Research Article Open Access
High Frequency of CYP3A4*1B among Opiate Dependent Patients in Malaysia
Nasir Mohamad1,2*, Nurfadhlina M2, Nazila T2, Ahmad A2, Nor Hidayah Abu Bakar3, Hussein H4, Khafidz I5 and Ismail R2,61Department of Emergency Medicine, School of Medical Sciences, Universiti Sains Malaysia, Kelantan, Malaysia
2Pharmacogenetics Research Group, Institute for Research in Molecular Medicine (INFORMM), Universiti Sains Malaysia, Kelantan, Malaysia
3Department of Pathology, Hospital Raja Perempuan Zainab II, Kelantan, Malaysia
4Department of Psychological Medicine, Universiti of Malaya, Kuala Lumpur, Malaysia
5Klinik Dr Khafidz, 43000 Semenyih Bandar Teknologi Kajang, Selangor, Malaysia
6Pharmacogenetics Research Group, Institute for Research in Molecular Medicine (INFORMM), Universiti Sains Malaysia, Kelantan, Malaysia
- *Corresponding Author:
- Nasir Mohamad
Department of Emergency Medicine/Institute for
Research in Molecular Medicine (INFORMM), School of Medical Sciences
Health Campus, Universiti Sains Malaysia
16150 Kubang Kerian, Kelantan, Malaysia
Tel: 609-7672407
Fax: 609-7657267
E-mail: drnasirmohamadkb@yahoo.com
Received July 14, 2012; Accepted July 24, 2012; Published July 27, 2012
Citation: Mohamad N, Nurfadhlina M, Nazila T, Ahmad A, Abu Bakar NH, et al. (2012) High Frequency of CYP3A4*1B among Opiate Dependent Patients in Malaysia. J Addict Res Ther 3:130. doi:10.4172/2155-6105.1000130
Copyright: © 2012 Mohamad N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Addiction Research & Therapy
Abstract
The sharing of injection needles among drug user is a leading cause for the spread of HIV/AIDS. Malaysia introduced methadone as a management of heroin dependents to reduce HIV spread. Methadone has variable pharmacokinetics and CYP3A4 has been implicated in its metabolism. The objective of this study therefore was to determine if polymorphisms exist with CYP3A4 among opiate users in Malaysia. This study was approved by Ethics Committees at University of Malaya and Universiti Sains Malaysia. Control subjects comprised blood donors, students and residents of a village. Opiate-dependents were from methadone clinics and drop-in centers. They signed a written-informed consent to participate and gave blood for DNA CYP3A4 genotyping. DNA was extracted using QIAgen DNA mini kit. A nested two-step allele specific PCR method was developed to detect CYP3A4*1B, CYP3A4*3, CYP3A4*4, CYP3A4*5, CYP3A4*6, CYP3A4*7, CYP3A4*8, CYP3A4*9, CYP3A4*10, CYP3A4*11, CYP3A4*12, CYP3A4*13, CYP3A4*14, CYP3A4*15 and CYP3A4*16. Normal controls comprised Malays, Chinese and Indians but opiate-dependent subjects were majority Malay males. Control subjects all carried the wild-type gene. Mutant CYP3A4*1B allele was found in 2.17% of opiate-dependent subjects. Our results revealed that CYP3A4 was not polymorphic among Malaysian Malays, Chinese and Indians who were not opiate-dependent. To date, we are not aware of any study to associate CYP3A4 polymorphism and heroin addiction. It is conceivable that altered CYP3A4 function may contribute towards addiction liabilities in subsets of individuals. We conclude that CYP3A4 is polymorphic among heroin-dependent individuals. The mutation, CYP3A4*1B is not silent. This may have implications on heroin addiction liability as well as on dose requirements for MMT and HAART.
Keywords
CYP3A4; Heroin addiction; Methadone; Anti-retrovirals; Endogenous metabolism
Introduction
Heroin use and the sharing of injection needles among drug users is a leading cause for the spread of HIV/AIDS in Malaysia and several other South East Asian countries. In 2006, Malaysia introduced methadone as a management of heroin dependents in an effort to break the viscous cycles of heroin addiction and HIV/AIDS. Methadone is taken orally and it prevents withdrawal and reduces illicit drug use. It is a vital public health strategy for HIV/AIDS risk reduction [1]. Variability in methadone clearance, susceptibility to drug interactions, and a long elimination half-life can however be major impediments to optimal methadone use [2,3]. In in vitro drug metabolism studies, CYP3A4 has been implicated in methadone metabolism [4]. Authors have suggested dosing guidelines for methadone and warned about the potential for CYP3A4-mediated interactions and suggested the need to adjust doses accordingly [2,3,5-11].
