Genomic Profiling of Pre-Clinical Models of Inflammatory Breast Cancer Identifies a Signature of Epithelial Plasticity and Suppression of TGF�?² Signaling
Received: 26-Jun-2012 / Accepted Date: 26-Jul-2012 / Published Date: 28-Jul-2012 DOI: 10.4172/2161-0681.1000119
Abstract
Abstract
Study background: Inflammatory Breast Cancer (IBC) is the most metastatic variant of breast cancer. Although
IBC is recognized as a distinct variant of breast cancer, the molecular basis for the rapid progression of IBC remains
largely undefined, in part due to the lack of preclinical models that recapitulate the human disease as well as a lack of
comprehensive analysis of the preclinical models of IBC that are available.
Methods: All available 7 pre-clinical models of IBC, including 2 new models, FC-IBC01 and FC-IBC02 developed
from pleural effusion, were used to identify genes and molecular pathways that are selectively altered compared to non
IBC breast tumor models. Laser capture micro dissection of biopsy tissue from core biopsy and skin punch biopsies
were also analyzed by whole transcriptome analysis.
Results: Whole transcriptome analysis defined 7 pre-clinical models of IBC as being within either the triple
negative or ErbB2/Her-2 expressing subtypes, similar to the prevalence of these subtypes of breast cancer observed
in IBC patients. Comparative analysis of the FC-IBC01, FC-IBC02 and Mary-X models of IBC demonstrated that each
of these recapitulate the formation of tumor emboli with encircling lymphovasculogenesis. The majority (6/7) of the
pre-clinical models of IBC express CDH1, which encodes for E-cadherin, which was associated with a loss of ZEB1,
a transcriptional repressor of E-cadherin.
The lack of ZEB1 expression was validated in a limited set of 4 skin punch biopsy samples from IBC patients that
were isolated by laser capture micro-dissection, demonstrating concordance with loss of ZEB1 in pre-clinical models of
IBC. Expression of other transcription factors involved in acquisition of a cancer stem cell phenotype, including Snai1,
which encodes for Snail, SNAI2, which encodes for Slug and TWIST1, was retained in pre-clinical models of IBC.
Maintenance of E-cadherin in pre-clinical models of IBC was associated with a loss of genes within the transforming
growth factor beta (TGFβ signaling pathway, with expression of SMAD6, a known repressor of TGFβ. This is similar
to a recent study reporting the persistence of E-cadherin and loss of TGFβ signaling in IBC patient tumors based on
gene profiling of 3 independent data sets.
Conclusion: The present studies provide first time comparison of gene signatures of 7 pre-clinical models of IBC,
including our 2 newly developed pre-clinical models, FC-IBC01 and FC-IBC02, that recapitulate formation of tumor
emboli with encircling lymphatic vessels, similar to that observed in biopsy tissues of IBC patients. We demonstrate
that E-cadherin expression was associated with both loss of ZEB1 and diminished expression of multiple genes within
the TGFβ signaling pathway, with retention of expression of transcription factors and surface markers consistent with
maintenance of a cancer stem cell phenotype, as has been reported to be a characteristic of IBC tumors. Collectively,
these observations provide first time characterization of the molecular signatures of all available pre-clinical models
of IBC, and suggest that IBC has a signature of epithelial plasticity, with characteristics of their ability to undergo the
mesenchymal to epithelial reverting transition. The loss of genes within the TGFβ signaling is also consistent with the
tight cell: cell aggregation of IBC tumor cells within tumor emboli that exhibit “cohesive invasion”. The new pre-clinical
models of IBC that recapitulate the human disease will serve as useful tools to accelerate our understanding of the
molecular underpinnings and therapeutic targets of IBC as the most lethal form of breast cancer.
Keywords: Inflammatory breast cancer; Tumor emboli; E-cadherin; ZEB1; Epithelial
311594Abbreviations
APC: Allophycocyanin; BFGF: Basic Fibroblast Growth Factor; DMEM: Dulbecco’s Minimal Essential Medium; DSC2: Desmocolin-2; EDTA: Ethylene Diamine Tetraacetic Acid; EGF: Epidermal Growth Factor; EGFR: Epidermal Growth Factor Receptor; EMT: Epithelial-mesenchymal Transition; ER: Estrogen Receptor; FBS: Fetal Bovine Serum; FN: Fibronectin; IBC: Inflammatory Breast Cancer; IL6: Interleukin6; IRB: Institutional Review Board; JUP/γ catenin: Junction Plakoglobin/Gamma Catenin; MEGM: Mammary Epithelial Growth Medium; mm: Millimeter; MErT: Mesenchymal to Epithelial Reverting Transition; MMP2: Matrix Metalloproteinase-2; PBS: Phosphate Buffered Saline; PE: Phycoerythrin; PR: Progesterone Receptor; RT-PCR: Reverse Transcription Polymerase Chain Reaction; TGFβ: Transforming Growth Factor Beta; U: Units; VIM: Vimentin; ZEB: Zinc Finger E-box Binding Homeobox
Introduction
Of the clinically distinct types of breast cancer, the most lethal variant is Inflammatory Breast Cancer (IBC) [1]. Although primary IBC is less commonly diagnosed than other types of breast cancer, accounting for an estimated 2-5% of all breast cancers in the United States and an estimated 13% worldwide, IBC is responsible for a disproportionate number of breast cancer-related deaths (7%) that occur each year world-wide [1,2]. IBC is a form of locally advanced breast cancer with involvement of skin and/or chest wall and common lymph node metastasis at the time of first diagnosis. IBC is a distinct clinic-pathologic entity characterized by rapid metastasis. The clinical diagnosis of IBC is based on the combination of the physical appearance of the affected breast, a careful medical history, physical examination, and pathological findings from a skin biopsy and/or needle or core biopsy to confirm the diagnosis of carcinoma [3]. The symptoms of IBC include a rapid onset of changes in the skin overlying the involved breast, including edema, redness and swelling involving over one half to two thirds of the breast, which may include a wrinkled, orange peel appearance, defined as “peaud’orange” [4,5]. The changes in the skin of the involved breast of IBC patients are the first clinical signs of IBC and are due to the presence of tumor cells that are tightly aggregated to form multicellular nests of cells, defined as tumor emboli, that are lodged within dermal lymphatic vessels [6,7]. The presence of tumor emboli is one of the classical histopathological findings in IBC [4,5] and, while their presence is not a 10 requirement for a diagnosis of IBC, approximately 75% of IBC patients have tumor emboli and they serve as one of most distinctive characteristic signatures of IBC [4-6].
Interestingly, IBC tumor emboli retain expression of E-cadherin [7,8], while in the majority of other tumor types with some exceptions, E-cadherin is lost and N-cadherin is gained, as part of the process of the Epithelial-Mesenchymal Transition (EMT) [9,10]. The loss of E-cadherin occurs with a change from an epithelial phenotype to favor a mesenchymal motile phenotype and acquisition of characteristics of cancer stem cells associated with expression of specific transcription factors such as TWIST1 and SNAI1, [9-11]. Although E-cadherin has been demonstrated to be necessary for tumorigenesis and survival of IBC tumor cells [12,13], little is known about the regulation of E-cadherin in IBC nor have the transcription factors that are involved in mediating retention of E-cadherin in IBC been defined.
Research to elucidate the molecular mechanisms that underlie the rapid metastasis exhibited by IBC has been hampered by the relatively rarity of the disease, and an associated lack of preclinical models that recapitulate the human disease. The present studies describe the first whole unbiased transcriptional analysis of all of the available preclinical models of IBC, including 2 new models of IBC, FC-IBC01 and FC-IBC02, that we have recently developed and which we demonstrate here to recapitulate the skin involvement observed in IBC patients. The present studies are the first to define the transcriptional pathways that are either gained or lost in all available pre-clinical models 11 of IBC, with the goal of identifying the molecular basis for the retention of an epithelial phenotype in this most metastatic variant of breast cancer.
