evaluation.
The removal of high molecular weight proteins (HMWPs) from the blood serum was performed before the ESI-DIMS analyses. This treatment was absolutely essential because HMWPs overshadow lower weight proteins, which are the molecules commonly targeted as potential biomarkers [28-32]. Different protein extraction/precipitation procedures, such as using a 60% trichloroacetic acid solution (TCA), 100% acetonitrile and acetonitrile containing 0.1% trifluoroacetic acid (TFA), were performed to remove the HMWPs. It was discovered that proteins precipitated using 60% TCA did not adequately separate, and a gelatinous aspect was observed in these samples. Consequently, the samples could not be infused into the mass spectrometer.
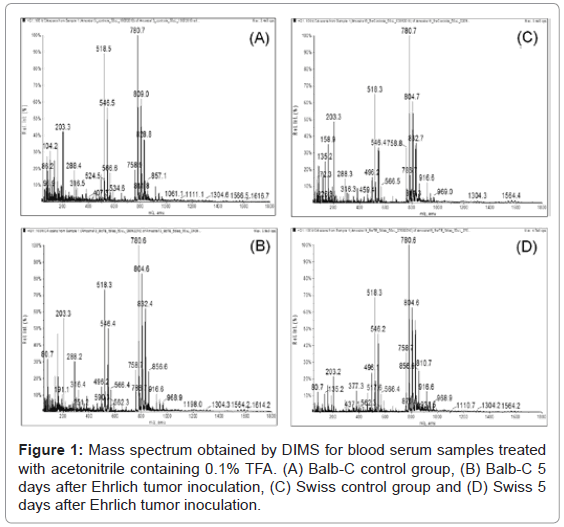
The use of 100% acetonitrile and acetonitrile in the presence of 0.1% TFA as the extracting/precipitating agent provided a rapid precipitation of the HMWPs and formed a dense precipitate that was easily removed by centrifugation. However, ions in the range between m/z 600 to 800 Da were only detected at high intensities in samples treated with acetonitrile containing 0.1% TFA (Figure 1). Therefore, acetonitrile containing 0.1% TFA was used for serum sample preparation.
After the HMWP depletion, analyses were conducted by directly infusing the samples into the mass spectrometer (DIMS). For each mass spectrum, approximately 17,000 ions were found, which makes visually distinguishing between the control and experimental groups impossible (Figure 1). Chemometric analyses were conducted using PCA to help interpret the intensity signals for each m/z in the mass spectra using scores plots for data classification and loadings plots to indicate the ions responsible for differentiation.
Sample classification and biomarker discovery
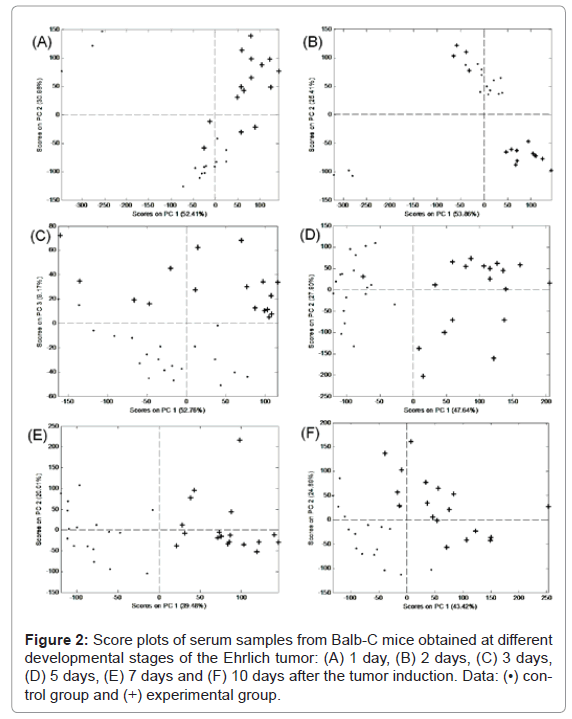
The PCA score plots obtained from the mass spectra for the Balb-C control vs. the experimental groups at different stages of Ehrlich tumor development are presented in figure 2. All data points refer to individual mice. PCA analysis was unable to separate the experimental and control groups both 1 and 2 days after the tumor induction (Figures 2A and 2B). However, a good differentiation between these groups was achieved after 3 days (Figures 2C-2F).