CYP3A4 is the most abundant CYPs in human. Substrates of CYP3A4 include methadone, anti-depressants, immunosuppressive agents, macrolide antibiotics, benzodiazepines, calcium channel blockers [12], and several antiretroviral [4,13,14] used in HIV/AIDS. Also importantly, CYP3A4 is involved in the metabolism of endogenous substances that include testosterone [15], progesterone [16], cortisol [17], and 17β-estradiol [18], and this may be important in the patho physiology of diseases including drug dependence. CYP3A4 exhibits genetic polymorphism and as with other polymorphic enzymes, the identification of molecular variants in the CYP3A4 gene is a major focus of pharmaco genetic studies.
Malaysia is a multiethnic country where genetic polymorphisms of several drug metabolizing enzymes have been previously described [19-26]. Although our earlier studies failed to detect polymorphism at the CY3A4 locus, mutant alleles have recently been described in Malaysia [27]. The objective of this study therefore was to determine if polymorphisms exist with CYP3A4 among opiate users in Malaysia given its role in endogenous metabolism and in the metabolism of methadone.
Methods
Recruitment of subjects
The protocols for this study received the approval of the Ethics Committees at the University of Malaya in Kuala Lumpur and Universiti Sains Malaysia in Kelantan. For the normal controls, subjects comprised blood donors at Universiti Malaysia Medical Centre, Universiti Sains Malaysia Hospital, students at the two universities and residents of a Malaysian Indian community in Kuala Krai, Kelantan. For opiate-dependent individuals, subjects were enrolled from several methadone clinics and drop-in centres for drug users in Kuala Lumpur and in Kota Bharu, Kelantan. They were given an explanation about the study and were invited to participate if they were willing to sign a writteninformed consent. They were administered standard questionnaires to obtain demographic data and to establish opiate dependence. Five milliliter of blood was then obtained from them for DNA extraction and CYP3A4 genotyping.
| No. | Primer Name | Sequence (5’ to 3’) |
|---|---|---|
| 1 | ProFw | GTT CAG GGA AAC AGG CGT GGA |
| 2 | ProRv | ACA GAT AAG GGA AAG AGA GGC |
| 3 | Ex5Fw | CCA CAC AAA TAC ATC CCA GGA C |
| 4 | Int6Rv | GGT CAC TGG AAT AAC CCA ACA G |
| 5 | Ex3Fw | CCT CTA ACT GCC AGC AAG |
| 6 | Ex3Rv | GCA TGC AGA TTC CCA TTG C |
| 7 | Ex7Fw | GTT GCA TGC ATA GAG GAA GGA TGG |
| 8 | Ex7Rv | GAT GAC AGG GTT TGT GAC AGG GG |
| 9 | Ex9Fw | GAG CCA TCT CAC ATG ATA GC |
| 10 | Ex9Rv | CAA ACA TGT GTC GTT CTG C |
| 11 | Ex11Fw | GCA CCA CCC ACC TAT GAT AC |
| 12 | Ex11Rv | CTT GAA CCA GGC TGG TTC AG |
| 13 | Ex12Fw | GTG GAA CCA GAT TCA GCA AG |
| 14 | Ex12Rv | CTG TGT TTC TTT ACA AGG TTT G |
| 15 | CYP3A4*1B Wt | CTA TTA AGT CGC CTC TCT CT |
| 16 | CYP3A4*1B Mt | CTA TTA AGT CGC CTC TCT CC |
| 17 | CYP3A4*8 Wt | GAA GAA TGG AAG AGA TTA GC |
| 18 | CYP3A4*8 Mt | GAA GAA TGG AAG AGA TTA GA |
| 19 | CYP3A4*15 Wt | GGT GAG AAA TCT GAG GCG |
| 20 | CYP3A4*15 Mt | GGT GAG AAA TCT GAG GCA |
| 21 | CYP3A4*5 Wt | ATA CTT ATT GAG AGA AAG AAT G |
| 22 | CYP3A4*5 Mt | ATA CTT ATT GAG AGA AAG AAT C |
| 23 | CYP3A4*9 Wt | CTT ACT CTT TCA AGG TGA C |