Materials and Methods
Cell lines and culture conditions
The IBC cell lines SUM149, SUM190 and the non-IBC cell line SUM159 were all purchased from Asterand (Detroit MI). MDAIBC- 3 cells were obtained from Dr. WA Woodward (The University of Texas MD Anderson Cancer Center, Houston, TX). KPL-4 cells were obtained from Dr. N.T. Ueno (MD Anderson). The MCF-7 and MDA-MB-231 non-IBC human breast cancer cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA). All cell lines were authenticated by short tandem-repeat profiling. The SUM149, SUM190, SUM159, MDA-IBC-3 and KPL-4 cells lines were maintained in F12 medium supplemented with 10% fetal bovine serum (FBS) (Invitrogen Life Technologies, Carlsbad, CA), insulin (1 mg/ml; Sigma Aldrich, St Louis, MO), and hydrocortisone (1 mg/ml; Sigma- Aldrich). MCF-7 and MDA-MB-231 cells were cultured in Dulbecco’s minimal essential medium (DMEM)/F12 medium (Invitrogen Life Technologies) supplemented with 10% heat-inactivated FBS (Invitrogen Life Technologies). All cell lines were incubated at 37°C in 5% CO2-air atmosphere, with constant humidity. Mary-X IBC tumor model was generously provided by Dr. Sanford H. Barsky. We recently developed two new models of IBC, designated as FCIBC01 and FCIBC02. Both of these cell lines and xenografts models were derived from tumor cells isolated in pleural effusion of 2 IBC patients with disease progression using an IRB approved protocol with patient consent. Mary-X, FC-IBC01 and FC-IBC02 cells propagate optimally when cultured under low adherent culture conditions. Mary-X, FC-IBC01 and FCIBC02 cells were maintained as 3 dimensional tumor spheroids in a serumfree defined medium consisting of mammary epithelial growth medium (MEGM, BioWhittaker, Lancaster, MA) supplemented with B27 (Invitrogen/Life Technologies), 20 ng/ml epidermal growth factor (EGF), 40 ng/ml basic fibroblast growth factor (bFGF; BD Biosciences, San Jose, CA), 4 mg/ml heparin (SigmaAldrich) Lglutamine, and 100 U/ml penicillin and streptomycin. All cell lines were maintained at 37°C under 5% CO2 with constant humidity.
Growth of Mary-X, FC-IBC01 and FC-IBC02 xenografts
All experiments involving animals were conducted in accordance with protocols approved by the University of Texas MD Anderson Cancer Center Institutional Animal Care and Use Committee (IACUC). Prior to injection into mice, tumor spheroids derived from Mary-X, FC-IBC01 and FC-IBC02 were washed twice, resuspended in a 1: 2 mixture of sterile phosphate buffered saline (PBS; Hyclone, Logan, UT) and Matrigel (BD Biosciences, San Jose, CA). Naïve female 7-9 week old NOD. Cg-Prkdcscid Il2rgtm1Wj1/SzJ mice (Jackson Laboratories, Bar Harbor, ME) were anesthetized using 2.5% isoflurane (Baxter Healthcare, Deerfield, IL) prior to injection. Tumor spheroids were injected subcutaneously into the right hind flanks of the mice in a volume of 100 μl sterile PBS mixed with Matrigel (BD Biosciences). Tumor growth was monitored two-three times weekly using direct caliper measurements. Tumor volumes were calculated using the formula (width)2*(length)*0.52. Tumors were isolated when they reached a volume of 500 mm3 or greater. Mice were euthanized via carbon dioxide inhalation followed by cervical dislocation, tumors were removed and portions of each tumor were fixed and processed for paraffin embedment, sectioning and staining as described below.
Immunofluorescence and confocal microscopy imaging of tumor emboli enwrapped by lymphatic vessels in Mary-X, FC-IBC01 and FC-IBC02 tumor xenografts
Using tissue sections of Mary-X, FC-IBC01 and FC-IBC02 tumor xenografts, E-cadherin, as a marker of IBC tumor emboli, and podoplanin, as a marker of lymphatic endothelial cells were visualized using triple label immunofluorescence staining combined with confocal microscopy. Staining was performed using tissue sections that were deparaffinized in xylene and rehydrated in decreasing concentrations of ethanol. Sections were treated in target antigen retrieval solution (DAKO, Carpinteria, CA) in a steamer for 40 min, allowed to cool for 20 min and then rinsed in PBS. After treatment with 0.1% Triton X-100 solution for 5 min, tissue sections were incubated with 5% normal donkey serum for 1 hr followed by incubation with a mixture of rabbit anti-E-cadherin primary antibody (Cell Signaling, Danvers, MA) at a 1:100 dilution and goat anti-mouse podoplanin primary antibody (R&D Systems, Minneapolis, MN) at a dilution of 1:50. Slides of tissue were incubated overnight at 40ºC. Tissue sections were then washed three times in PBS for 5 min each, followed by incubation with secondary antibodies including donkey anti-rabbit Daylight 488 (Jackson Immuno Research), which has a green fluorescence signal, used for detection of E-cadherin, at a dilution of 1:200 and Alexa Fluor 568 donkey anti-goat (Invitrogen, Life Technologies, Carlsbad, CA), which has a red fluorescence signal, used for detection of podoplanin, at a dilution of 1:200 for 2 hrs at room temperature, followed by a wash with PBS. Sections were then stained with Topro-3(Invitrogen/ Life Technologies) at a 1:300 dilution for 15 min to label nuclear DNA. Cover slips were placed on stained slides using anti-fade mounting medium (Molecular Probes/Invitrogen, Carlsbad, CA). For negative controls, tissue was processed in the same way with the omission of primary antibody. Images were examined and captured using a LSM 510 confocal laser scanning system (Carl Zeiss, Thornwood, NY) at 25x magnification.
cDNA microarray and heat map analysis of cell lines
Total RNA was isolated from individual cell lines and xenograft tumor tissue using TRIzol® reagent (Invitrogen/Life Technologies). The quality of RNA was determined using a Nanodrop 2000 instrument (Thermo Fisher Scientific, Hanover Park, IL). Using GeneChip® One- Cycle cDNA Synthesis Kit (Affymetrix, Santa Clara, CA), total RNA was reverse transcribed to cDNA and synthesized to cRNA using the GeneChip® IVT Labeling Kit (Affymetrix,) and biotin labeled. The cRNA was then purified using GeneChip® Sample Cleanup Module kits (Affymetrix) and fragmented before hybridization. The hybridization cocktail containing the fragmented target cRNAs and probe array controls using the GeneChip® Hybridization Control Kit (Affymetrix) was hybridized to the probe array. The arrays were washed and stained according to protocol and scanned using the GeneChip® Scanner 3000 (Affymetrix). Raw signal intensities of gene expression data were analyzed using DNA-Chip Analyzer software (dChip; Harvard School of Public Health, Boston, MA; http://www.dchip.org). Ingenuity Pathway Analysis software (Ingenuity Systems, Burlingame, CA) was used to identify the genes and pathways present in cell lines examined.
cDNA microarray and analysis of cultured cells and IBC core and skin punch biopsy tissues isolated by laser capture microdissection
Tumor tissues were isolated using laser capture micro-dissection of core and skin punch biopsy of IBC patients using an IRB approved protocol. Cell lines and tumor spheroids, in the case of Mary-X cells, were cultured as described above and isolated from culture flasks or culture dishes by trypsinization. Total RNA was extracted from tumor tissue using RN easy kits (Qiagen; Gaithersburg, MD) based on manufacturer instructions. RNA integrity was determined using an Agilent Bioanalyzer 2100 (Agilent Technologies, Inc; Santa Clara, CA), with RNA-Integrity-Numbers ranging from 10.0-8.0. Whole transcriptome analysis of IBC and non-IBC samples was performed using the Genechip® U133 plus 2 microarray platform (Affymetrix, Inc; Santa Clara CA). The U133 plus 2 platform consists of >56,000 probe sets representing both annotated (i.e., Reference genes) and purported genes (i.e., Expressed Sequence tags; ESTs). Generation of biotin-labeled amplified-RNA suitable for application to microarrays, and the subsequent microarray hybridizations were performed by the Non-Coding RNA core facility at the MD Anderson, University of Texas. RNA concentrations utilized for the current study and sample quality metrics were standardized as recommended by Affymetrix. Briefly, 1 mg total RNA was amplified using the Affymetrix Genechip® 3’ IVT Express kit according to manufacturer’s suggestions, before processing and application to the Genechip®. Samples were hybridized to microarrays in triplicate following the manufacturers’ suggestions. Microarrays were scanned and signal intensities corresponding to transcript gene expression levels were determined. Differences between the expression of ZEB1 between IBC cell lines and non-IBC cell lines, samples of IBC tumor emboli and IBC core biopsy samples, IBC core biopsy samples and IBC cell lines and IBC tumor emboli samples and IBC cell lines was analyzed using the “Student’s” unpaired t test. The results of the statistical analysis are shown in Table 1.
| Non- IBC cells vs IBC Cells | IBC Cells vs IBC CORE | IBC Cells vs IBC EMBOLI | IBC CORE vs IBC EMBOLI |
|---|---|---|---|
| P<0.0007 | P<0.004 | N.S. | N.S. |
Table 1: Statistical analysis of the comparative differences in ZEB1 gene expression from cells and tissues from studies shown in Figure 5.
