Research Article Open Access
Immobilization of P22 Bacteriophage Tailspike Protein on Si Surface for Optimized Salmonella Capture
Sarang Dutt1,2, Jamshid Tanha3 , Stephane Evoy1,2, and Amit Singh1,2,4*
1Electrical and Computer Engineering, University of Alberta, Canada
2National Institute for Nanotechnology, University of Alberta, Canada
3Human Health Therapeutics, National Research Council Canada, Canada
4Department of Pharmaceutical Sciences, Northeastern University, USA
- *Corresponding Author:
- Amit Singh
Department of Pharmaceutical Sciences
Northeastern University
Boston, MA, 02176 USA
Tel: +1 617 373 3127
E-mail: am.singh@neu.edu
Received date: February 27, 2013; Accepted date: April 22, 2013; Published date: April 24, 2013
Citation: Dutt S, Tanha J, Evoy S, Singh A (2013) Immobilization of P22 Bacteriophage Tailspike Protein on Si Surface for Optimized Salmonella Capture. J Anal Bioanal Techniques S7:007. doi: 10.4172/2155-9872.S7-007
Copyright: © 2013 Dutt S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Bacteriophage based technology has gained interest in developing pathogen detection platforms for biosensing applications. In this study, P22 phage tail spike proteins (TSPs) have been immobilized on Si surfaces for optimized capture of host Salmonella enteric serovar Typhimurium. It was then demonstrated that roughening of the Si surface before the TSP immobilization improves the bacterial capture 2-fold compared to a flat Si surface. Coarse, medium, fine and superfine size ridges were patterned on the Si surface using block copolymer layer and plasma etching and each surface was functionalized by TSPs for bacterial capture. The capture density increased with decreasing size of the ridge until it reached an optimum for fine ridges; the capture density decreased when the surface ridges were superfine and deep. This method shows a 22-fold and 3-fold increase in bacterial capture density compared to a Cys- and a His6-tag based oriented TSP immobilization, respectively. Bovine serum albumin (BSA) was used as a surface protective layer to prevent non-specific binding of bacteria and E. coli cells were used as control to demonstrate the specificity of recognition. Negligible binding was observed for control bacteria in presence of TSPs and the host bacteria in the absence of TSP on the surfaces.
Keywords
Bacteriophage; Tail spike protein; Salmonella; Block copolymer; Plasma etching
Abbreviations
TSP: Tail Spike Protein; BSA: Bovine Serum Albumin; N-Cys: N-terminal Cysteine; MEMS: Micro-electro Mechanical Systems; NEMS: Nano-electro Mechanical Systems; C-Cys: C-terminal Cysteine; GST: Glutathione S-transferase; cfu: Colonyforming units
Introduction
Sensitive and selective detection of bacterial pathogens is an area of interest due to increasing concerns over food, water, environment and public health safety [1]. Conventional microbiological and serological assay are reliable for pathogen identification and detection but are time consuming, labor intensive and cost ineffective [2]. Some reports suggest that selective and differential plating method may not always give an accurate identification of the bacteria in real life samples [3]. Biosensors have been looked upon as alternative for detecting pathogen, their spores and toxin using different transduction platforms. A biosensor has two key components; a biological probing element (e.g. protein, peptide, deoxyribonucleic acid, ribonucleic acid, whole organism), which imparts specificity of recognition, and a transducer, which converts the recognition event into a measurable signal. Different probing elements and transducer platforms have been integrated to improve the recognition specificity and detection sensitivity of a pathogen biosensor.
Mass perturbance [4], electrochemical [1], flow cytometry [5], optical [1,6] and mechanical resonator-based [7,8] sensors have been explored for developing a sensitive pathogen detection platform. Antibodies/peptides [4], DNA [9], RNA [10], aptamers [11], peptide nucleic acid (PNA) [12] and carbohydrate [13] have similarly been used as recognition elements to detect different pathogens. Antibodies particularly have been extensively used as molecular probe for pathogen detection [14]. However, antibodies suffer from temperature and pH instability and are prone to protease degradation [15]. Polyclonal antibodies further suffer from cross-reactivity. Bacteriophages therefore have been looked upon as a robust alternative biological probe for pathogen detection. Phages are bacterial viruses that recognize and bind to specific receptor on the host bacterial surface. Many phages use their tail spike proteins for host recognition and binding [16]. The phage recognition is highly specific and is thus used for bacterial typing. This specificity and selectivity makes intact phages an excellent candidate as probing element in a biosensor. Orientation of the phages is pertinent to an optimized capture of the target on the sensor platform, and use of various tags has been shown to improve the density of bacterial capture by phages [17,18]. Many reports have described application of intact phages in detecting bacterial pathogens using different transducer platforms [17-22]. In a previous study, we have shown that chemical anchoring of phages on gold-coated substrates leads to improved host bacterial capture compared to physically adsorbed phages [23]. We have further demonstrated that phages lysate can be purified by column chromatography to remove host bacterial contaminants [24] and that such purified T4 phage lysate could be chemically anchored on surface plasmon resonance substrates for a sensitive and selective detection of host E. coli K12 cells [25].
