Editorial Open Access
In-Cell NMR Spectroscopy–In vivo Monitoring of the Structure, Dynamics, Folding, and Interactions of Proteins at Atomic Resolution
Thallapuranam K Suresh Kumar*, Ryan Thurman and Srinivas Jayanthi
Department of Chemistry and Biochemistry, University of Arkansas, Fayetteville, Arkansas 72701, USA
- *Corresponding Author:
- Thallapuranam K Suresh Kumar
Associate Professor, Room #235
Department of Chemistry and Biochemistry
University of Arkansas, Fayetteville
Arkansas 72701, USA
Tel: 1-479-575-5646
Fax: 1-479-575-4049
E-mail: sthalla@uark.edu
Received date: December 26, 2012; Accepted date: December 27, 2012; Published date: January 02, 2013
Citation: Suresh Kumar TK, Thurman R, Jayanthi S (2013) In-Cell NMR Spectroscopy–In vivo Monitoring of the Structure, Dynamics, Folding, and Interactions of Proteins at Atomic Resolution. J Anal Bioanal Tech 4:e112. doi: 10.4172/2155-9872.1000e112
Copyright: © 2013 Suresh Kumar TK. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Multidimensional nuclear magnetic resonance spectroscopy (NMR) is a very versatile technique which is routinely used for the characterization of 3D structure, backbone dynamics, and intermolecular interactions of proteins in solution at atomic resolution. NMR spectroscopy as an analytical technique has come of age. Availability of higher field spectrometers, equipped with cryoprobes, has drastically increased the sensitivity and resolution of NMR data. In recent years, several new pulse sequences have been developed which have significantly diminished the perceived molecular-size limitation of NMR spectroscopy. Currently, determination of 3D solution structures of proteins, with molecular mass in the 100 kDa range, is no longer impossible.
Like X-ray crystallography and electron microscopy, multidimensional NMR spectroscopy has till recently been only conceived as an in vitro technique which can help in the structural characterization of proteins. However, several interesting questions pertaining to the physiological relevance of the protein structures determined using these in vitro techniques have been raised. For example, are the 3D structures of isolated proteins determined under in vitro conditions different than when they are present in a crowded cellular environment with other proteins and biomolecules? Do proteinligand structural interactions characterized under in vitro conditions bear resemblance to those which occur in the cellular milieu? Most importantly, do post-translational modifications which occur during protein maturation cause significant change in the conformation of proteins? These intriguing questions can be satisfactorily addressed by the recently developed in-cell NMR spectroscopy.
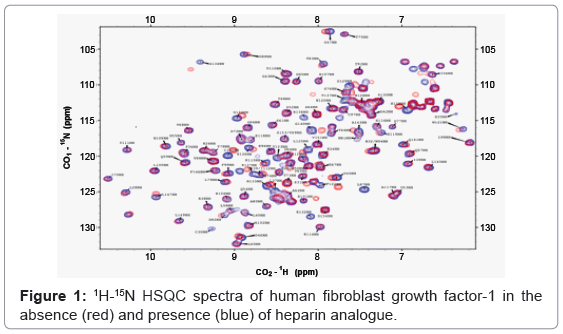
In-cell NMR spectroscopy is an emerging bioanalytical technique which can be employed to obtain atomic-level information on the structure, folding, dynamics, and interactions of proteins in a cell. Two-dimensional correlation experiments involving NMR sensitive atomic nuclei (such as, 1H, 13C and 15N) are primary to the characterization of protein structure and their interactions. 1H-15N/1H- 13C HSQC (Heteronuclear Single Quantum Coherence) is the most informative and widely employed 2D correlation experiment used in in-cell NMR spectroscopy. 1H-15N HSQC is preferred over the 1H-13C HSQC experiment due to its simplicity and significantly lower costs of preparation of 15N-enriched protein samples. Each 1H-15N correlated peak in a 1H-15N HSQC spectrum represents an amino acid in a particular backbone conformation of a protein. Therefore, 1H-15N HSQC spectrum of a protein can be expected to be unique (Figure 1). Although the total number of 1H-15N resonance crosspeaks in a given HSQC spectrum remains the same, local changes in chemical microenvironment of the NMR sensitive atomic nuclei (1H and 15N), effectedeither due to ligand binding or due to conformational changes induced in a protein, causes difference(s) in the chemical shift value of definite atomic nuclei. Differences in the chemical shift values of the participating nuclei, measured as a composite chemical shift value [Δδcomposite=Δδ1H+Δ15N/13C], provide valuable information on specific amino acid residues involved in ligand binding or in a conformational change. This rationale applies to both in vitro and in vivo experiments.
There are unlimited applications for in-cell NMR spectroscopy. Three-dimensional structures of proteins can be determined in a cellular environment and these 3D structures would bear more relevance than those which are solved under in vitro conditions. Similarly, protein structures are not plastic because proteins exhibit interesting backbone as well as side-chain dynamics in the picoseconds to micro/ millisecond time scale. In-cell NMR is an apt technique to monitor these dynamic protein motions. In addition, proteins are targets for most of the pharmaceutical drugs and the inherent capability of incell NMR to characterize molecular interactions at atomic resolution is immensely valuable towards efforts to design novel, specific, and potent therapeutic principles against multitude of diseases. Further, protein aggregation is currently plaguing pharmaceutical industries and billions of dollars are lost due to the problem of formation of inclusion bodies (protein aggregates) during over expression of recombinant proteins in heterologous hosts. In-cell NMR spectroscopy provides a unique avenue to study in vivo molecular mechanisms that govern the partition between productive folding of proteins to their biologically active conformation and unproductive pathways leading to aggregated state(s). Post-translational modifications of proteins either serve as cellular regulatory mechanism(s) that critically alters protein-protein interactions or they facilitate the specific targeting of proteins in to different sub-cellular compartments. In-cell NMR spectroscopy is an invaluable technique to trace the different steps of protein maturation and understand how they shape the folding and function of proteins. Lastly, in recent years, there has been increased interest in the characterization of intrinsically disordered proteins (IDPs). This class of proteins lack elaborate structure(s) but serve as hub(s) for proteinprotein interactions that regulate key cellular processes. Recently, incell NMR spectroscopy has been successfully employed to track the structure and folding of IDPs in a cellular environment. In conclusion, the scope of applications of in-cell NMR seems limitless and future development(s) in the NMR spectrometer hardware, pulse sequences and methods of overexpression of isotope enriched proteins will help unravel the full potential of this fascinating technique.
Acknowledgements
This publication was supported by NIH (P30 GM 103450), DOE (DE-02- 01ER15161) and the Arkansas Bioscience Institute grants to TKSK.
--Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 15731
- [From(publication date):
February-2013 - Dec 08, 2025] - Breakdown by view type
- HTML page views : 10928
- PDF downloads : 4803