Flow cytometric analysis of CD44/CD24 and CD133
For analysis of expression of CD44/CD24 and CD133 surface markers, tumor cells were treated with trypsin-Ethylenediaminetetraacetic acid (EDTA) (trypsin 0.25%; EDTA 0.02%) for 5 minutes at 37ºC to obtain single cell suspensions, then were transferred to 12 × 75 mm polystyrene test tubes and washed twice with PBS. Cells were then resuspended in PBS containing 0.1% sodium azide and 2% FBS. Cells were incubated at 4ºC for 15~20 min with either a combination of anti-human CD24- PE and anti-human CD44-APC (BD Bioscience, San Jose, CA, USA) or anti-human CD133-APC (Miltenyl Biotec, CA, USA), followed by washing twice with PBS. Anti-mouse IgG2a-Phycoerythrin (PE) and anti-mouse IgG2b-Allophycocyanin (APC) were used as isotypic control antibodies (BD Biosciences). Flow cytometry was performed on a BD LSR Fortessa, and data were analyzed using FlowJo software (Tree Star, OR, USA).
Results
Defining the distinct characteristics of pre-clinical models of IBC
Subtypes of pre-clinical models of IBC
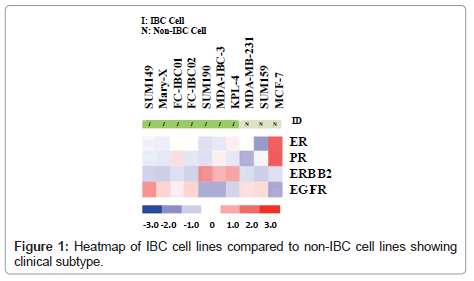
Whole transcriptome analysis was performed to define the molecular subtypes of preclinical models of IBC. Figure 1 is a heatmap showing the comparative analysis of Estrogen Receptor (ER), Progesterone Receptor (PR), the ERBB2/Her-2 oncogene and Epidermal Growth Factor Receptor (EGFR) in each of the available 7 pre-clinical models of IBC compared to the non-IBC MCF-7, MDAMB- 231 and SUM159 cell lines. There are four IBC cell lines that are classified as triple negative based on their lack of expression of ER, PR and the ErBB2/Her-2 oncogene. These cell lines include the most well characterized IBC cell line, SUM149, Mary-X cells, and our two newly developed.
IBC cell lines, FC-IBC01 and FC-IBC02. Note that each of these IBC cell lines/systems expresses EGFR, as do the non-IBC triple negative cell lines, MDA-MB-231 and SUM159. The SUM190, MDA-IBC-3 and KPL-4 cell lines all express the Her-2 oncogene. The prevalence of triple negative and Her-2+ IBC models mirrors the prevalence of these subtypes in IBC patients in general. This heat map also demonstrates that there is no IBC cell line that is of the luminal a subtype, and expresses only ER.
Histological characteristics of pre-clinical models of IBC that form tumor emboli invivo
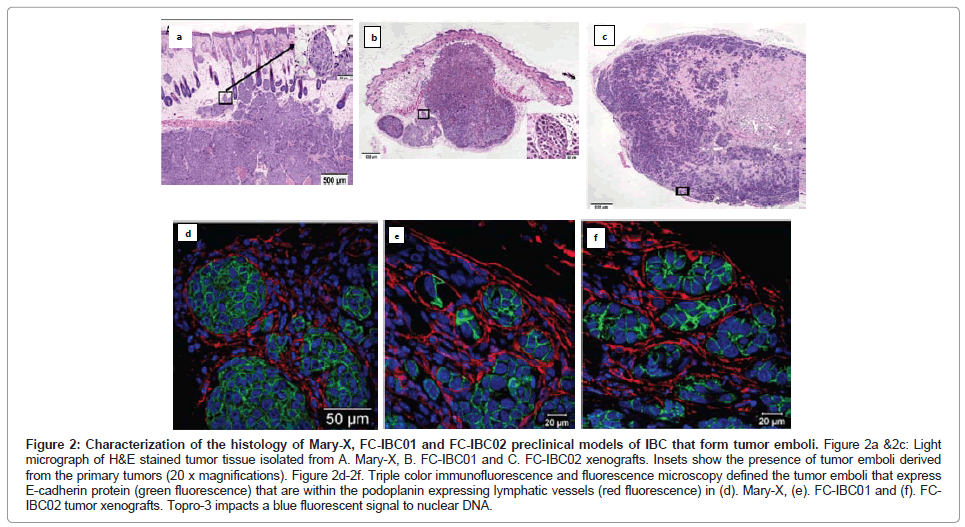
The Mary-X IBC model is a stable transplantable xenograft model developed by Dr. Sanford H. Barsky from the primary tumor of an IBC patient with triple negative basallike breast cancer [12,14,15]. Mary-X was developed by serially passaging tumors and then isolating tumor cells from xenografts [12,14]. When Mary-X tumor spheroids are injected into immune compromised mice in vivo, tumor emboli which stain for E-cadherin can be identified within the dermal layer of the skin that are encircled by lymphatic endothelium, which stain for podoplanin, a selective marker of lymphatic vessels [15] (Figures 2A and 2D).
Figure 2: Characterization of the histology of Mary-X, FC-IBC01 and FC-IBC02 preclinical models of IBC that form tumor emboli. Figure 2a &2c: Light micrograph of H&E stained tumor tissue isolated from A. Mary-X, B. FC-IBC01 and C. FC-IBC02 xenografts. Insets show the presence of tumor emboli derived from the primary tumors (20 x magnifications). Figure 2d-2f. Triple color immunofluorescence and fluorescence microscopy defined the tumor emboli that express E-cadherin protein (green fluorescence) that are within the podoplanin expressing lymphatic vessels (red fluorescence) in (d). Mary-X, (e). FC-IBC01 and (f). FCIBC02 tumor xenografts. Topro-3 impacts a blue fluorescent signal to nuclear DNA.
Prior to our studies developing the FC-IBC01 and FC-IBC02 models of IBC, Mary-X was the only pre-clinical model of IBC that recapitulated the formation of tumor emboli and encircling lymphovasculogenesis. We recently developed 2 new models of IBC, designated as FC-IBC01 and FC-IBC02, which were derived from tumor cells isolated following thoracentesis of IBC patients who had developed metastatic pleural effusions [16,17]. As was observed with Mary-X cells, FC-IBC01 and FC-IBC02 cells spontaneously form 3 dimensional tumor spheroids in vitro and are optimally propagated for short periods of time in low adherence culture. As such, the FC-IBC01, FC-IBC02 and Mary-X tumor spheroids provide a convenient in vitro surrogate for IBC tumor emboli that form in vivo. As with Mary-X tumor cells, neither FC-IBC01 nor FC-IBC02 cells can be successfully passaged or propagated long term as adherent cultures on a plastic substrate. Since these IBC cell systems cannot be cultured as adherent cultures, there is no method to determine their doubling time, but rather we routinely calculate their clonogenicity when cultured is soft agar, which is a measure of their self renewal. All three of these IBC models have very robust clonogenic growth.
When FC-IBC01 and FC-IBC02 tumor cells are injected into immunecompromised NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice, tumors rapidly grow, with the formation of IBC tumor emboli that invade into the skin (Figures 2B and 2C). Both FC-IBC01 (Figures 2B and 2E) and FC-IBC02 (Figures 2C and 2F) form tumor emboli that express E-cadherin protein (green fluorescence) and are encircled by lymphatic vessels that express podoplanin, used as a selective marker of lymphatic endothelium (red fluorescence). Topro-3 was used as a marker of nuclear DNA (blue fluorescence).