After 3 days of analyses, three principal components (PCs) were responsible for 61.93% of data variance. After 5, 7 and 10 days, only two PCs were necessary to explain 75.54%, 59.49% and 68.31% of data variance, respectively. These results demonstrated that ESI-DIMS-PCA is highly efficient at obtaining data and sample classification.
After the group classification, the loading plots were constructed to identify the ions differentiating the control and experimental groups. These loadings plot were constructed for the PCs differentiating the groups, for example, positive PC3 loadings for the day 3 analysis. The positive loading data for PC3 shows that the ions most relevant to distinguishing the experimental and control groups were m/z 518.5, 546.5, 780.1, 804.2 and 805.0 Da. The most relevant ions demonstrated for days 5, 7 and 10 with positive loadings of PC1 were m/z 518.5, 546.5, 780.1, 804.2, 828.2 and 832.2 Da.
ESI-DIMS-PCA was also used for the early diagnosis of cancer in groups of Swiss mice receiving i.p. injections of Ehrlich tumor. Experiments were conducted to validate the use of ESI-DIMS-PCA to detect cancer despite genetic variability. Because the Balb-C experiments demonstrated that ESI-DIMS-PCA was unable to separate the experimental and control groups after only 1 and 2 days post tumor induction, the tests with Swiss mice were conducted 3, 5, 7 and 10 days after tumor challenge.
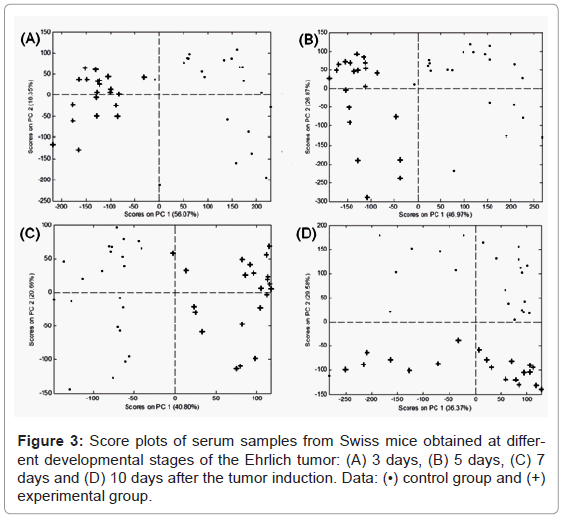
The PCA score plots of the Swiss mice serum samples demonstrated a clear distinction between the control and experimental groups for the 3, 5, 7 and 10 days analyses (Figure 3). Two PCs were capable of explaining 74, 73, 62 and 66% of the data variance for days 3, 5, 7 and 10 post tumor induction, respectively. These results led to the conclusion that ESI-DIMS-PCA was able to classify the appropriate groups despite the genetic variability.
Loadings were then plotted to verify which ions differentiated between the groups. Negative PC1 loadings for days 3 and 5 and positive PC1 loadings for day 7 indicated the most representative ions distinguishing the experimental and control groups were m/z 780.6, 804.6 and 828.6 Da. Negative PC2 loadings indicated that the most relevant ions for the day 10 analysis were the same mentioned above plus m/z 856.8 Da.
Considering the survival time of the animals inoculated with Ehrlich tumor was 15 ± 1 days and 18 ± 1 days for the Balb-C and Swiss strains, respectively, the detection of cancer 3 days after the induction of the tumor cells was considered an excellent result because this time corresponds to 1/5 (one fifth) and 1/6 (one sixth) of the mean survival time of the animals, respectively. Therefore, the employed method allowed for the early classification of the samples.