| 24 | CYP3A4*9 Mt | CTT ACT CTT TCA AGG TGA T |
| 25 | CYP3A4*13 Wt | GCC TGA GAA GTT CCT CCC |
| 26 | CYP3A4*13 Mt | GCC TGA GAA GTT CCT CCT |
| 27 | CYP3A4*16 Wt | CAC TCC AAA TGA TGT GCT AG |
| 28 | CYP3A4*16 Mt | CAC TCC AAA TGA TGT GCT AC |
| 29 | CYP3A4*11 Wt | GAC ATG GTG GTG AAT GAA AC |
| 30 | CYP3A4*11 Mt | GAC ATG GTG GTG AAT GAA AT |
| 31 | CYP3A4*14 Wt | CAC CAG GCT GAC AGC CA |
| 32 | CYP3A4*14 Mt | CAC CAG GCT GAC AGC CG |
| 33 | CYP3A4*6 Wt | CCT TCA GCT GAT GAT TGA C |
| 34 | CYP3A4*6 Mt | CCT TCA GCT GAT GAT TGA AC |
| 35 | CYP3A4*7 Wt | TTG TTT CTC CTC CCA GGG |
| 36 | CYP3A4*7 Mt | TTG TTT CTC CTC CCA GGA |
| 37 | CYP3A4*10 Wt | GCC TGT CAC CTT GAA AG |
| 38 | CYP3A4*10 Mt | GCC TGT CAC CTT GAA AC |
| 39 | CYP3A4*12 Wt | TAT TCC CAA TTG CTA TGA GAC |
| 40 | CYP3A4*12 Mt | TAT TCC CAA TTG CTA TGA GAT |
| 41 | CYP3A4*3 Wt | CCA GAA ACT GCA TTG GCA T |
| 42 | CYP3A4*3 Mt | CCA GAA ACT GCA TTG GCA C |
| 43 | CYP3A4*4 Wt | CAT CCT CAG CTA TAG AGA T |
| 44 | CYP3A4*4 Mt | CAT CCT CAG CTA TAG AGA C |
Table 1: Primer sequences used in nested two-step allele specific PCR of CYP3A4 allele.
Isolation of DNA and PCR genotyping
Genomic DNA was extracted from subjects’ blood by using QIAgen DNA mini kit (QIAGEN, Hilden) according to the protocols recommended by the manufacturer. The quantity and quality of the extracted DNA was determined on the spectrophotometer with measurements done at 260 and 280 nm.
A nested two-step allele specific PCR method was developed to detect CYP3A4*1B, CYP3A4*3, CYP3A4*4, CYP3A4*5, CYP3A4*6, CYP3A4*7, CYP3A4*8, CYP3A4*9, CYP3A4*10, CYP3A4*11, CYP3A4*12, CYP3A4*13, CYP3A4*14, CYP3A4*15 and CYP3A4*16. The first PCR used the primers listed in Table 1 to isolate the CYP3A4 gene to improve on specificity. The products were used as templates for three allele specific second PCR using primers also listed in Table 1 to detect the alleles of interest. All PCR reactions were performed using Bio-Rad MyCycler™ Thermal Cycler.
In the first PCR, the reaction mixture consisted of 1X Biotools Reaction Buffer, 3.0 mM Biotools MgCl2, 0.2 mM of each Biotools dNTP Mix, 0.2 pmol of each primer which were divided into two groups; Set A and Set B as listed in Table 4, 1.0 U Biotools DNA Polymerase, and 20-100 ng of DNA in a total volume of 25 μL. The cycle condition consisted of pre-denaturation for 5 min at 94°C, followed by 10 cycles of denaturation at 94°C for 45 s, annealing at 65°C for 45 s with touchdown from 65°C to 60.5°C (reduce by 0.5°C every cyle) and extension at 72°C for 30 s. This was followed by another 25 cycles of denaturation at 94°C for 45 s, annealing at 60°C for 45 s and extension at 72°C for 30 s. A final extension 72°C for 5 min was also included.
Subsequent to successful first PCR, the products were subjected to two parallel allele-specific second PCR, one with wild-type specific primers and the other with mutation-specific primers, were carried out in separate PCR reactions. In this second PCR, five set of reaction mixtures and cycle condition were required to detect all the alleles as listed in Table 2 and Table 3. The predicted PCR products as listed in Table 4 were analyzed on 2.5 % agarose gel in 1X TBE buffer run at 90 V for 75 min.