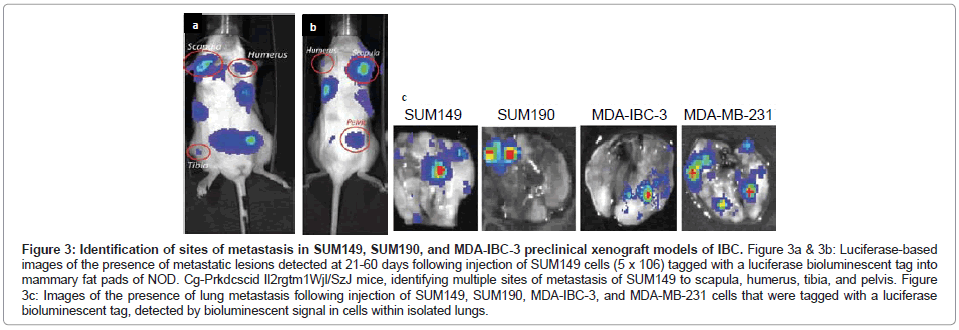
The SUM149 and SUM190 IBC cell lines, developed by Dr. Steven Ethier and colleagues [18-20], have been used for the vast majority of studies to define the molecular signatures of IBC. Both of these cell lines were developed from patients with invasive ductal carcinoma prior to receiving chemotherapy [18]. Both SUM149 and SUM190 cells express cytokeratins 8, 18 and 19 and are chromosomally abnormal [18]. SUM190 cells were characterized as having amplified Her-2 and cyclin D-1 [18,19], while SUM149 cells were characterized as triple negative, lacking ER, PR and the Her-2oncogene [19,20], which results from the present study of gene profiling of the IBC cell lines validates (Figure 1). SUM149 cells are independent of EGF, and as demonstrated in the present study, SUM149 cells express EGFR (Figure 1) and have an amphiregulin autocrine loop that contributes to their aggressive phenotype [21]. SUM149 cells have a very short doubling time (~21 hrs) and, although they do not form tumor emboli when grown as xenografts in immunocompromised mice in vivo, SUM149 cells rapidly form primary tumors as well as commonly form metastatic lesions at multiple sites, including lymph nodes, lung, liver, soft tissue and bone when injected into immunocompromised mice, which is detected when SUM149 cells are labeled with luciferase to allow for in vivo live imaging (Figures 3A-3C).
Figure 3: Identification of sites of metastasis in SUM149, SUM190, and MDA-IBC-3 preclinical xenograft models of IBC. Figure 3a & 3b: Luciferase-based images of the presence of metastatic lesions detected at 21-60 days following injection of SUM149 cells (5 x 106) tagged with a luciferase bioluminescent tag into mammary fat pads of NOD. Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice, identifying multiple sites of metastasis of SUM149 to scapula, humerus, tibia, and pelvis. Figure 3c: Images of the presence of lung metastasis following injection of SUM149, SUM190, MDA-IBC-3, and MDA-MB-231 cells that were tagged with a luciferase bioluminescent tag, detected by bioluminescent signal in cells within isolated lungs.
SUM190 cells proliferate at a much slower rate compared with SUM149 cells, with a doubling time of ~ 42 hrs, however when they are cultured under low adherence conditions supporting formation of tumor spheroids and then injected into immunocompromised mice, SUM190 produce primary tumors as well as metastatic lesions, primarily to the lung, which can be identified using luciferase based live in vivo imaging of isolated lungs of tumor bearing mice (Figure 3C).
The KPL-4 cell line was derived from tumor cells isolated from pleural effusion of a patient with inflammatory skin metastasis [22]. KPL-4 has been characterized as expressing EGFR, Her-2 and ERBB3 family of genes, with 15-fold amplification of Her-2.
In the present study, we did not detect expression of EGFR by KPL-4 cells but we did detect robust expression of the Her-2 oncogene (Figure 1). When injected into mice, KPL-4 cells induced cachexia, which was found to be associated with production of copious amounts of Interleukin-6 [22-24]. The KPL-4 model has primarily been used to examine the effects of therapeutic agents on IL-6 production [22-24]. More recent studies have used the KPL-4 model to demonstrate the value of combining trastuzumab (Herceptin®; Genentech, South San Francisco, CA) with fluoropyrimidines or a taxane in circumstances where trastuzumab resistance has developed [25,26]. Due to the very high passage number of KPL-4, with their lack of expression of E-cadherin, considered to be a characteristic of IBC cell lines and the disease in patients, this cell line is seldom used in studies to identify the molecular basis of IBC.
MDA-IBC-3 cells were developed from an IBC patient with pleural effusion by serially transplantation from which the cell line was derived [27]. MDA-IBC-3 cells have been defined as expressing Her-2 and lack ER and PR, consistent with the present gene profiling studies (Figure 1). MDA-IBC-3 cells have a very slow doubling time (~76 hrs) when cultured as adherent cultures on plastic substrates, however they readily form tumor spheroids when placed in low adherence conditions in serum free defined medium. MDA-IBC-3 cells grow as xenografts when injected into immunocompromised mice and in our hands, spontaneously form metastatic lung tumors (Figure 3C), however they do not form tumor emboli when grown as xenografts.
In addition to these 7 IBC cell lines, the WIBC model is another model of IBC. Unfortunately, the WIBC model has not been made widely available for studies and therefore is not included in the present studies. The WIBC model of IBC was initially developed by serially transplanting tumor tissue from an IBC patient into immunocompromised mice [28]. The tissue isolated from the WIBC xenografts was characterized by a lack of central necrosis or fibrosis, angiogenesis, hyper-vascularity and high microvascular density. Interestingly, the WIBC tumor cells were observed to form tube like structures, in a process defined as vascular mimicry and vasculogenesis [28-33]. Since we have not had access to the WIBC model of IBC, we studied the ability of IBC cell lines to undergo vascular mimicry. In our hands, we find that the only cell lines that exhibit vascular mimicry are the SUM149 IBC cell line and the non-IBC MDAMB-231 cell line, which are both triple negative breast cancer cell lines. We have previously performed studies to characterize this activity, which are described in previous publications [34].
Analysis of CD44/CD24 and CD133 in preclinical models of IBC
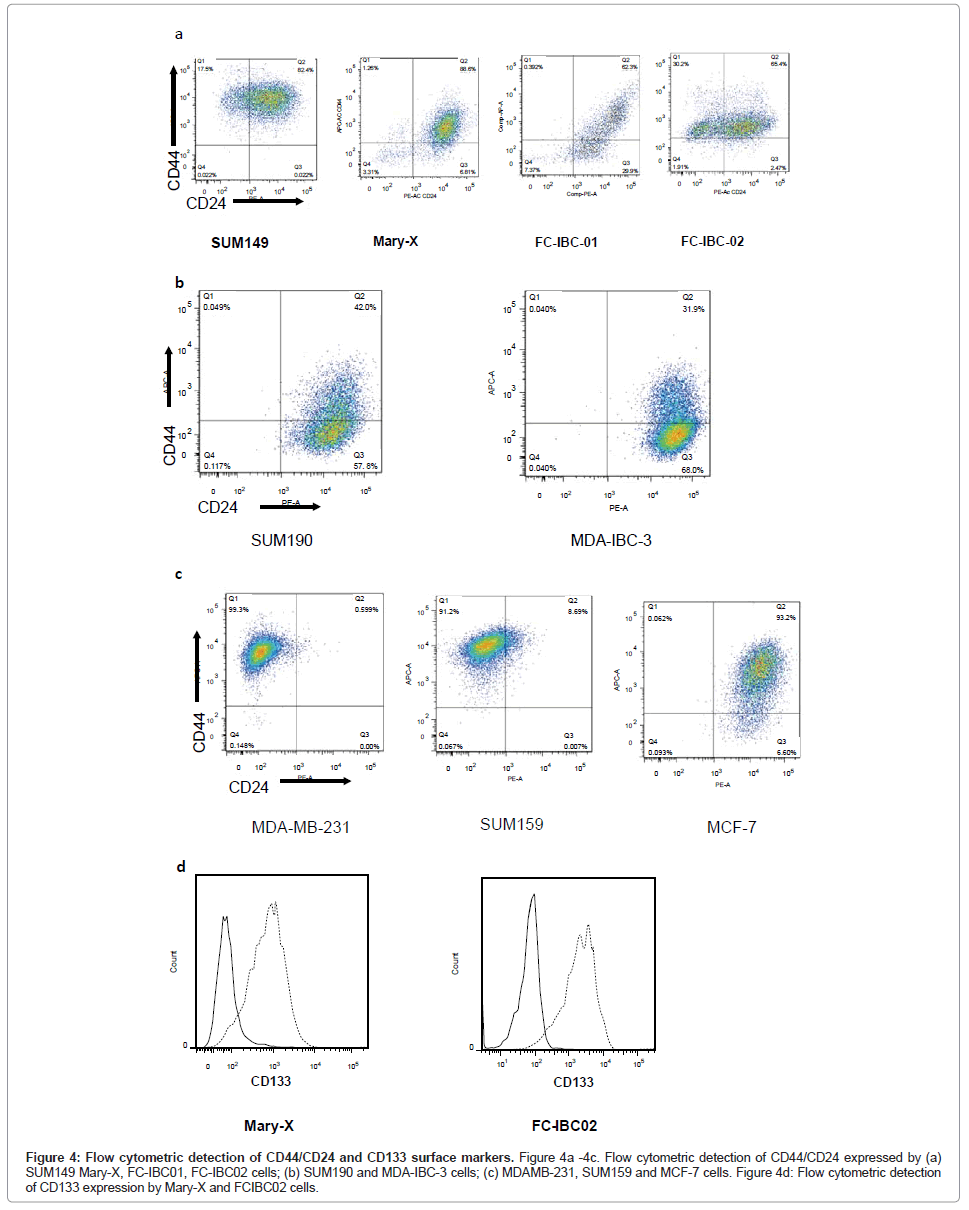
Flow cytometric analysis of IBC cells demonstrated that SUM149, Mary-X and FC-IBC02 were enriched for cells that express CD44+/ CD24-/low, while FC-IBC01, MDA-IBC-3 and SUM190 cells did not have significant expression of these surface markers (Figures 4A and 4B). The KPL-4 cells were not analyzed for their expression of CD44/ CD24. The triple negative non-IBC MDA-MB-231 and SUM159 breast tumor cells had >90% of cells that that expressed CD44+/CD24-/low (Figure 4C). The Mary-X and FC-IBC02 tumor cells also expressed CD133 (Figure 4D), which was not expressed by any other IBC cell line nor was it expressed by non-IBC cell lines examined (data not shown).