Intact phage-based approach however suffers from some drawbacks such as drying effect and lysis of captured bacteria on over exposure [26]. In a previous study, we have shown that use of the receptor recognizing and binding proteins at the tail of the phages could be used instead of the whole phage to overcome these limitations. Recombinant P22 phage tailspike proteins (TSPs), lacking the head-binding domain, were cloned, genetically engineered and over expressed to yield high quantities of the mutant protein [26]. P22 phage TSP recognizes the O-antigen polysaccharide from the serotype A, B and D1 of Salmonella having identical backbone of mannose, rhamnose and galactose sugars. The P22 TSP also shows endorhamnosidase enzymatic activity, which hydrolytically cleaves the O-antigen on the bacterial surface [27,28]. The TSPs used here have mutations that essentially eliminate their endorhamnosidase activity without changing their binding property [26,29,30]. These TSPs are highly stable against pH and temperature changes and resistant to trypsin as well. They also do not show any drying effect and bacterial lysis unlike intact phages [26]. Besides, genetic modification allows expression of desired tags on the TSPs to enable immobilization on biosensor surfaces. Recombinant P22 TSPs were expressed in two forms: one which has a cysteine tag at the C-terminus and a poly histidine tag (His6) at the N-terminus (C-Cys TSPs) and another which has both th ecysteine and His6 tags at the N-terminus (N-Cys TSPs). We have shown that the Cys tag could be exploited to immobilize these TSPs on gold surface using thiol chemistry for biosensor development, with N-Cys giving a preferred orientation compared to C-Cys TSP [26].
However, use of thiol chemistry limits the use of these TSPs to gold and silver based detection platforms. Beside the metal surface-based, silicon-based substrate interface are extremely popular for biosensor applications. Si based surfaces are primarily used in micro- and nano-fabrication for developing micro-electro mechanical systems (MEMS) and nano-electro mechanical system (NEMS) devices [31] and thus it is important to obtain a standardized method of TSP immobilization for such surfaces. It is a well-established fact that chemical anchoring of the probes provides stable monolayer for biosensor application [24-26]. However, right orientation of the probe is relevant for a receptor-based recognition and subsequent capture of the analyte [26]. In order to determine the most optimal method for generating TSP-derivitized Si-based biosensor platform, we functionalized the flat Si surfaces by 3 different methods to achieve random and oriented immobilization of TSPs on the surface to test the target bacterial capture efficiency. In the work presented here, we have also demonstrated that increase in surface area of Si substrate by roughening improves the immobilization of TSPs on the surface and subsequently improves the bacterial capture. Surface ridges of coarse, medium, fine and superfine sizes were etched on the Si surface using block copolymer template. We obtained maximum bacterial capture on fine ridge surface, which was similar to the capture density obtained on gold surfaces [26]. BSA-protected Si surfaces showed insignificant binding of control bacteria in the presence of TSPs and host bacteria in the absence of TSPs. We also demonstrate that roughened Si surfaces give better bacterial capture than the flat Si surfaces using random and oriented immobilization of TSPs. Thus, an optimized bacterial capture has been demonstrated on Si surfaces which could be extended to Si surface based biosensor platforms such as cantilevers, nanowires and nanobeams.
Materials and Methods
Materials
Silicon substrates were purchased from University Wafers Inc. (Boston, MA, US). 3-Aminopropyltriethoxysilane (APTES), 3-Mercaptopropyltriethoxysilane (MPTES), glutaraldehyde, tris(2-carboxyethyl) phosphine (TCEP), BSA, glycidyloxypropyltrimethoxysilane (GPTMS), N-(5-amino-1-carboxypentyl)iminodiacetic acid, sodium bicarbonate (NaHCO3), nickel chloride (NiCl2), sodium chloroplati- nite (Na2PtCl4) and glycine were purchased from Sigma-Aldrich (St. Louis, MO, USA) and were used as received. Tween-20 was purchased from MP Biomedicals, Inc. (Solon, OH, USA). All experiments were performed using deionized MilliQ® water as solvent. Phosphate buff- ered saline (PBS) was prepared by mixing one BupH phosphate buff- ered saline pack to 500 ml of Milli-Q water yielding a solution of 0.1 M phosphate, 0.15 M NaCl, pH 7.2 (Fisher Canada, Napean, ON). All other solvents and reagents were analytical grade and were used with- out further purification. All Si surfaces were cleaned in piranha solu- tion (3:1 H2SO4:H2O2) for 15 min followed by 2 washes in water prior to any functionalization.