| Set 1 | Set 2 | Set 3 | Set 4 | Set 5 | |
|---|---|---|---|---|---|
| Reaction Mixture (reaction volume – 25µl ) | |||||
| Reaction Buffer (10X)a | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| MgCl2 (50mM)a | 2.0 | 2.0 | 2.0 | 2.0 | 1.5 |
| dNTP Mix (each) (10mM)a | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 |
| DNA Polymerase (1U)a | 1.5 | 1.0 | 1.0 | 1.0 | 1.0 |
| Primers (each) (nM) | Refer Table 3 | ||||
| First-PCR product (mL) | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 |
| Cycle Condition | |||||
| Pre-denaturation (°C:min) | 94:4 | 94:4 | 94:4 | 94:4 | 94:4 |
| Denaturation (°C:sec) | 94:30 | 94:30 | 94:30 | 94:30 | 94:30 |
| Annealing (°C:sec) | 60b:30 | 58c:30 | 64:30 | 58c:30 | 58:30 |
| Extension (°C:sec) | 72:30 | 72:30 | 72:30 | 72:30 | 72:30 |
| Number of cycles | 16 | 16 | 15 | 16 | 15 |
| Final extension (°C:min) | 72:7 | 72:7 | 72:7 | 72:7 | 72:7 |
| Hold (°C:min) | 20:∞ | 20:∞ | 20:∞ | 20:∞ | 20:∞ |
| a Biotools Company (Spain) b Touchdown from 60°C to 53.6°C (reduce by 0.4°C every cycle) c Touchdown from 58°C to 52°C (reduce by 0.4°C every cycle) |
|||||
Table 2: Reaction mixtures and cycle condition for allele-specific second PCR.
| Set 1 | Conc. (pmol) |
Set 3 | Conc. (pmol) |
Set 5 | Conc. (pmol) |
|---|---|---|---|---|---|
| Pro Fw (5 pmol) | 0.2 | Pro Fw (5 pmol) | 0.20 | Ex12 Rv (5pmol) | 0.15 |
| Int6 Rv (5 pmol) | 0.2 | Ex11 Rv (5 pmol) | 0.25 | Ex5 Fw (5pmol) | 0.3 |
| CYP3A4*1B Wt/ CYP3A4*1B Mt (5 pmol) | 0.15 | CYP3A4*14 Wt/ CYP3A4*14 Mt (5 pmol) | 0.15 | CYP3A4*3 Wt/ CYP3A4*3 Mt (5pmol) | 0.15 |
| CYP3A4*15 Wt/ CYP3A4*15 Mt (5 pmol) | 0.2 | CYP3A4*11 Wt/ CYP3A4*11 Mt (5 pmol) | 0.30 | CYP3A4*4 Wt/ CYP3A4*4 Mt (5pmol) | 0.3 |
| CYP3A4*8 Wt/ CYP3A4*8 Mt (5 pmol) | 0.3 | Set 4 | Conc. (pmol) |
||
| Set 2 | Conc. (pmol) |
Ex3 Rv (5 pmol) | 0.4 | ||
| Ex7 Fw (5 pmol) | 0.3 | Ex9 Rv (5 pmol) | 0.4 | ||
| Ex11 Rv (5 pmol) | 0.25 | Int6 Rv (5 pmol) | 0.3 | ||
| Ex5 Fw (5 pmol) | 0.4 | Ex11 Rv (5 pmol) | 0.2 | ||
| CYP3A4*9 Wt/ CYP3A4*9 Mt (5 pmol) | 0.4 | CYP3A4*7 Wt/ CYP3A4*7 Mt (5 pmol) | 0.25 | ||
| CYP3A4*5 Wt/ CYP3A4*5 Mt (5 pmol) | 0.25 | CYP3A4*6 Wt/ CYP3A4*6 Mt (5 pmol) | 0.25 | ||
| CYP3A4*16 Wt/ CYP3A4*16 Mt (5 pmol) | 0.1 | CYP3A4*12 Wt/ CYP3A4*12 Mt (5 pmol) | 0.2 | ||
| CYP3A4*13 Wt/ CYP3A4*13 Mt (5 pmol) | 0.3 | CYP3A4*10 Wt/ CYP3A4*10 Mt (5 pmol) | 0.3 |
Table 3: Final concentration of each primers used for each Set in allele-specific second PCR.
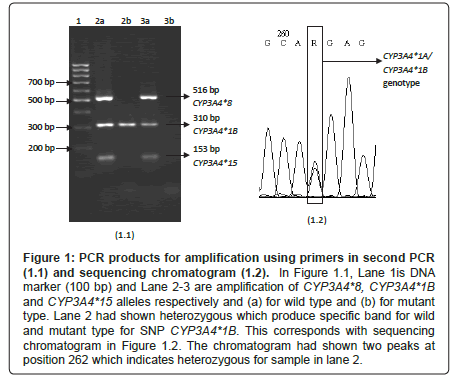
Figure 1: PCR products for amplification using primers in second PCR (1.1) and sequencing chromatogram (1.2). In Figure 1.1, Lane 1is DNA marker (100 bp) and Lane 2-3 are amplification of CYP3A4*8, CYP3A4*1B and CYP3A4*15 alleles respectively and (a) for wild type and (b) for mutant type. Lane 2 had shown heterozygous which produce specific band for wild and mutant type for SNP CYP3A4*1B. This corresponds with sequencing chromatogram in Figure 1.2. The chromatogram had shown two peaks at position 262 which indicates heterozygous for sample in lane 2.