Figure 4: Flow cytometric detection of CD44/CD24 and CD133 surface markers. Figure 4a -4c. Flow cytometric detection of CD44/CD24 expressed by (a) SUM149 Mary-X, FC-IBC01, FC-IBC02 cells; (b) SUM190 and MDA-IBC-3 cells; (c) MDAMB-231, SUM159 and MCF-7 cells. Figure 4d: Flow cytometric detection of CD133 expression by Mary-X and FCIBC02 cells.
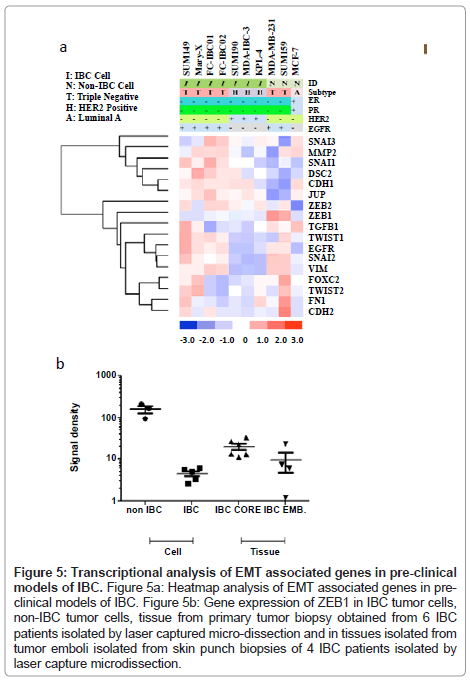
Analysis of genes involved in the process of EMT in preclinical models of IBC
Based on observations that tumor emboli from IBC patients commonly have robust expression of E-cadherin, which is a characteristic of epithelial cells, we next wanted to characterize the genes involved in the process of EMT in the pre-clinical models of IBC. Figure 5A shows the heatmap of the genes known to be primarily involved in EMT in all of the pre-clinical models of IBC. Since IBC patient tumors retain expression of Ecadherin, this was the first gene analyzed. All of the IBC cells express the CDH1 gene, which encodes for E-cadherin, with the exception of the KPL-4 cells. Interestingly, the SUM149 and FC-IBC01 cell lines express both CDH1 and CDH2, which encodes for Ncadherin, usually associated with cells undergoing the process of EMT and usually occurs with a loss of CDH1. While the SUM159 and MDA-MB-231 non-IBC breast cancer cell lines both lack CDH1 gene expression, only the SUM159 non-IBC triple negative cell line expresses CDH2. The expression of CHD1 by IBC cell lines was associated with a lack of expression of zinc finger E-box binding homeobox 1 (ZEB1), while both of the non-IBC triple negative cell lines, MDA-MB-231 and SUM159, expressed ZEB1 and lacked CDH1 expression.
A number of transcription factors implicated in the process of EMT and retention of a cancer stem cell phenotype were also examined. SNAI1, which encodes for Snail, was expressed by all IBC cell lines regardless of subtype, with lower levels of gene expression by SUM190 and MDA-IBC-3. In contrast, none of the non-IBC cell lines expressed SNAI1. All triple negative cell lines, regardless of whether they were derived from IBC patients, expressed SNAI2, which encodes for Slug. Cell lines, with the exception of SUM149, Mary-X and MDA-IBC-3, expressed SNAI3, which encodes for Snail 3. Interestingly, TWIST1 was expressed by triple negative IBC cells, including SUM149, Mary-X, FCIBC01, FC-IBC02 and non-IBC SUM159 cells but was not expressed by MDA-MB-231 cells. Another gene, VIM, which encodes for vimentin, was examined. This gene, known to be involved in the process of EMT with a specific role in tumor cell invasion, vimentin, was expressed by all cells with a triple negative subtype, regardless of whether or not they were derived from IBC patients.
In addition to CDH1, other genes involved in regulating the tight cell: cell aggregation exhibited by IBC tumor emboli was examined. All of the IBC cell lines, including the KPL-4 cells which did not express CDH1, expressed junction plakoglobin/gamma catenin (JUP/γ catenin), which functions with E-cadherin as part of the adherens junction complex. Another gene, DSC2, which encodes for Desmocolin-2 that is also involved in cell: cell adherence, was expressed by all of the IBC cells, with the exception of MDAIBC-3.
It is interesting to note that the heatmap shown in Figure 5A demonstrates a distinct clustering of genes associated with cell: cell adherence and an epithelial phenotype, including DSC2, CDH1 and JUP, as well as MMP2, SNAI1, SNAI3, that it very distinct from the clustering of other genes that have been identified as being involved in the process of EMT, including ZEB1, ZEB2, TGFβ1, TWIST1, TWIST2, EGFR, SNAI2, VIM, FOXC2, FN1 and CDH2. While all of the IBC cell lines express the majority of the genes identified in the cluster containing CDH1, which encodes for E-cadherin, the triple negative IBC cell lines also express genes associated with the EMT process.
Figure 5: Transcriptional analysis of EMT associated genes in pre-clinical models of IBC. Figure 5a: Heatmap analysis of EMT associated genes in preclinical models of IBC. Figure 5b: Gene expression of ZEB1 in IBC tumor cells, non-IBC tumor cells, tissue from primary tumor biopsy obtained from 6 IBC patients isolated by laser captured micro-dissection and in tissues isolated from tumor emboli isolated from skin punch biopsies of 4 IBC patients isolated by laser capture microdissection.
Analysis of ZEB1 gene expression in pre-clinical models of IBC compared with ZEB1 gene expression in IBC core biopsy tissue and tumor emboli within skin punch biopsies isolated by laser capture microdissection
To validate the loss of ZEB1 in pre-clinical models of IBC and in a limited set of IBC patient tissues, RT-PC analysis was performed using SUM149, Mary-X, SUM190, MDAIBC-3 and KPL-4 cells, MDAMB- 231 and SUM159 triple negative non-IBC breast cancer cell lines, and a set of 6 each samples of IBC patient core biopsy tissues and a set of 4 samples of tumor emboli from skin punch biopsies of IBC patients using an IRB approved protocol and consented patients, isolated by laser capture micro-dissection. Figure 5B shows a dot plot analysis of the levels of ZEB1 gene expression in each of these 4 groups. Table 2 provides the statistical analysis of the differences between the groups analyzed using the “Student’s” unpaired t test. ZEB1 was detected at very low abundance in cultured IBC cell lines, which was significantly lower compared with the level of ZEB1 expression by non-IBC cell lines MDA-MB-231 and SUM159 cells (p<0.0007). There were no significant difference in ZEB1 gene expression in IBC cell lines compared with IBC tumor emboli in skin punch biopsies, there was a statistically significant difference between ZEB1 expression in IBC cells compared with the IBC core biopsy tissues (p<0.004) (Figure 5B and Table 1). There was no significant difference in ZEB1 gene expression between IBC core biopsy tissues and IBC tumor emboli isolated by laser capture micro-dissection.