Cultures and bacterial cell preparation
Salmonella enteric serovar Typhimurium bacteria (ATCC19585) were purchased from American Type Culture Collection (Manassas, VA). Luria Bertani (LB) medium was purchased from Quelabs (Montreal, QC, Canada) and was prepared by dissolving 25 g of the LB powder in 1 lt of distilled water. LB-agar medium was prepared by adding 6 g of agar in 400 ml of LB media. Nutrient broth (NB) powder was purchased from Difco™ (Sparks, MD, USA) and was prepared by dissolving 8 g powder in 1 lt of distilled water. All the above reagents were purchased from Sigma-Aldrich. LB and NB medium were autoclaved prior to use according to standard protocols. Bacterial enumeration was done by plate count method and was expressed in colony-forming units (cfu)/ml.
An aliquot from frozen stock of Salmonella enteric serovar Typhimurium and E. coli K12 culture were streaked onto a Nutrient agar plate and incubated overnight at 37°C. Single colonies from each plate were inoculated into 4 ml LB medium and were grown at 37°C in a shaker. 1 ml aliquots of the bacteria cultures were centrifuged in a microfuge at 13,000 rpm for 1 min and pellet was resuspended in 1 ml PBS for all bacterial capture assays. All TSP functionalized surfaces were incubated in 1 mg/ml solution of BSA to block the free substrate surfaces and prevent non-specific adsorption of bacteria.
Preparation of His- and Cys-tagged P22 TSP
The TSPs were cloned and purified at Dr. Tanha’s lab (National Research Council Canada, Ottawa, ON, Canada). Cloning, expression and purification of the His- and Cys-tagged TSPs has been described elsewhere [26,32,33]. The recombinant P22 TSPs used in the present work (N-Cys TSP) consists of a cysteine, a His6 tag, followed by a TSP sequence, which has binding activity but lacks enzymatic activity. All TSP samples were treated with 50 mM TCEP at 60°C for 30 min to break the inter-molecular disulfide bonds and were functionalized at a concentration of 2 mg/ml in PBS buffer for all substrates.
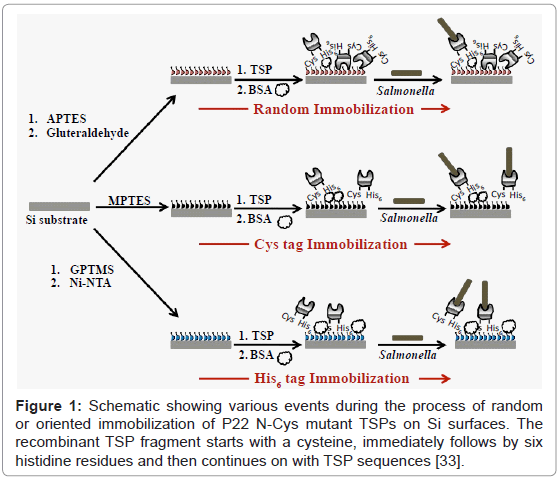
Randomly oriented immobilization of P22 TSPs on Si surface
A 2% solution of APTES in ethanol (95%) was prepared and the pH was adjusted to 5 using glacial acetic acid. This solution was kept at room temperature for 15 min after which the piranha cleaned silicon substrates were immersed for 15 min. The substrates were then incubated at 110°C for 10 min to cure the silane monolayer. The substrates were finally washed twice in 95% ethanol for 5 min each followed by washing twice in water for 5 min [32]. The amine group APTES was activated by incubation in 2% glutaraldehyde for 30 min followed by two washes in PBS. The glutaraldehyde activated Si substrates were finally incubated in TCEP reduced N-Cys mutant TSPs for 2 h to allow protein immobilization. The TSP immobilized substrates were washed twice in PBS followed by blocking free surface with BSA and exposure to Salmonella capture according to the method described above [33].
Oriented immobilization of P22 TSPs on flat Si using His6 tag
The protocol was adapted from a previous report of immobilizing His-tagged proteins on silicon nanowires [34]. The piranha cleaned substrates were immersed in 0.01% acetic acid containing 2% (v/v) 3-glycidyloxypropyl-trimethoxysilane (GPTMS) (Sigma) for 3 h at 90°C followed by two washes with water for 5 min each. The substrates were then incubated in 0.01 M NaHCO3 (pH 10.0) containing 10% (w/v) N-(5-amino-1-carboxypentyl)-iminodiacetic acid (AB-NTA) (Sigma) for 16 h at 60°C followed by two washes with water for 5 min each. The substrates were then incubated in 10 mM NiCl2 (or NiSO4) and 5 mM glycine (pH 8.0) for 2 h at room temperature followed by two washes with water for 5 min each. These surfaces were incubated with 1 mg/ml solution of N-Cys mutant TSPs overnight at 40°C to allow the His6 tag at N-terminal to bind to the Ni ions. The TSP immobilized surfaces were blocked with BSA and exposed to Salmonella binding according to the procedure described above.