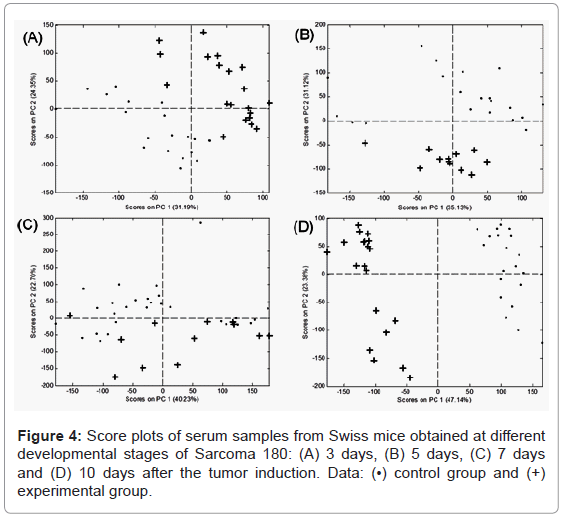
Because ESI-DIMS-PCA allowed for the early classification with the Ehrlich tumor, the method was used to analyze another type of tumor cell. Therefore, Swiss mice received i.p. injections of Sarcoma 180 (S180) cells. Data from PCA demonstrated that, for 3, 5, 7 and 10 days, two PCs differentiating the control and experimental groups corresponded to 56, 66, 71 and 63% of the cumulative variance, respectively. The score plots related to the mass spectra of the experimental and control groups at different stages of S180 development are shown in figure 4.
Analysis of the loading plots demonstrated that, for days 3, 5 and 7, the ions differentiating between the experimental and control groups were m/z 518.5, 780.6, 808.8 and 828.6. At day 10, the highlighted ions were m/z 780.6, 828.6 and 856.8. Considering the survival time of Swiss mice inoculated with S180 was 20.0 ± 2 days, a determination after 3 days corresponded to 1/6 (one sixth) of the mean survival time of the mice, which was also considered a precocious diagnosis. Therefore, ESI-DIMS-PCA was able to detect distinct neoplasms in their early stages.
Biomarker identification
The loading plot analyses demonstrated that the ions responsible for differentiating between the experimental and control groups were those with m/z between 500 and 850 Da (Ehrlich and S180 tumor cells). Data from the literature indicated that these ions were metabolites related to a particular class of phospholipids, specifically phosphatidylcholines (PC) and lysophosphatidylcholines (LPC) [33].
Researchers have demonstrated the importance of LPC as biomarkers for pathophysiological processes. The study by Okita et al. [34] is an example that shows a change in LPC ratio of 18:2 and 16:0 in the plasma of patients with ovarian cancer. In addition, LPC have been used as markers in renal and liver metabonomic studies [22,35]. These studies reported that tumor cells consume more LPC than normal cells, and therefore, patients with a malignant disease have a significantly changed plasma phospholipid pattern. This fact has been confirmed in groups of patients with different types of cancer, which all decreased the phospholipids overall [36].
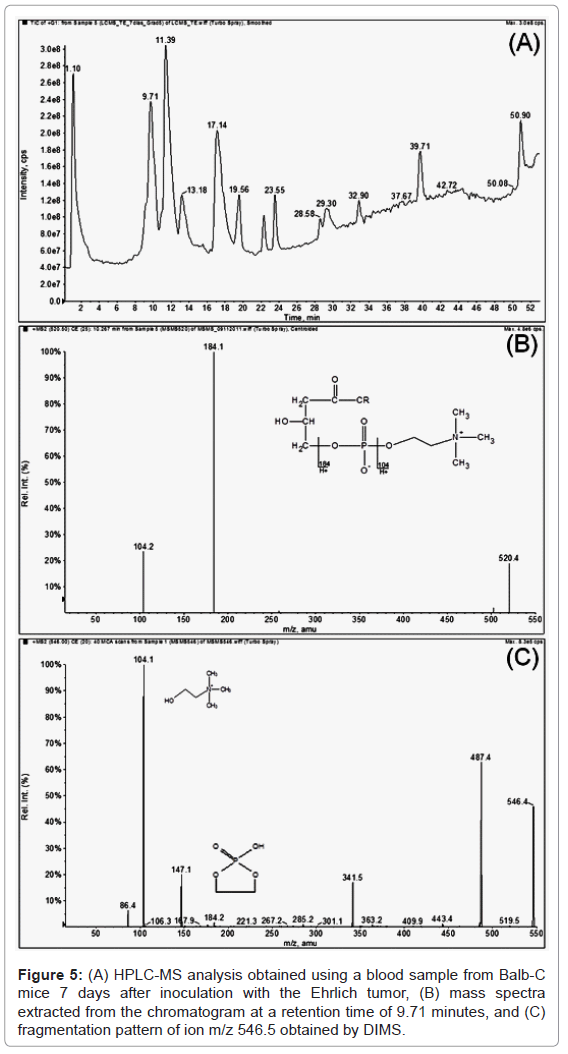