| Symbol | Entrez Gene Name | Fold Change |
|---|---|---|
| PAI-1 | serpin peptidase inhibitor, clade E | -3.0973451327433628 |
| GSC | goosecoid homeobox | -3.021276595744681 |
| Bcl-2 | B-cell CLL/lymphoma 2 | -2.1538461538461537 |
| MAPK12 | mitogen-activated protein kinase 12 | -1.7128712871287128 |
| Runx2 | runt-related transcription factor 2 | -1.5742753623188408 |
| TGFB1 | transforming growth factor, beta 1 | -1.5596868884540114 |
| AMHR2 | anti-Mullerian hormone receptor, type II | -1.5166666666666666 |
| Nkx2.5 | NK2 homeobox 5 | -1.5108225108225108 |
| TCF | hepatocyte nuclear factor 4, alpha | -1.5104895104895106 |
| BMP2K | BMP2 inducible kinase | -1.4548872180451127 |
| INHBA | inhibin, beta A | 1.2581344902386116 |
| BMPR1B | bone morphogenetic protein receptor, type IB | 1.2714285714285716 |
| VDR | vitamin D (1,25- dihydroxyvitamin D3) receptor | 1.292483660130719 |
| TGIF | TGFB-induced factor homeobox 1 | 1.2964509394572026 |
| MAPK13 | mitogen-activated protein kinase 13 | 1.3747680890538034 |
| TLX2 | T-cell leukemia homeobox 2 | 1.3917525773195878 |
| MAP2K6 | mitogen-activated protein kinase kinase 6 | 1.4218749999999998 |
| Smad6 | SMAD family member 6 | 1.5411764705882354 |
| MAPK11 | mitogen-activated protein kinase 11 | 1.6130952380952381 |
| Runx3 | runt-related transcription factor 3 | 2.760330578512397 |
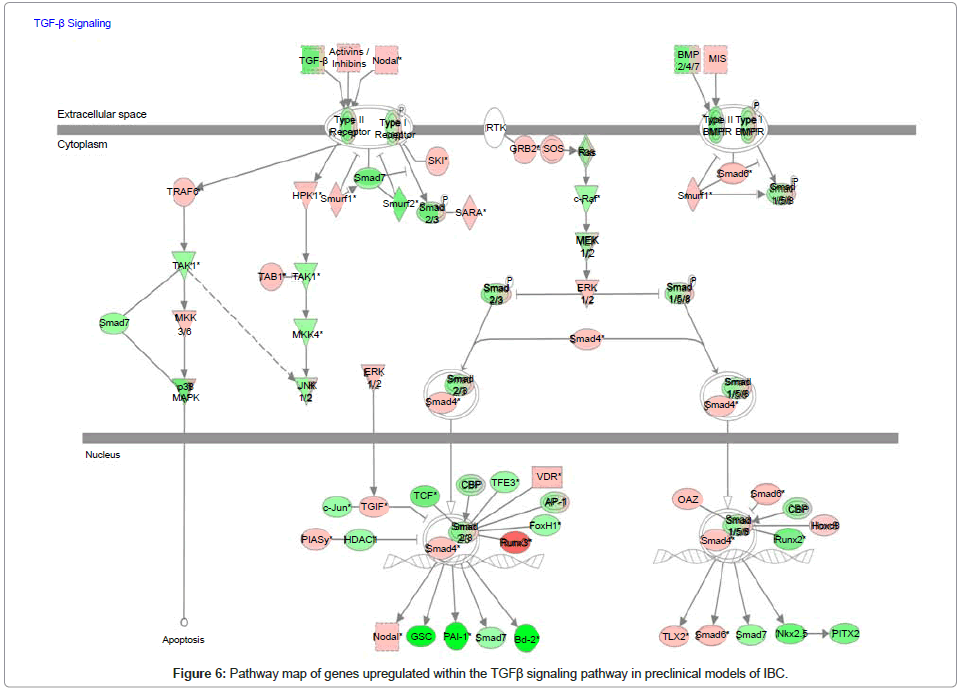
Table 2: List of genes showing the relative difference in expression of genes within the TGFβ signaling pathway in pre-clinical models of IBC.
Analysis of genes within the TFGβ signaling pathway in preclinical models of IBC
Since our collaborative studies recently revealed that IBC patient tumors were characterized as having a loss of expression of genes within the transforming growth factor beta (TGFβ) signaling pathway [35], we used whole transcriptome analysis and Ingenuity Pathway Analysis mapping software to analyze the gains and losses of genes within this signaling pathway in pre-clinical models of IBC. Figure 6 is the pathway map showing the relative levels of expression of genes associated with the TGFβ signaling pathway and Table 2 is the gene list showing the levels of change in expression of the specific genes of interest within the TGFβ signaling pathway. A common feature of the pre-clinical models of IBC, regardless of subtype, was a loss of genes that positively regulate the TGFβ signaling pathway, with the gain of genes that suppress TGFβ signaling. Most notably, SMAD6, a repressor of TGFβ signaling by its ability to block SMAD4, was significantly upregulated in pre-clinical models of IBC (Figure 6, Table 2).
Discussion
Although IBC is recognized as a distinct clinical entity [1,3,4], the molecular underpinnings for the retention of an epithelial phenotype by IBC tumor cells, while simultaneously displaying an accelerated program of metastasis, remain largely undefined. The lack of understanding of IBC has been, at least in part, due to the lack of pre-clinical models with which to define the specific pathways that mediate the retention of an epithelial phenotype, while allowing for rapid spread of tumor emboli via the lymphatic route.
The present report provides first time analysis of all of the available pre-clinical models of IBC, including 2 new models that we have recently developed FC-IBC01 and FCIBC02. The present studies demonstrate that both FC-IBC01 and FC-IBC02, which are both triple negative breast cancers, recapitulate the specific histopathology characteristic of IBC, similar to the Mary-X model of IBC, which also has a triple negative subtype.
As with non-IBC breast cancers, IBC tumors are transcriptionally heterogeneous and our recent studies divided IBC tumors into intrinsic subtypes including luminal A, luminal B, ErbB2/Her-2+ and triple negative breast cancers lacking ER/PR and ERBB2/Her-2, similar to the subtypes found in non-IBC tumors. Although IBC tumors can be of any subtype, the most prevalent molecular subtypes for IBC patients are either triple negative or ERBB2+ tumors [35-38], which is reflected in the pre-clinical models of IBC. The present study represents the first comparative analysis of these important characteristics of all available pre-clinical models of IBC and demonstrates the prevalence of triple negative or ERBB2/Her-2+, which is consistent with the subtypes observed in IBC patients.
Surprisingly, recent studies using an expanded database of IBC patient tumors reported that IBC patients with luminal A subtype (ER+/PR+/Her-2-) have shorter, distant metastases- free survival intervals compared with their non-luminal IBC counterparts [35]. These observations are in direct contrast to observations in non-IBC breast cancers, in which patients with luminal A tumors have improved survival [39]. While the pre-clinical models of IBC are reflective of the most prevalent subtypes of this variant of breast cancer in IBC patients, the observation that the worst prognosis of IBC patients is associated with those with a luminal A subtype of underscores the need for further development of pre-clinical models of luminal A IBC. The development of pre-clinical models of Luminal A IBC is critical to the ability to define the molecular basis for the observation of the differential lower overall survival of those patients with luminal A subtype IBC.
The FC-IBC01 and FC-IBC02 models of IBC were derived from individual IBC patients with triple negative breast cancer who had developed metastatic disease progression manifested by pleural effusion [16,17]. The present studies demonstrate that both the FCIBC01 and FC-IBC02 models of IBC recapitulate the human disease and form tumor emboli encircled by lymphatic vessels. These models are similar in gene signatures to the Mary-X pre-clinical model of IBC, which is also triple negative and also forms tumor emboli with robust lymphangiogenesis [12,14,15].
Based on whole unbiased transcriptome analysis, the present results demonstrate that pre-clinical models of IBC, with the exception of the KPL-4 cells, retain robust expression of E-cadherin. E-cadherin is a primary characteristic of IBC tumor emboli and is considered to be a hallmark of the skin involvement of IBC [1,3-8,12,14], with few studies that have gone beyond this observation. Although IBC is well recognized as being the most metastatic variant of breast cancer [1,3,4], the robust expression of E-cadherin as a primary characteristic of this aggressive variant of breast cancer is apparently paradoxical to the current hypothesis that tumor cells undergo an EMT process, with the loss of E-cadherin, and the expression of transcription factors such as TWIST1 and SNAI1 and SNAI2, which encode for Snail and Slug transcription factors, respectively, that are known to be involved in tumor cell acquisition of a cancer stem cell phenotype [9-11], which is a prerequisite for invasion and metastasis.