Oriented immobilization of P22 TSPs on flat Si using Cys tag
The piranha cleaned substrates were silanized using 2% solution of mercapto-propyl triethoxysilane (MPTES) in acetone for 2 h. The silanized surfaces were washed twice in acetone for 5 min each followed by blow drying in dry N2. These surfaces were incubated at 110°C for 10 min to cure and crosslink the silanized surface. These were incubated overnight in TCEP-reduced 1 μg/ml solution of N-Cys mutant TSP in PBS. The N-CysTSP attaches to the thiol functionality of the silane monlayer by forming a disulfide linkage. The TSP immobilized surfaces were blocked with BSA and exposed to Salmonella as per the procedure described above.
Preparation of rough Si surface
Silicon wafers were cleaned with RCA 1 (1:1:5 mixture of NH4OH:H2O2:H2O) at 90°C for 10 min followed by RCA 2 (1:1:6 mixture of HCl:H2O2:H2O) at 90°C for 10 min. The clean wafers were spin-coated with block copolymer solution (1% solution of poly(styrene-b-2-vinyl pyridine) in toluene) to obtain a film of polystyrene containing poly-2-vinyl pyridine. This polymer film was used to template the binding of metal salts. The wafers were immersed in a solution of 20 mM Na2PtCl4 in 3% HCl for 3 h. The metal salt binds to 2-vinyl pyridine to form localized masks of high metal salts. The wafers were cleaned in water for 5 min to remove excess salt. These wafers were exposed to oxygen plasma (55 mTorr, 30W RF) for 35 s followed by argon plasma (100 mTorr, 50W RF) for 5 s to reduce the metal salts and wash away the polymer matrix. These wafers were exposed to 1:2.5 mixture of O2 and SF6 plasma (10 m Torr, 20W RF) to etch patterns followed by exposure to O2 plasma to make the wafers hydrophilic. These wafers were used to immobilize N-Cys TSP and subsequently study host and non-host bacterial capture. The functionalization of ridged Si substrates was achieved by following the immobilization protocol described in section 2.6 to prepare covalently linked oriented N-Cys TSP based surfaces.
Bacterial binding and surface density calculation
The BSA protected surfaces were exposed to the host or non-host bacteria for 20 min to facilitate their binding. The substrates were then washed in 0.05% (v/v) Tween-20 solution in PBS for 5 min followed by two washes in PBS for 5 min each. The bound bacteria were fixed by incubating surfaces in 2% glutaraldehyde solution in water for 1 h. The fixed surfaces were washed twice with water for 5 min followed by sequential incubation in 50, 60, 70, 80, 90 and 100% ethanol for 5 min each. Finally, these substrates were dried overnight in an incubator at 42°C and were imaged in Hitachi S-4800 scanning electron microscope (SEM). The bacterial capture density was enumerated from the high magnification SEM images using the cell counter plugin of Image J software (NIH, USA). Eight images for each sample were counted to obtain the capture density and the standard deviation for the data set. Same procedure was applied for all the TSP immobilized samples.
Results and Discussion
Bacterial capture on flat Silicon
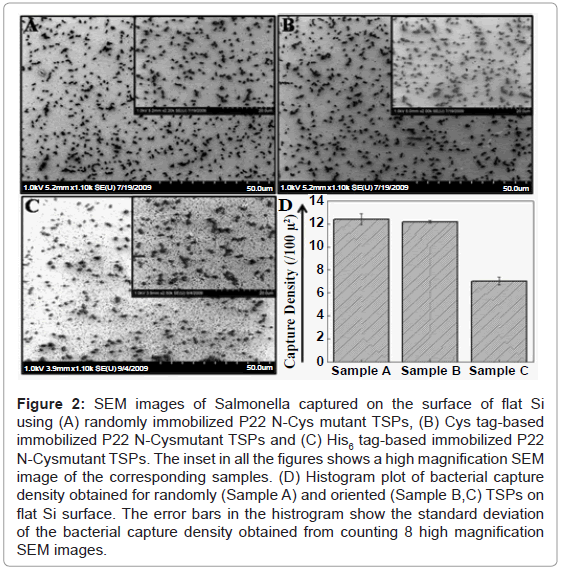
The P22 mutant TSPs were immobilized on flat silicon in random as well as oriented fashion and the host Salmonella capture was analyzed using SEM images. All the samples were exposed to 109 cfu/ml bacteria suspended in PBS. Figure 1 is a schematic showing various possible events during the random and oriented immobilization of TSPs on Si surfaces and the subsequent bacterial capture. Figure 2A shows the SEM image of the host bacteria captured on flat Si substrates using randomly immobilized N-Cys mutant TSPs. The inset shows a representative high magnification SEM image of the sample. A bacterial capture density of 12.43 ± 0.48 bacteria/100 μ2, was calculated for randomly immobilized TSPs (Figure 2D). The higher density of bacterial capture by the sample could be explained by the more stable chemical immobilization of the TSPs on the Si surface though the capture density was still nearly 2-fold lesser than that on gold surface with an oriented immobilization [25]. The inset shows a representative high magnification SEM image of the sample.