In the present study, HPLC-MS experiments were performed to identify possible biomarkers for Ehrlich and S180 tumors in mice. The mass spectra were extracted from each peak observed in the chromatogram using the full scan mode, and a comparison to those ions obtained via DIMS analysis and highlighted in the loading plots (m/z 518.5, 542.6, 546.5, 758.7, 780.6, 804.6, 808.8, 828.6, 832.5 and 856.8) was performed. The mass spectra obtained from the HPLC-MS analysis showed that the ions appeared as [M+H]+ and were apt to form sodium ions adducts [M+Na]+ in the DIMS analysis (Figure 5). Considering this fact, the correlated ions found by the HPLC-MS and DIMS analyses could have been released via specific ion fragmentations, i.e., the specific fragmentation of the polar head of the LPC in the positive ionization mode provides the characteristic product ions m/z 104.1 (choline) and 184.1, while ions m/z 104.1 and m/z 147.0 have been described in the literature as being characteristic of sodiated LPC regioisomers [17,37,38]. Therefore, the identification of these metabolites was assessed via tandem mass spectrometry experiments (MS/MS), and these data were compared to those described in the literature (Table 1) [17,38-40]. The MS/MS analysis confirmed the presence of LPC (18:2 (9Z, 12Z)), LPC (16:0), LPC (18:0), LPC (18:1), LPC (20:3), (PC) C46H78NO8P, (PC) C46H82NO8P, (PC) C44H78NO8P, (PC) C48H82NO8P, (PC) C48H78NO8P and (PC) C50H82NO8P. These results agree with those of others obtained using high-resolution mass spectrometer, which suggests the results are reliable.
| RT(a)(min) | HPLC-MS/MS | MS/MS (DIMS) | Identification | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ÍON(m/z) | Product Ions | CE(b) | CXP(c) | ÍON (m/z) | Product Ions | CE | CXP | ||
| 9,71 | 520,5 [M+H]+ | 104,1/ 184,1 | 25 | 2,0 | 542,6 [M+Na]+ | 104,1/147,0 | 20 | 2,0 | LPC (18:2 (9Z, 12Z)) |
| 11,4 | 496,5 [M+H]+ | 86/104,1/ 184,1 | 40 | 2,0 | 518,5 [M+Na]+ | 104,1/147,0 | 15 | 3,8 | LPC (16:0) |
| 13,2 | 522,4 [M+H]+ | - | - | - | - | - | - | - | LPC (18:1) |
| 17,1 | 524,3 [M+H]+ | 104,1/ 184,1 | 40 | 546,6 [M+Na]+ | 104,1/ 147,0 | 20 | 2,0 | LPC (18:0) | |
| 19,5 | 546,6 [M+H]+ | 104,1/ 487,1 | 25 | 3,0 | - | - | - | - | LPC (20:3) |
| 19,5 | 804,6 [M+H]+ | 104,1/ 184,1 | 25 | 3,0 | 804,6 [M+H]+ | 104,1/ 184,1 | 20 | 2,0 | (PC) C46H78NO8P |
| 19,5 | 808,8 [M+H]+ | 104,1/ 184,1 | 25 | 3,0 | 808,8 [M+H]+ | 104,1/ 184,1 | 20 | 2,0 | (PC) C46H82NO8P |
| 23,5 | 758,7 [M+H]+ | 184,1 | 40 | 2,0 | 780,6[M+Na]+ | 147,2 / 184,3 | 25 | 2,0 | (PC) C44H78NO8P |
| 23,5 | 832,5 [M+H]+ | 104,1/ 184,1 | 25 | 2,0 | 832,5 [M+H]+ | 104,1/ 184,1 | 25 | 2,0 | (PC) C48H82NO8P |
| 51,0 | 828,6 [M+H]+ | - | - | - | 828,6 [M+H]+ | 104,1/ 184,1 | 25 | 2,0 | (PC) C48H78NO8P |
| 51,0 | 856,8 [M+H]+ | - | - | - | 856,8 [M+H]+ | 104,1/ 184,1 | 20 | 2,0 | (PC) C50H82NO8P |
(a)Retention Time
(b)Collision Energy
(c)Collision Cell Exit Potential
Table 1: Qualitative identification of phospholipids found in serum from Balb-C and Swiss mice inoculated with Ehrlich and S180 tumor cells.