Results of the whole transcriptome analysis of pre-clinical models of IBC demonstrate that the expression of E-cadherin is associated with a lack of expression of ZEB1, which is known to be a transcriptional repressor of E-cadherin [9,10]. The loss of ZEB1 expression was validated in a limited set of core biopsy and skin punch biopsy samples of patients with primary IBC. Our previous studies demonstrated that E-cadherin expression in SUM149 and SUM190 IBC cell lines was associated with the expression of microRNA (miR) 200 c [40]. Given previous reports that the presence of miR 200c itself is sufficient to inhibit ZEB1 expression and restore E-cadherin [41], this may be one of the molecular mechanisms underlying the retention of E-cadherin by IBC tumor cells. Further studies using IBC patient tumor samples and pre-clinical models of IBC will be needed to confirm this mechanism as well as to elucidate other molecular underpinnings of retention of the epithelial phenotype that is mediated, in part, through E-cadherin expression, in IBC.
In contrast to the lack of ZEB1 expression in pre-clinical models of IBC and in IBC patient tumor tissues, there was differential expression of other transcription factors that have previously been associated with the process of EMT and/or the acquisition of a cancer stem cell phenotype, including SNAI1, SNAI2, and TWIST1. In the case of SNAI1, which encodes for Snail, its expression was limited to IBC cells, while expression of SNAI2, which encodes for Slug, was restricted to triple negative breast cancer cells, regardless of whether they originated from IBC or not. In a similar fashion, the present study demonstrated that TWIST1 was expressed by human breast tumor cells that were identified as being of the triple negative tumor subtype, with the exception of MDA-MB-231 cells, which we show have high numbers of CD44+/CD24 low cells indicative of a cancer stem cell phenotype, suggesting that there are other transcription factors that have overlapping activities with TWIST1 in regulating acquisition of a cancer stem cell phenotype. The expression of TWIST1 was previously demonstrated to regulate acquisition of a cancer stem cell phenotype, which occurred during the process of EMT [11]. While the MDAMB- 231 cells had a high percentage of CD44+ expressing cells, they lacked expression of both TWIST1 and E-cadherin. In contrast, the IBC cell lines that were triple negative, including SUM149, Mary-X and FC-IBC02 had CD44+ cells, expressed TWIST1 but retained E-cadherin gene expression. These studies suggest that acquisition of a cancer stem cell phenotype is not mutually exclusive with retention of E-cadherin expression in IBC. The relevance of these observations is underscored by the recent reports that IBC has been characterized as a disease enriched for cancer stem cells [42,43]. Additionally, these studies suggest that there is redundancy in the role of transcription factors TWIST1, SNAI1 and SNAI2, with respect to acquisition and maintenance of a cancer stem cell phenotype in triple negative breast cancers in general, regardless of whether they were derived from IBC patients.
Collectively, the present results demonstrate that, at least in preclinical models of IBC that are of the triple negative subtype, the gene signature is a mixture of those genes involved in maintenance of an epithelial phenotype, characterized by expression of Ecadherin, JUP/γ catenin and DSC2, while simultaneously retaining expression of genes associated with characteristics of mesenchymal cells, with expression transcription factors, such as TWIST1, associated with acquisition of a cancer stem cell phenotype and VIM, which encodes for vimentin, which is involved in tumor cell invasion.
The present studies are the first to report that pre-clinical models of IBC, like IBC patient tumors [35], have a loss of expression of genes involved in TGFβ signaling. Interestingly, recent studies demonstrated reported that TGFβ is a key factor in the reversible regulation of motility by single cells, which in its absence, allows cells to revert to the process of “cohesive invasion” [44]. In addition to retention of E-cadherin, IBC tumor cells are characterized by formation of highly motile aggregates of tumor cells that have been defined as tumor emboli. Therefore, it is perhaps not surprising that E-cadherin gene expression that is associated with the retention of an epithelial phenotype by IBC tumor cells occurs with a loss of genes within the TGFβ signaling pathway, with an increased expression of the inhibitory SMAD6 gene. This may be the molecular basis for the invasion of aggregates of IBC tumor cells within lymphatic vessels that we demonstrate in this study to be observed using microscopic examination of tissue sections in patient tissues and in the Mary-X, FC-IBC01 and FC-IBC02 pre-clinical models of IBC.
Interestingly this same study reported that cells exhibiting collective invasion could invade into lymphatic vessels but were incapable of hematogenous metastasis [44]. These results are consistent with the observations that IBC tumor emboli primarily invade into dermal lymphatic vessels that we use immunofluorescence and confocal microscopy to visualize in the present studies. The adaptive ability of tumor cell aggregates that comprise the IBC tumor emboli provide a route of metastasis to local lymph nodes which is a commonly observed in IBC patients. Collectively, the results of the present studies suggest that IBC tumor cells display plasticity in their gene signature that allows them to retain the epithelial phenotype, while regulating specific signaling pathways that program them to a specific lymphatic route of metastasis. Based on the high degree of plasticity exhibited in IBC tumor cells, it may be that they are capable of expressing genes within different signaling pathways, depending upon whether they display a propensity for a lymphatic or blood borne route of metastasis.
Metastasis is a multi-stage process characterized by discreet events that ultimately ends with the re-establishment of colonies of tumors with an epithelial phenotype in a site distant from the site of the primary tumor [45]. During the process of EMT, E-cadherin is lost, the miR 200 family is suppressed and ZEB1 is transcriptionally activated [9-11]. While loss of E-cadherin has been associated with the phenotypic program of EMT characterized by increased motility and invasion similar to that of fibroblasts, a reversion from a mesenchymal to an epithelial phenotype is now believed to be necessary for colonization to form distant metastasis, which is heavily dependent on the microenvironment. In one study using MDA-MB-231 cells, the microenvironment of the lung induced re-expression of E-cadherin associated with what was defined as mesenchymal to epithelial reverting transition (MErT) [46]. This phenotypic change of the MDAMB- 231 cells, which are usually mesenchymal cells, was associated with altered cell behavior and was concluded to be a potentially critical step in survival at sites of metastasis. A recent review pointed out that there are a number of tumor types including IBC, ovarian carcinoma, and glioblastoma, that are characterized by retention of E-cadherin and exhibiting cohesive invasion. Collectively, each of these tumor types have an accelerated program of metastasis, suggesting that E-cadherin may have what has been defined as a “dark side” in terms of its role in metastasis, which is currently unappreciated [47].
Thus, the apparent dichotomy of the gene signature of IBC that includes genes known to be involved in both the process of EMT while retaining an epithelial phenotype may be explained by the extreme plasticity exhibited by IBC tumor cells as one of their adaptive mechanisms for survival and accelerated rates of metastasis to multiple sites. The lack of ZEB1 expression and the loss of genes involved in the TGFβ signaling pathway that we find is a characteristic of all of the pre-clinical models of IBC also support maintenance of an epithelial phenotype, while the expression of a specific repertoire of transcription factors, including Snail and TWIST1, allows maintenance of a cancer stem cell phenotype, which may confer a survival advantage in the face of chemotherapy and radiation. Taken together, the simultaneous expression of genes that support an epithelial phenotype with suppression of expression of genes associated with a mesenchymal phenotype such as ZEB1 and TGFβ allows IBC tumor emboli to migrate as aggregates of cells into lymphatic vessels, providing a conduit for IBC tumor emboli to rapidly colonize regional lymph nodes. This program of simultaneous gain and loss of specific gene programs may be the basis for the metastatic phenotype exhibited in IBC patient, which is recapitulated in pre-clinical models of IBC, especially those that are triple negative and can be accurately characterized as having a high degree of epithelial plasticity.
In summary, the present studies are among the first to identify retention of E-cadherin expression by pre-clinical models of IBC which was associated with the suppression of genes within the TGFβ signaling pathway and lack of or low expression of the ZEB1 transcription factor that are both known to be involved in the process of EMT. These results are among the first to shed light on molecular mechanisms underlying the retention of E-cadherin observed in IBC patient tumors and on a signaling pathway that supports the retention of an epithelial phenotype, in the face of enrichment of cells with a cancer stem cell phenotype and a program of accelerated metastasis that is observed in IBC. Additionally, the observations for the suppression of expression of genes within the TGFβ signaling pathway are consistent with similar findings in whole transcriptome analysis of IBC patient tumors [38]. Collectively, these data provide first time evidence that a signature of epithelial plasticity characterizes pre-clinical models of IBC, similar to the IBC patient tumors.