Figure 2: SEM images of Salmonella captured on the surface of flat Si using (A) randomly immobilized P22 N-Cys mutant TSPs, (B) Cys tag-based immobilized P22 N-Cysmutant TSPs and (C) His6 tag-based immobilized P22 N-Cysmutant TSPs. The inset in all the figures shows a high magnification SEM image of the corresponding samples. (D) Histogram plot of bacterial capture density obtained for randomly (Sample A) and oriented (Sample B,C) TSPs on flat Si surface. The error bars in the histrogram show the standard deviation of the bacterial capture density obtained from counting 8 high magnification SEM images.
The flat Si substrates were also tested for oriented immobilization of the TSPs using the Cys tag on N terminal and His6 tag on N terminal of N-Cys TSP. Figures 2B and 2C show the SEM image of bacteria captured on flat Si using the Cys tag and the His6 tag of N-Cys mutant TSP, respectively. The inset in both images shows the high magnification images of the corresponding samples. The bacterial capture density using Cys tag and His6 tag was found to be 12.21 ± 0.26 bacteria/100 μ2 and 7.03 ± 0.36 bacteria/100 μ2, respectively (Figure 2D). The inset in both images shows the high magnification images of the corresponding samples. The difference in the capture density between these two samples could be due to the fact that Cys tag-based immobilization chemistry involves strong covalent disulfide linkage between MPTMS and thiol group of cysteine while the His6 tag based immobilization results from weaker coordination linkage with Ni ions. Similarly, a more stable chemical immobilization of TSPs on the Si surfaces could account for the higher bacterial capture density observed in the case of the random immobilization approach (Figures 2A and 2D). We however observed a 2-fold greater density of host bacteria capture on Cys tag based orientation of N-Cys TSP on gold surface (25.87 ± 0.61 bacteria/100 μ2) [25] compared to that on flat Si surface. Figure 2D shows the histogram plot of the host bacterial capture on flat Si using random immobilization (Sample A), Cys tag based immobilization (Sample B) and His6 tag based immobilization (Sample C) of N-Cys mutant TSP. The error bars in the histogram show the standard deviation of the bacterial capture density obtained from counting 8 high magnification SEM images.
Fabrication of ridged Si surfaces
Ridged Si surfaces were fabricated by plasma etching to enhance the surface area and hence improve the immobilization of the TSPs and subsequent bacterial capture. Four different types of surface patterns were fabricated called as coarse, medium, fine and superfine depending on the size of the ridges. The control on the ridge sizes was achieved by tailoring the ratio of styrene to 2-vinyl pyridine chain length used to form the block copolymer masking layer on the surface. Table 1 show the ratio of styrene to 2-vinyl pyridine chain length which was spin coated on the Si surface for eventual fabrication of ridges by plasma etching. Typically, a higher chain length block copolymer on the surface generates coarser surface ridges while a smaller chain length block copolymer gives finer surface ridges.
| Sample | Ratio (Styrene:Vinyl pyridine) |
|---|---|
| Coarse Ridge | 125,000:58,500 |
| Medium Ridge | 52,400:28,000 |
| Fine Ridge | 32,500:12,000 |
| Superfine Ridge | 25,500:23,500 |
Table 1: Table showing the ratio of styrene to 2-vinly pyridine chain length used to fabricate coarse, medium, fine and superfine ridged Si surfaces.
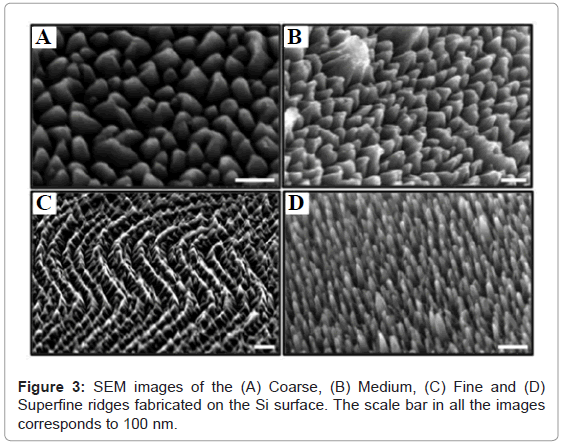
Figure 3 shows the SEM images of the surface ridges generated on the Si surface by plasma etching. Figures 3A-3D shows the coarse, medium, fine and superfine ridge pattern, respectively, generated on the Si surface. The coarse ridges were broader at the base while the depth of etched Si was smaller (Figures 3A and 3B) compared to the finer ridges, which were narrow at the base and the depth of etched Si was greater (Figures 3C and 3D). It was anticipated that while the fine and superfine structures will result in better surface functionalization and subsequent TSP immobilization, the greater depth of etch might interfere with the interaction of surface bound TSPs with the host bacteria.