Upon successful PCR, three heterozygous samples were sent for sequencing. The PCR products were purified using QIAquick PCR Purification kit before being sent for DNA sequencing by standard kit of ABI PRISM Big Dye Terminator. The sequencing results were verified against the published sequence for CYP3A4 (Gene bank accession number: AF185589).
Results
Subjects
Control subjects comprised 270 Malays, 172 Chinese and 174 Indians and their average age are 29.4, 30.2, and 26.1 respectively. The control subject consist of 174 males and 96 females for Malays, 132 males and 40 females for Chinese and 123 males and 51 females Indian. Opiate dependent subjects comprised 114 Malays with average age of 39.57. Out of the 114 subjects, 112 are males and 2 are females.
PCR genotyping
Figure 1.1 shows a gel picture from a successful Set 1 for second PCR for two representative samples from two different subjects. CYP3A4*1B was detected in five subjects who carried heterozygous CYP3A4*1A/CYP3A4*1B genotype. The remaining subjects did not carry any mutant allele. The heterozygous subjects then were validated using DNA sequencing and the chromatogram is shown in Figure 1.2. Double peaks was produced at position 262 which indicates the presence of heterozygous CYP3A4*1A/CYP3A4*1B genotype.
| Forward | Reverse | Product size (bp) | |
|---|---|---|---|
| First PCR | |||
| Set A | ProFw | ProRv | 831 |
| Ex5Fw | Int6Rv | 757 | |
| Set B | Ex3Fw | Ex3Rv | 360 |
| Ex7Fw | Ex7Rv | 447 | |
| Ex9Fw | Ex9Rv | 412 | |
| Ex11Fw | Ex11Rv | 276 | |
| Ex12Fw | Ex12Rv | 173 | |
| Allele-specific second PCR | |||
| Set 1 | CYP3A4*8 Wt/ CYP3A4*8 Mt | Int6Rv | 516 |
| ProFw | CYP3A4*1B Wt/ CYP3A4*1B Mt | 310 | |
| CYP3A4*15 Wt/ CYP3A4*15 Wt | Int6Rv | 153 | |
| Set 2 | Ex5Fw | CYP3A4*9 Wt/ CYP3A4*5 Mt | 663 |
| Ex7Fw | CYP3A4*5 Wt/ CYP3A4*5 Mt | 221 | |
| Ex7Fw | CYP3A4*16 Wt/ CYP3A4*16 Wt | 120 | |
| CYP3A4*13 Wt/ CYP3A4*13 Wt | Ex11Rv | 73 | |
| Set 3 | ProFw | CYP3A4*14 Wt/ CYP3A4*14 Wt | 742 |
| CYP3A4*11 Wt/ CYP3A4*11 Wt | Ex11Rv | 234 | |
| Set 4 | CYP3A4*7 Wt/ CYP3A4*7 Wt | Ex3Rv | 261 |
| CYP3A4*12 Wt/ CYP3A4*12 Wt | Ex11Rv | 206 | |
| CYP3A4*6 Wt/ CYP3A4*6 Wt | Ex9Rv | 178 | |
| CYP3A4*10 Wt/ CYP3A4*10 Wt | Int6Rv | 117 | |
| Set 5 | Ex5Fw | CYP3A4*4 Wt/ CYP3A4*4 Wt | 242 |
| CYP3A4*3 Wt/ CYP3A4*3 Wt | Ex12Rv | 101 | |
Table 4: Primer combinations for amplification of first and allele-specific second PCR of CYP3A4.
| SNPs | Observed genotype frequency (%) ± 95% Confidence Interval (CI) | Predicted genotype frequency (%) by Hardy-Weinberg Law ± 95% Confidence Interval (CI) |
|---|---|---|
| CYP3A4*1B | ||
| CYP3A4*1A/ *1A | 95.65 ± 2.82 | 95.70 ± 2.80 |
| CYP3A4*1A/ *1B | 4.35 ± 2.82 | 4.25 ± 2.79 |
| CYP3A4*1B/ *1B | 0 | 0.05 ± 0.30 |
| n | 115 | |
Table 5: Genotype frequency according to Hardy-Weinberg equilibrium in opiatedependent patients.
The most common allele carried by subjects was CYP3A4*1A that occurred at a frequency of 97.83%. CYP3A4*1B was found in 2.17% of subjects and another fourteen alleles were not detected. In terms of observed genotypes, 95.65 % were CYP3A4*1A/*1A and 4.35% CYP3A4*1A/*1B. However in terms of predicted genotype frequency by Hardy-Weinberg Law, 95.70 % were CYP3A4*1A/*1A, 4.25% CYP3A4*1A/*1B and 0.05% CYP3A4*1B/*1B. Both observed and predicted genotype frequency is shown in Table 5 with 95% confidence interval.