Cancer development prediction capabilities
To create a training set for building a PLS model, 80% of the data obtained by DIMS analysis were extracted from each group. The remaining data formed the independent prediction set and were used to evaluate the developed model. The number of latent variables (LV) was chosen to account for the minimization of the root mean standard error of cross validation (RMSECV) value; a parameter calculated from the results of a venetian blind cross-validation routine. For all of the mice groups, the presence of outliers was evaluated by examining the Studentized residuals versus the leverage graphs. Using limiting values of ± 2.5 for the Studentized residuals and 3 (LV)/n for leverage, no anomalies were observed in the training sets for the developed models. In addition, the values of the root mean square errors (RMSEP) were <1.0 for the independent prediction set, which indicated the models were capable of predicting unknown samples at different stages of cancer development (Table 2).
| Ehrlich tumor | Sarcoma 180 | ||
|---|---|---|---|
| Balb-C | Swiss | Swiss | |
| Number of samples used in model | 67 | 65 | 65 |
| Number of samples used for prediction | 29 | 27 | 35 |
| Number of latent variables (LV) | 8 | 5 | 4 |
| Variance in X (%) | 81 | 79 | 63 |
| Variance in Y (%) | 99 | 98 | 97 |
| RMSEP(*) | 0.8009 | 0.4892 | 0.6422 |
(*)Root mean square error of prediction
Table 2: Results obtained for the constructed models for predicting the developmental stage of the cancer.
Conclusions
The early detection of cancer is a key factor in its successful treatment; therefore, using noninvasive, reliable methods capable of detecting the early stages of the disease is an attractive option. The present work demonstrates the inedited use of low-resolution tandem mass spectrometry with chemometric analysis for diagnosing cancer in mice. ESI-DIMS-PCA was able to detect distinct neoplasms 3 days post induction with Ehrlich and S180 tumors. Considering that the survival time observed for animals inoculated with Ehrlich and S-180 tumors was >15 days, cancer detection after only three days of the tumor challenge was considered an early diagnosis.
It is known that, as DIMS samples are infused together in the mass spectrometer, matrix effects are inevitable, which can reduce the sensitivity and capability of metabolite identification. Furthermore, the isomeric compounds cannot be distinguished by DIMS analysis using only accurate mass. Despite these limitations, in this work, ESI-DIMS-PCA was capable of detecting distinct neoplasms in their early stages. In addition, ESI-DIMS-PCA demonstrated highly sensitive, reproducible and low-cost analysis relative to those obtained using high-resolution mass spectrometers.
The ions most relevant to differentiating between the experimental and control groups were those related to phospholipids, particularly phosphatidylcholines (PC) and lysophosphatidylcholines (LPC). Furthermore, partial least squares (PLS) analysis revealed that it was possible to predict the stage of cancer development using ESI-DIMS-PCA.
Therefore, ESI-DIMS with chemometric analysis is a promising technique for diagnosing early-stage cancer without requiring invasive procedures.
Acknowledgements
The authors would like to thank Dr. Maria Regina Orofino Kreuger (Universidade do Vale do Itajaí, SC, Brazil) for donating the Ehrlich and S180 tumor cells, the National Council of Technological and Scientific Development (CNPQ) for its financial support and the Coordination of Improvement of Higher Education Personnel (CAPES) for providing scholarships.
References
- Zhang X, Li L, Wei D, Yap Y, Chen F (2007) Moving cancer diagnostics from bench to bedside. Trends Biotechnol 25: 166-173.