Bacterial capture analysis on ridged Si surface
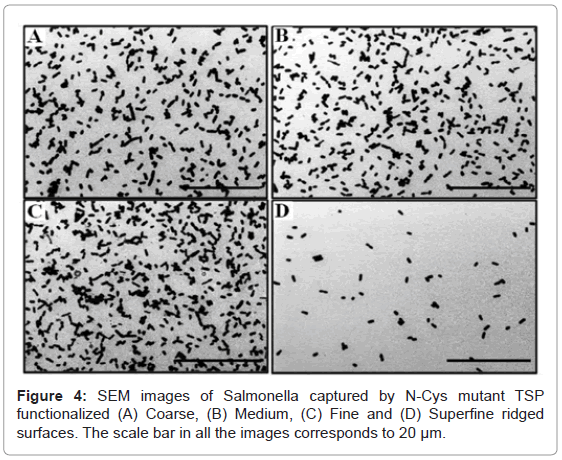
The N-Cys mutant TSPs were randomly immobilized on the ridged Si surfaces by the procedure described above and such surfaces were exposed to 109 cfu/ml concentration of host Salmonella bacteria. The bacterial capture was confirmed and capture density was enumerated using SEM images of the surface. Figures 4A-4D shows the capture of host bacteria on the surface of coarse, medium, fine and superfine ridged Si substrates, respectively. The coarse, medium, fine and superfine ridged structures show a bacterial capture density of 13.75 ± 2.09, 16.97 ± 1.98, 23.85 ± 1.32 and 1.78 ± 0.23 bacteria/100 μ2, respectively. The bacteria capture analysis shows an increase with decreasing size of surface ridges and increasing depth of etched Si till it reaches a maximum for fine etched Si surface. There is a sharp decline in bacterial capture density on the TSP immobilized superfine ridges which could be attributed to the deep etches on the Si surface. As indicated in previous section, these deep edges would result in interfering with the interaction of surface TSPs to the host bacteria resulting in decreased capture. Thus, an optimized host bacterial capture was achieved on the ridged Si surfaces using fine size surface ridges. The bacterial capture density obtained on fine ridged Si surface is similar to that achieved by oriented immobilization of N-Cys mutant TSPs on gold surface [25]. We therefore demonstrated that ridged Si could be used to achieve high bacterial capture even by random immobilization of the TSPs on the surface. The result further confirms that increasing the surface area of the Si greatly improves the TSP immobilization on the surface and subsequent bacterial capture.
Surface specificity and selectivity analysis
Bovine serum albumin (BSA) was used as surface protective layer in all experiments to check non-specific binding of bacteria to the surface, and E. coli K12 was used as positive non-host control bacteria. N-Cys mutant TSPs immobilized on flat as well as coarse and fine ridged Si surfaces were checked for capture of the non-host bacteria which showed insignificant binding on the surface. Alternatively, host Salmonella was exposed to BSA blocked coarse and fine ridged surfaces in the absence of TSPs which also did not show any bacterial binding. These results confirmed that the BSA protective layer successfully checks the non-specific binding of any bacteria to the surface and thus all the bacterial capture densities obtained for different samples were indeed due to specific and selective recognition of the TSPs. Table 2 shows the capture density on different surfaces using host (h) and non-host (nh) bacteria in the presence or absence of TSPs. The density comparison clearly indicates that we have successfully demonstrated a specific and optimized capture of host Salmonella bacteria on Si surfaces using TSPs as recognition elements.
| Sample | P22 TSP | Bacteria Target | Capture Density |
|---|---|---|---|
| Coarse Ridge Si | Yes | Salmonella (h*) | 13.75 ± 2.09 |
| Medium Ridge Si | Yes | Salmonella (h*) | 16.97 ± 1.98 |
| Fine Ridge Si | Yes | Salmonella (h*) | 23.85 ± 1.32 |
| Superfine Ridge Si | Yes | Salmonella (h*) | 1.78 ± 0.23 |
| Flat Si | Yes | E. coli (nh**) | 0.12 ± 0.07 |
| Coarse Ridge Si | Yes | E. coli (nh**) | 0.22 ± 0.1 |
| Fine Ridge Si | Yes | E. coli (nh**) | 0.05 ± 0.04 |
| Coarse Ridge Si | No | Salmonella (h*) | 0.07 ± 0.06 |
| Fine Ridge Si | No | Salmonella (h*) | 0.14 ± 0.1 |
h*=host, nh**=non-host
Table 2: Table showing the calculated bacterial capture density for different samples using Salmonella as well as E. coli K12 bacteria in the absence or presence of N-CysTSPs.