Discussion
Malaysia is facing a double challenge of illicit drug use and a rapid spread of HIV/AIDS. As a “harm-reducing” measure, the Malaysian Government introduced Methadone Maintenance Therapy (MMT) for heroin users in Malaysia. In our previous study in 52 Malay male subjects given a single 8 mg dose of methadone, we found a wide variability in its clearance, consistent with other studies previously reported [28]. The variability in our 52 subjects could not be explained by polymorphisms at the CYP2D6 locus, a very polymorphic gene in our population [19-21], another enzyme implicated in methadone metabolism [29]. This led us to investigate the genetic polymorphism at the CYP3A4 locus among heroin drug users. Apart from being involved in the metabolism of methadone, CYP3A4 is also involved in the metabolism of several anti-retrovirals used in the treatment of AIDS [13], thus our interest. We were also led to study this because of the role of CYP3A4 in endogenous metabolism, especially involving testosterone, as a possible contributing factor to heroin addiction. Heroin addiction in Malaysia is a predominantly male disease. It would therefore be interesting if an association can be found between altered CYP3A4 testosterone metabolism and heroin addiction.
Our results revealed that CYP3A4 was not polymorphic among Malaysian Malays, Chinese and Indians who were not opiate-dependent. Many previous studies failed to show significant polymorphism at the CYP3A4 locus. Rebbeck, et al. (1998) reported a 9.6% frequency for CYP3A4*1B in healthy White subjects [30] but Lamba, et al. (2002) reported a very low percentage among their healthy Caucassian, African Americans, Mexican, Pacific Islander and Middle Eastern subjects [12]. Mutant alleles, with some exceptions, appeared to occur only at low percentages. This probably implies an important role of CYP3A4 in endogenous metabolism and therefore mutations would most likely be selected out. It is assumed that evolutionary pressure acts against coding polymorphisms and environment tends to wipe them out.
Heroin addiction is a complex disease. It probably results from deleterious interactions between genes and the environment. Many genes have been studied for potential roles in heroin addiction and to date, we are not aware of any study to associate CYP3A4 polymorphism and heroin addiction. It was interesting therefore to find a 2.17 % frequency for CYP3A4*1B among our opiate-dependent subjects, against the background of an absence of polymorphism among Malaysian non-drug dependent subjects. P450 enzymes in liver microsomes have been reported to play important roles in the metabolism of steroids, fatty acids, fat soluble enzymes and prostaglandins [16] and CYP3A4 has been shown to be a major form that catabolised steroids in human livers. It is conceivable that altered CYP3A4 function may contribute towards addiction liabilities in subsets of individuals, either directly through altered metabolism of some putative, yet unknown endogenous substance(s) or indirectly through its effects on steroid and prostaglandin metabolism.
CYP3A4*1B allele is also known as CYP3A4-V and has an A→G substitution at position -392 on the 5’ promoter region. It was first identified by Rebbeck et al. in 1998 [30]. The functional significance of CYP3A4*1B mutation has recently been reported. CYP3A4*1B/ CYP3A4*1B genotype has been reported to reduce the absorption of indinavir in patients initiating Highly Active Anti-Retroviral Treatment (HAART) naïve patients [14]. This SNP has also been associated with the more aggressive forms and advanced clinical stages of prostate cancer in African-Americans but not in Portugese victims [31]. It also linked to increase transcriptional activity. The finding of a 2.17% frequency for CYP3A4*1B among our heroin using population was therefore interesting and the functional significance of this mutation may need to be elucidated. Future studies on its association should also probably include family data to allow for a transmission disequilibrium test to be performed. CYP3A4 is known to be involved in endogenous metabolism of steroid hormones, both testosterone (2β-, 6β-, or 15β-hydroxytestosterone) and estrogen (4- and 16α-hydroxylation) [32-35]. As far as addiction is concerned, stress influences the pathophysiology of addiction, to be linkages with drug dependence.
We conclude that CYP3A4 is polymorphic among heroindependent individuals. The mutation, CYP3A4*1B is not silent. This may have implications on heroin addiction liability as well as on dose requirements for MMT and HAART.
Funding Source
This work was supported by a USM grant under the “Research University Program” (Grant Number: 1001/PSK/8620014)
References
- Connock M, Juarez-Garcia A, Jowett S, Frew E, Liu Z, et al. (2007) Methadone and buprenorphine for the management of opioid dependence: a systematic review and economic evaluation. Health Technol Assess 11: 1–171.