- Veenstra TD, Conrads TP, Hood BL, Avellino AM, Ellenbogen RG, et al. (2005) Biomarkers: Mining the Biofluid Proteome. Mol Cell Proteomics 4: 409-418.
- Bedair M, Sumner LW (2008) Current and emerging mass-spectrometry technologies for metabolomics. TrAC Trends Anal Chem 27: 238-250.
- Breitling R, Pitt AR, Barrett MP (2006) Precision mapping of the metabolome. Trends Biotechnol 24: 543-548.
- Moco S, Bino RJ, De Vos RCH, Vervoort J (2007) Metabolomics technologies and metabolite identification. TrAC Trends Anal Chem 26: 855-866.
- Spratlin JL, Serkova NJ, Eckhardt SG (2009) Clinical Applications of Metabolomics in Oncology: a review. Clin Cancer Res 15: 431-440.
- Madsen R, Lundstedt T, Trygg J (2010) Chemometrics in metabolomics--A review in human disease diagnosis. Anal Chim Acta 659: 23-33.
- Want EJ, O'Maille G, Smith CA, Brandon TR, Uritboonthai W, et al. (2006) Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry. Anal Chem 78: 743-752.
- Weckwerth W (2003) Metabolomics in systems biology. Annu Rev Plant Biol 54: 669-689.
- Hojer-Pedersen J, Smedsgaard J, Nielsen J (2008) The yeast metabolome addressed by electrospray ionization mass spectrometry: Initiation of a mass spectral library and its applications for metabolic footprinting by direct infusion mass spectrometry. Metabolomics 4: 393-405.
- Dettmer K, Aronov PA, Hammock BD (2007) Mass spectrometry-based metabolomics. Mass Spec Rev 26: 51-78.
- Smedsgaard J, Frisvad JC (1996) Using direct electrospray mass spectrometry in taxonomy and secondary metabolite profiling of crude fungal extracts. J Microbiol Methods 25: 5-17.
- Castrillo JI, Hayes A, Mohammed S, Gaskell SJ, Oliver SG (2003) An optimized protocol for metabolome analysis in yeast using direct infusion electrospray mass spectrometry. Phytochemistry 62: 929-937.
- Allen J, Davey HM, Broadhurst D, Heald JK, Rowland JJ, et al. (2003) High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat Biotechnol 21: 692-696.
- Goodacre R, Vaidyanathan S, Bianchi G, Kell DB (2002) Metabolic profiling using direct infusion electrospray ionisation mass spectrometry for the characterisation of olive oils. Analyst 127: 1457-1462.
- Rashed MS (2001) Clinical applications of tandem mass spectrometry: ten years of diagnosis and screening for inherited metabolic diseases. J Chromatogr B Biomed Sci Appl 758: 27-48.
- Lin L, Yu Q, Yan X, Hang W, Zheng J, et al. (2010) Direct infusion mass spectrometry or liquid chromatography mass spectrometry for human metabonomics? A serum metabonomic study of kidney cancer. Analyst 135: 2970-2978.
- Lokhov PG, Kharybin ON, Archakov AI (2012) Diagnosis of lung cancer based on direct-infusion electrospray mass spectrometry of blood plasma metabolites. Int J Mass Spec 309: 200-205.
- Issaq HJ, Van QN, Waybright TJ, Muschik GM, Veenstra TD (2009) Analytical and statistical approaches to metabolomics research. J Sep Sci 32: 2183-2199.
- Scholz M, Gatzek S, Sterling A, Fiehn O, Selbig J (2004) Metabolite fingerprinting: detecting biological features by independent component analysis. Bioinformatics 20: 2447-2454
- Dagli MLZ, Guerra JL, Saldiva PHN (1992) An experimental study on the lymphatic dissemination of the solid Ehrlich tumor in mice. Braz J Vet Res Anim Sci 29: 97-103.
- Yang J, Zhao X, Liu X, Wang C, Gao P, et al. (2006) High performance liquid chromatography-mass spectrometry for metabonomics: Potential biomarkers for acute deterioration of liver function in chronic hepatitis B. J Proteome Res 5: 554-561.