Conclusion
This paper demonstrates an optimized approach for capturing host Salmonella enteric serovar Typhimurium on Si based surfaces using P22 phage tail spike proteins as probing element. A recombinant P22 TSP was genetically engineered to express a Cys residue tag and an immediately followed His6 tag at its N-terminus, which were exploited to immobilize iton Si surfaces. We have shown in our previous study that Cys tag based immobilization of N-Cys mutant TSPs give the protein a preferred orientation with high density of bacteria capture [26]. However, random as well as tag based oriented immobilization of N-Cys mutant TSPs on flat Si results in a poor bacterial capture. We fabricated coarse, medium, fine and superfine ridged Si surfaces to improve the surface area facilitating a better TSP immobilization and subsequent bacterial capture. All 4 types of ridged surfaces were functionalized with TSP and exposure to host Salmonella bacteria show that fine ridged surfaces show maximum bacterial capture with a density of 23.85 ± 1.32 bacteria/100 μ2 which was similar to that observed for Cys tag based immobilization on gold surface [26]. Thus, we could optimize the capture of the host bacteria on ridged Si surface using P22 phage TSPs as recognition element. We further demonstrated the specificity and selectivity of bacterial binding using BSA as surface protective layer and E. coli K12 as non-host positive control bacteria. Arguably, the relative lower binding densities obtained with E. coli K12 could have alternatively been related to the formation of capsules that would have impeded their binding to the substrate, rather than related to the specificity of the moiety. However, E. coli K12 grown under similar conditions have successfully been captured by surface-bound T4 phages with capture densities comparable to those obtained in the P22 TSPs-based Salmonella capture experiments reported here [23-25]. This suggests that capsule formation, if any, does not significantly impair capture of that species, and is thus unlikely to account for the hundred-fold lower binding densities observed in the negative control experiments reported here. BSA-protected surfaces did not show any significant binding to host and non-host bacteria. Thus, we have demonstrated an optimized method for binding and capture of the host Salmonella bacteria on Si based surfaces using P22 TSPs as molecular probes.
We have recently identified the receptor-binding protein (Gp48) of phage NCTC 12673 and over-expressed and purified it as glutathione S-transferase-Gp48 fusion protein (GST-Gp48) [35]. The GST-Gp48 protein can be immobilized on glutathione modified gold surface and their recognition specificity can be leveraged to capture the host Campylobacter jejuni bacteria. We have further demonstrated that GST-Gp48 fusion protein can be easily integrated onto gold surface-based biosensor platforms such as surface plasmon resonance and promises to be an excellent recognition element for Campylobacter detection [35]. A recent review comprehensively summarizes our work on functionalization of metal-based surfaces with phage proteins and their application in biosensor development [36,37]. These receptor-binding proteins with tags can be easily duplicated on Si based biosensing platforms such as microcantilevers or silica micro/nanoparticles to develop a specific and sensitive pathogen detection system.
Acknowledgements
We acknowledge University of Alberta and National Institute for Nanotechnology for the postdoctoral fellowship. We acknowledge the financial support received from the National Research Council Canada. E. coli K12 bacteria was kindly provided by Prof. Mansel Griffiths (Canadian Research Institute for Food Safety, University of Guelph, Guelph, ON, Canada).
References
- Lazcka O, Del Campo FJ, Muñoz FX (2007) Pathogen detection: a perspective of traditional methods and biosensors. Biosens Bioelectron 22: 1205-1217.
- Dwivedi HP, Jaykus LA (2011) Detection of pathogens in foods: the current state-of-the-art and future directions. Crit Rev Microbiol 37: 40-63.
- Cox NA, Berrang ME (2000) Inadequacy of selective plating media in field detection of Salmonella. J Appl Poultry Res 9: 403-406.
- Dover JE, Hwang GM, Mullen EH, Prorok BC, Suh SJ (2009) Recent advances in peptide probe-based biosensors for detection of infectious agents. J Microbiol Methods 78: 10-19.
- Theron J, Eugene Cloete T, de Kwaadsteniet M (2010) Current molecular and emerging nanobiotechnology approaches for the detection of microbial pathogens. Crit Rev Microbiol 36: 318-339.
- Homola J (2008) Surface plasmon resonance sensors for detection of chemical and biological species. Chem Rev 108: 462-493.
- Ilic B, Czaplewski D, Craighead HG, Neuzil P, Campagnolo C et al. (2000) Mechanical resonant immunospecific biological detector. Appl Phys Lett 77: 450-452.
- Ilic B, Czaplewski D, Zalalutdinov M, Craighead HG, Neuzil P et al. (2001) Single cell detection with micromechanical oscillators. J Vac Sci Technol B 19: 2825-2828.
- Velusamy V, Arshak K, Korostynska O, Oliwa K, Adley C (2010) An overview of foodborne pathogen detection: in the perspective of biosensors. Biotechnol Adv 28: 232-254.
- Elsholz B, Wörl R, Blohm L, Albers J, Feucht H, et al. (2006) Automated detection and quantitation of bacterial RNA by using electrical microarrays. Anal Chem 78: 4794-4802.
- Torres-Chavolla E, Alocilja EC (2009) Aptasensors for detection of microbial and viral pathogens. Biosens Bioelectron 24: 3175-3182.
- Sforza S, Corradini R, Tedeschi T, Marchelli R (2011) Food analysis and food authentication by peptide nucleic acid (PNA)-based technologies. Chem Soc Rev 40: 221-232.
- Jelinek R, Kolusheva S (2004) Carbohydrate biosensors. Chem Rev 104: 5987-6015.
- Skottrup PD, Nicolaisen M, Justesen AF (2008) Towards on-site pathogen detection using antibody-based sensors. Biosens Bioelectron 24: 339-348.