- Foster DJ, Somogyi AA, Dyer KR, White JM, Bochner F(2000) Steady-state pharmacokinetics of (R)- and (S)-methadone inmethadone maintenance patients. Br J Clin Pharmacol 50: 427-440.
- Ferrari A, Coccia CP, Bertolini A, Sternieri E (2004) Methadone–metabolism, pharmacokinetics and interactions. Pharmacol Res50: 551–559.
- Kharasch ED, Walker A, Whittington D, Hoffer C, Bedynek PS (2009) Methadone metabolism and clearance are induced by nelfinavir despite inhibition of cytochrome P450 (CYP3A) activity. Drug Alcohol Depend 101: 158-168.
- Eap CB, Buclin T, Baumann P (2002) Interindividual variability of the clinical pharmacokinetics of methadone: implications for the treatment of opioid dependence. Clin Pharmacokinet 41: 1153–1193.
- Shinderman M, Maxwell S, Brawand-Amey M, Golay KP, Baumann P, et al. (2003) Cytochrome P4503A4 metabolic activity, methadone blood concentrations, and methadone doses. Drug Alcohol Depend 69: 205–211.
- Wang J-S, DeVane CL (2003) Involvement of CYP3A4, CYP2C8, and CYP2D6 in the metabolism of (R)- and (S)-methadone in vitro. Drug Metab Dispos 31: 742–747.
- McCance-Katz EF (2005) Treatment of opioid dependence and coinfection with HIV and hepatitis C virus in opioid-dependent patients: the importance of drug interactions between opioids and antiretroviral agents. Clin Infect Dis 41: 89-95.
- Bruce RD, Altice FL, Gourevitch MN, Friedland GH (2006) Pharmacokinetic drug interactions between opioid agonist therapy and antiretroviral medica medications: implications and management for clinical practice. J Acquir Immune Defic Syndr 41: 563–572.
- Coller JK, Barratt DT, Dahlen K, Loennechen MH, Somogyi AA (2006) ABCB1 genetic variability and methadone dosage requirements in opioid-dependent individuals. Clin Pharmacol Ther 80: 682–690.
- Crettol S, Deglon JJ, Besson J, Croquette-Krokar M, Hammig R, et al. (2006) ABCB1 and cytochrome P450 genotypes and phenotypes: influence on methadone plasma levels and response to treatment. Clin Pharmacol Ther 80: 668–681.
- Lamba JK, Lin YS, Thummel K, Daly A, Watkins PB, et al. (2002) Common allelic variants of cytochrome P4503A4 and their prevalence in different populations. Pharmacogenetics 12: 121-132.
- Decker CJ, Laitinen LM, Bridson GW, Raybuck SA, Tung RD, et al. (1998) Metabolism of amprenavir in liver microsomes: role of CYP3A4 inhibition for drug interactions. J Pharm Sci 87: 803-807.
- Bertrand J, Treluyer JM, Panhard X, Tran A, Auleley S, et al. (2009) Influene of pharmacogenetics of indinavir disposition and short-term response in HIV patients initiating HAART. Eur J Clin Pharmacol 65: 667-678.
- Waxman DJ, Attisano C, Guengerich FP, Lapenson DP (1988) Human liver microsomal steroid metabolism: identification of the major microsomal steroid hormone 6ß-hydroxylase cytochrome P-450 enzyme. Arch Biochem Biophys 263: 424-436.
- Yamazaki H, Shimada T (1997) Progesterone and testosterone hydroxylation by Cytochromes P450 2C19, 2C9, and 3A4 in human liver microsomes. Arch Biochem Biophys 346: 161-169.
- Abel SM, Back DJ (1993) Cortisol metabolism in vitro-III. Inhibition of microsomal 6ß-hydroxylase and cytosolic 4-ene-reductase. J Steroid Biochem Mol Biol 46: 827-832.
- Kerlan V, Dreano Y, Bercovici JP, Beaune PH, Floch HH, et al. (1992) Nature of cytochrome P450 involved in the 2-/4-hydroxylation of estradiol in human liver microsomes. Biochem Pharmacol 44: 1745-1756.
- Ismail R, Teh LK (2001) Genetic polymorphism of CYP2D6: Malaysian Indians have the highest frequncy for CYP2D6*4 in Asia. Eur J Clin Pharmacol 57: 617-618.
- Teh LK, Ismail R, Yusoff R, Hussein A, Isa MN, et al. (2001) Hetergeneity of the CYP2D6 gene among Malays in Malaysia. J Clin Pharm Ther 26 : 205-211.