- Jia LW, Chen J, Yin PY, Lu X, Xu GW (2008) Serum metabonomics study of chronic renal failure by ultra performance liquid chromatography coupled with Q-TOF mass spectrometry. Metabolomics 4: 183-189.
- Maronpot RR, Flake G, Huff J (2004) Relevance of Animal Carcinogenesis Findings to Human Cancer Predictions and Prevention. Toxicol Pathol 32: 40-48.
- Hood BL, Zhou M, Chan KC, Lucas DA, Kim GJ, et al. (2005) Investigation of the mouse serum proteome. J Proteome Res 4: 1561-1568.
- Nascimento FR, Cruz GV, Pereira PV, Maciel MC, Silva LA, et al. (2006) Ascitic and solid Ehrlich tumor inhibition by Chenopodium ambrosioides L. treatment. Life Sci 78: 2650-2653.
- Ozaslan M, Karagoz ID, Kilic IH, Guldur ME (2011) Ehrlich ascites carcinoma. Afr J Biotechnol 10: 2375-2378.
- Dekker LJ, Bosman J, Burgers PC, van Rijswijk A, Freije R, et al. (2007) Depletion of high-abundance proteins from serum by immunoaffinity chromatography: A MALDI-FT-MS study. J Chromatogr B Analyt Technol Biomed Life Sci 847: 65-69.
- Rosenblatt KP, Bryant-Greenwood P, Killian JK, Mehta A, Geho D, et al. (2004) Serum proteomics in cancer diagnosis and management. Annu Rev Med 55: 97-112.
- Chertov O, Biragyn A, Kwak LW, Simpson JT, Boronina T, et al. (2004) Organic solvent extraction of proteins and peptides from serum as an effective sample preparation for detection and identification of biomarkers by mass spectrometry. Proteomics 4: 1195-1203.
- Chen YY, Lin SY, Yeh YY, Hsiao HH, Wu CY, et al. (2005) A modified protein precipitation procedure for efficient removal of albumin from serum. Electrophoresis 26: 2117-2127.
- Mrozinski P, Zolotarjova N, Chen H, Barrett W, Martosella J, et al. (2004) Removal of multiple high-abundant proteins from mouse plasma using immunoaffinity depletion. Mol Cell Prot 3: S222.
- Ackerstaff E, Glunde K, Bhujwalla ZM (2003) Choline phospholipid metabolism: A target in cancer cells? J Cell Biochem 90: 525-533.
- Okita M, Gaudette DC, Mills GB, Holub BJ (1997) Elevated levels and altered fatty acid composition of plasma lysophosphatidylcholine(lysoPC) in ovarian cancer patients. Int J Cancer 71: 31-34.
- Zhang F, Jia Z, Gao P, Kong H, Li X, et al. (2009) Metabonomics study of atherosclerosis rats by ultra fast liquid chromatography coupled with ion trap-time of flight mass spectrometry. Talanta 79: 836-844.
- Taylor LA, Arends J, Hodina AK, Unger C, Massing U (2007) Plasma lyso-phosphatidylcholine concentration is decreased in cancer patients with weight loss and activated inflammatory status. Lipids Health Dis 6: 17.
- Liebisch G, Drobnik W, Lieser B, Schmitz G (2002) High-throughput quantification of lysophosphatidylcholine by electrospray ionization tandem mass spectrometry. Clin Chem 48: 2217-2224.
- Dong J, Cai XM, Zhao LL, Xue XY, Zou LJ, et al. (2010) Lysophosphatidylcholine profiling of plasma: discrimination of isomers and discovery of lung cancer biomarkers. Metabolomics 6: 478-488.
- Lokhov PG, Dashtiev MI, Moshkovskii SA, Archakov AI (2010) Metabolite profiling of blood plasma of patients with prostate cancer. Metabolomics 6: 156-163.
- Jia LW, Wang C, Kong H, Cai Z, Xu G (2006) Plasma phospholipid metabolic profiling and biomarkers of mouse IgA nephropathy. Metabolomics 2: 95-104.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 15689
- [From(publication date):
February-2013 - Dec 05, 2025] - Breakdown by view type
- HTML page views : 10834
- PDF downloads : 4855