- Wang W, Singh S, Zeng DL, King K, Nema S (2007) Antibody structure, instability, and formulation. J Pharm Sci 96: 1-26.
- Kutter E, Sulakvelidze A (2004) Bacteriophages: Biology and Applications. CRC Press, Boca Raton, FL.
- Gervais L, Gel M, Allain B, Tolba M, Brovko L, et al. (2007) Immobilization of biotinylated bacteriophages on biosensor surfaces. Sens Actuators B 125: 615-621.
- Tolba M, Minikh O, Brovko LY, Evoy S, Griffiths MW (2010) Oriented immobilization of bacteriophages for biosensor applications. Appl Environ Microbiol 76: 528-535.
- Balasubramanian S, Sorokulova IB, Vodyanoy VJ, Simonian AL (2007) Lytic phage as a specific and selective probe for detection of Staphylococcus aureus--A surface plasmon resonance spectroscopic study. Biosens Bioelectron 22: 948-955.
- Edgar R, McKinstry M, Hwang J, Oppenheim AB, Fekete RA, et al. (2006) High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. Proc Natl Acad Sci U S A 103: 4841-4845.
- Nanduri V, Sorokulova IB, Samoylov AM, Simonian AL, Petrenko VA, et al. (2007) Phage as a molecular recognition element in biosensors immobilized by physical adsorption. Biosens Bioelectron 22: 986-992.
- Olsen EV, Sorokulova IB, Petrenko VA, Chen IH, Barbaree JM, et al. (2006) Affinity-selected filamentous bacteriophage as a probe for acoustic wave biodetectors of Salmonella typhimurium. Biosens Bioelectron 21: 1434-1442.
- Singh A, Glass N, Tolba M, Brovko L, Griffiths M, et al. (2009) Immobilization of bacteriophages on gold surfaces for the specific capture of pathogens. Biosens Bioelectron 24: 3645-3651.
- Naidoo R, Singh A, Arya SK, Beadle B, Glass N, et al. (2012) Surface-immobilization of chromatographically purified bacteriophages for the optimized capture of bacteria. Bacteriophage 2: 15-24.
- Arya SK, Singh A, Naidoo R, Wu P, McDermott MT, et al. (2011) Chemically immobilized T4-bacteriophage for specific Escherichia coli detection using surface plasmon resonance. Analyst 136: 486-492.
- Singh A, Arya SK, Glass N, Hanifi-Moghaddam P, Naidoo R, et al. (2010) Bacteriophage tailspike proteins as molecular probes for sensitive and selective bacterial detection. Biosens Bioelectron 26: 131-138.
- Baxa U, Steinbacher S, Miller S, Weintraub A, Huber R, et al. (1996) Interactions of phage P22 tails with their cellular receptor, Salmonella O-antigen polysaccharide. Biophys J 71: 2040-2048.
- Israel V, Rosen H, Levine M (1972) Binding of bacteriophage P22 tail parts to cells. J Virol 10: 1152-1158.
- Danner M, Fuchs A, Miller S, Seckler R (1993) Folding and assembly of phage P22 tailspike endorhamnosidase lacking the N-terminal, head-binding domain. Eur J Biochem 215: 653-661.
- Miller S, Schuler B, Seckler R (1998) Phage P22 tailspike protein: removal of head-binding domain unmasks effects of folding mutations on native-state thermal stability. Protein Sci 7: 2223-2232.
- Fritz J (2008) Cantilever biosensors. Analyst 133: 855-863.
- Wood MA, Riehle M, Wilkinson CDW (2002) Patterning colloidal nanotopographies. Nanotechnology13: 605-609.
- Waseh S, Hanifi-Moghaddam P, Coleman R, Masotti M, Ryan S, et al. (2010) Orally administered P22 phage tailspike protein reduces salmonella colonization in chickens: prospects of a novel therapy against bacterial infections. PLoS One 5: e13904.
- Liu YC, Rieben N, Iversen L, Sørensen BS, Park J, et al. (2010) Specific and reversible immobilization of histidine-tagged proteins on functionalized silicon nanowires. Nanotechnology 21: 245105.
- Kropinski AM, Arutyunov D, Foss M, Cunningham A, Ding W, et al. (2011) Genome and proteome of Campylobacter jejuni bacteriophage NCTC 12673. Appl Environ Microbiol 77: 8265-8271.
- Singh A, Arutyunov D, McDermott MT, Szymanski CM, Evoy S (2011) Specific detection of Campylobacter jejuni using the bacteriophage NCTC 12673 receptor binding protein as a probe. Analyst 136: 4780-4786.
- Singh A, Arutyunov D, Szymanski CM, Evoy S (2012) Bacteriophage based probes for pathogen detection. Analyst 137: 3405-3421.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 15659
- [From(publication date):
specialissue-2013 - Apr 03, 2025] - Breakdown by view type
- HTML page views : 11020
- PDF downloads : 4639