- Ismail R, Teh LK, Amir J, Alwi Z, Lopez CG (2003) Genetic polymorphism of CYP2D6 in Chinese subjects in Malaysia. J Clin Pharm Ther 28: 279-284.
- Muthiah YD, Lee WL, Teh LK, Ong CE, Ismail R (2005) Genetic polymorphism of CYP2C8*2 in three Malaysian ethnics: CYP2C8*2 and CYP2C8*3are found in Malaysian Indians. J Clin Pharm Ther 30: 487-4902.
- Nurfadhlina M, Foong K, Teh LK, Tan SC, Mohd Zaki S, et al. (2006) CYP2A6 polymorphisms in Malays, Chinese and Indians. Xenobiotica 36: 684-692.
- Zainuddin Z, Teh LK, Suhaimi AW, Ismail R (2006) Malaysian Indians are genticaly similar to Caucasians: CYP2C9 polymorphism. J Clin Pharm Ther 31: 187-191.
- Teh LK, Lee WL, Amir J, Salleh MZ, Ismail R (2007) Single step PCR for detection of allelic variation of MDR1 gene (P-glycoprotein) among three ethnic groups in Malaysian. J Clin Pharm Ther 32: 313-319.
- Ngow HA, Wan Khairina WM, Teh LK, Lee WL, Harun R, et al. (2009) CYP2C9 polymorphism: prevalance in healthy and warfarin-treated Malay and Chinese in Malaysia. Singapore Med J 50: 490-493.
- Ruzilawati AB, Mohd Suhaimi AW, Gan SH (2007) Genetic polymorphisms of CYP3A4*18 allele is found in five healty Malaysian subjects. Clinica Chemica Acta 383: 158-162.
- Oda Y, Kharasch ED (2001) Metabolism of methadone and levo-a-acetlymethadol (LAAM) by human intestinal cytochrome P450 3A4 (CYP3A4): Potential contribution of intestinal metabolism to presystemic clearance and bioactivation. J Pharm and Experimental Therapeutics 298: 1021-1032.
- Wang J-S, Devane L (2003) Involvement of CYP3A4, CYP2C8, and CYP2D6 in the metabolism of (R)- and (S)-Methadone in vitro. Drug Metab Dispos 31: 742-747.
- Rebbeck TR, Jaffe JM, Walker AH, Wein AJ, Malkowicz SB (1998) Modification of clinical presentation of prostate tumors by a novel genetic variant in CYP3A4. J Natl Cancer Inst 90: 1225-1229.
- Nogal A, Coelho A, Catarino R, Morais A, Lobo F, et al. (2007). The CYP3A4*1B polymorphism and prostate cancer susceptibility in a Portuguese population. Cancer Genetics and Cytogenetics 177: 149-152.
- Niwa T, Yabusaki Y, Honma K, Matsuo N, Tatsuta K, et al. (1998) Contribution of human hepatic cytochrome P450 isoforms to regioselective hydroxylation of steroid hormones. Xenobiotica 28: 539-547.
- Shou M, Korzekwa KR, Brooks EN, Krausz KW, Gonzalez FJ, et al. (1997) Role of human hepatic cytochrome P450 1A2 and 3A4 in the metabolic activation of estrone. Carcinogenesis 18: 207–14.
- Rendic S, Di Carlo FJ (1997) Human cytochrome P450 enzymes: a status report summarizing their reactions, substrates, inducers, and inhibitors. Drug Metab Rev 29: 413–580.
- Waxman DJ, Attisano C, Guengerich FP, Lapenson DP (1988) Human liver microsomal steroid metabolism: identification of the major microsomal steroid hormone 6 beta-hydroxylase cytochrome P-450 enzyme. Arch Biochem Biophys 263: 424–36.
Relevant Topics
- Addiction Recovery
- Alcohol Addiction Treatment
- Alcohol Rehabilitation
- Amphetamine Addiction
- Amphetamine-Related Disorders
- Cocaine Addiction
- Cocaine-Related Disorders
- Computer Addiction Research
- Drug Addiction Treatment
- Drug Rehabilitation
- Facts About Alcoholism
- Food Addiction Research
- Heroin Addiction Treatment
- Holistic Addiction Treatment
- Hospital-Addiction Syndrome
- Morphine Addiction
- Munchausen Syndrome
- Neonatal Abstinence Syndrome
- Nutritional Suitability
- Opioid-Related Disorders
- Relapse prevention
- Substance-Related Disorders
Recommended Journals
Article Tools
Article Usage
- Total views: 15349
- [From(publication date):
August-2012 - Dec 21, 2025] - Breakdown by view type
- HTML page views : 10538
- PDF downloads : 4811