References
- Robertson FM, Bondy M, Yang W, Yamauchi H, Wiggins S, et al. (2010) Inflammatory breast cancer: the disease, the biology, the treatment. CA Cancer J Clin 60: 351-375.
- Levine PH, Veneroso C (2008) The epidemiology of inflammatory breast cancer. Semin Oncol 35: 11-16.
- Dawood S, Merajver SD, Viens P, Vermeulen PB, Swain SM, et al. (2011) International expert panel on inflammatory breast cancer: consensus statement for standardized diagnosis and treatment. Ann Oncol 22: 515-523.
- Cristofanilli M, Valero V, Buzdar AU, Kau SW, Broglio KR, et al. (2007) Inflammatory breast cancer (IBC) and patterns of recurrence: understanding the biology of a unique disease. Cancer110: 1436-1444.
- Resetkova E (2008) Pathologic aspects of inflammatory breast carcinom: part 1. Histomorphology and differential diagnosis. Semin Oncol 35: 25-32.
- Vermeulen PB, van Golen KL, Dirix LY (2010) Angiogenesis, lymphangiogenesis, growth pattern and tumor emboli in inflammatory breast cancer: a review of the current knowledge. Cancer 116: 2748-2754.
- Kleer CG, van Golen KL, Braun T, Merajver SD (2001) Persistent E-cadherin expression in inflammatory breast cancer. Mod Pathol 14: 458-464.
- Van den Eynden GG, Van der Auwer I, Van Laere S, Colpaert CG, van Dam P, et al. (2004) Validation of a tissue microarray to study differential protein expression in inflammatory and non-inflammatory breast cancer. Breast Cancer Res Treat 85: 13-22.
- Thiery JP, Acloque H, Huang RY, Nieto MA (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139: 871-890.
- Kalluri R, Weinberg RA (2009) The basics of epithelial-mesenchymal transition. J Clin Invest 119: 1420-1428.
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, et al. (2008) The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133: 704-715.
- Tomlinson JS, Alpaugh ML, Barsky SH (2001) An intact overexpressed E-cadherin/alpha beta-catenin axis characterizes the lymphovascular emboli of inflammatory breast carcinoma. Cancer Res 61: 5231-5241.
- Dong HM, Liu G, Hou YF, Wu J, Lu JS, et al. (2007) Dominant-negative E-cadherin inhibits the invasiveness of inflammatory breast cancer cells in vitro. J Cancer Res Clin Oncol 133: 83-92.
- Alpaugh ML, Tomlinson JS, Shao ZM, Barsky SH (1999) A novel human xenograft model of inflammatory breast cancer. Cancer Res 59: 5079-5084.
- Mahooti S, Porter K, Alpaugh ML, Ye Y, Xiao Y, et al. (2010) Breast carcinomatous tumoral emboli can result from encircling lymphovasculogenesis rather than lymphovascular invasion. Oncotarget 1:131-147.
- Chu K, Mu Z, Alpaugh RK,Fernandez S, Freiter EM, et al. (2011) Development and Comparative Characterization of Metastasis in Newly Developed Pre-clinical Models of Inflammatory Breast Cancer. Cancer Res 71: 439s.
- Fernandez SV, Zhaomei Mu, Aburto L, Xiaoshen D, Khoi C, et al. (2012) FC-IBC-02: A new in vitro-in vivo model of inflammatory breast cancer (IBC) Cancer Res 72.
- Forozan F, Veldman R, Ammerman CA, Parsa NZ, Kallioniemi A, et al. (1999) Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br J Cancer 81: 1328-1334.
- Ignatoski KM, Ethier SP (1999) Constitutive activation of pp125fak in newly isolated human breast cancer cell lines. Breast Cancer Res Treat 54: 173-182.
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, et al (2006) A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell10: 515-527.
- Willmarth NE, Ethier SP (2006) Autocrine and juxtacrine effects of amphiregulin on the proliferative invasive and migratory properties of normal and neoplastic human mammary epithelial cells. J Biol Chem 281: 37728-37737.
- Kurebayashi J, Otsuki T, Kurosumi M, Yamamoto S, Tanaka K, et al. (1999) Isolation and characterization of a new human breast cancer cell line, KPL-4, expressing the Erb B family receptors and interleukin-6. Br J Cancer 79: 707-717.
- Kurebayashi J, Yamamoto S, Otsuki T, Sonoo H (1999) Medroxyprogesterone acetate inhibits interleukin 6 secretion from KPL-4 human breast cancer cells both in vitro and in vivo: a possible mechanism of the anticachectic effect. Br J Cancer 79: 631-636.
- Kurebayashi J (2000) Regulation of interleukin-6 secretion from breast cancer cells and its clinical implications. Breast Cancer 7: 124-129.
- Fujimoto-Ouchi K, Sekiguchi F, Tanaka Y (2002) Antitumor activity of combinations of anti-HER-2 antibody trastuzumab and oral fluoropyrimidines capecitabine/5'-dFUrd in human breast cancer models. Cancer Chemother Pharmacol 49: 211-216.
- Fujimoto-Ouchi K, Sekiguchi F, Yamamoto K, Shirane M, Yamashita Y, et al. (2010) Preclinical study of prolonged administration of trastuzumab as combination therapy after disease progression during trastuzumab monotherapy. Cancer Chemother Pharmacol 66: 269-276.
- Klopp AH, Lacerda L, Gupta A, Debeb BG, Solley T, et al. (2010) Mesenchymal stem cells promote mammosphere formation and decrease E-cadherin in normal and malignant breast cells. PLoS One 5: e12180.
- Shirakawa K, Tsuda H, Heike Y, Kato K, Asada R, et al. (2001) Absence of endothelial cells, central necrosis and fibrosis are associated with aggressive inflammatory breast cancer. Cancer Res 61: 445-451.
- Shirakawa K, Kobayashi H, Heike Y, Kawamoto S, Brechbiel MW, et al. (2002) Hemodynamics in vasculogenic mimicry and angiogenesis of inflammatory breast cancer xenograft. Cancer Res 62: 560-566.
- Shirakawa K, Furuhata S, Watanabe I, Hayase H, Shimizu A, et al. (2002) Induction of vasculogenesis in breast cancer models. Br J Cancer 87: 1454-1461.
- Shirakawa K, Wakasugi H, Heike Y, Watanabe I, Yamada S, et al. (2002) Vasculogenic mimicry and pseudo-comedo formation in breast cancer. Int J Cancer 99: 821-828.
- Shirakawa K, Kobayashi H, Sobajima J, Hashimoto D, Shimizu A, et al. (2003) Inflammatory breast cancer: vasculogenic mimicry and its hemodynamics of an inflammatory breast cancer xenograft model. Breast Cancer Res 5:136-139.
- Shirakawa K, Shibuya M, Heike Y, Takashima S, Watanabe I, et al. (2002) Tumor-infiltrating endothelial cells and endothelial precursor cells in inflammatory breast cancer. Int J Cancer 99: 344-351.
- Robertson FM, Simeone AM, Lucci A, McMurray JS, Ghosh S, et al. (2010) Differential regulation of the aggressive phenotype of inflammatory breast cancer cells by prostanoid receptors EP3 and EP4. Cancer 116: 2806-2814.
- Van Laere SJ, Ueno NT, Finetti P, Vermeulen PB, Lucci A, et al. (2011) An Integrated Analysis of Three distinct IBC/nIBC Affymetrix Gene Expression Data Sets Further Unveils the Molecular Biology of IBC. Cancer Res 71: 33s.
- Iwamoto T, Bianchini G, Qi Y, Cristofanilli M, Lucci A, et al. (2011) Different gene expressions are associated with the different molecular subtypes of inflammatory breast cancer. Breast Cancer Res Treat 125: 785-795.
- Van Laere S, Van der Auwera I, Van den Eynden G, G Trinh X Van Hummelen P, et al (2007) Confirmation of the distinct molecular phenotype of inflammatory breast cancer compared to non-inflammatory breast cancer using Affymetrix based genome-wide gene expression analysis. J Clin Oncol 25: 21055.
- Van Laere SJ, Ueno NT, Finetti P, Vermeulen, PB, Lucci A, et al. (2011) An integrated analysis of three distinct IBC/non-IBC affymetrix gene expression data sets to study the transcriptional heterogeneity both between IBC and non-IBC and within IBC. J Clin Oncol 29: 10571.
- Voduc KD, Cheang MC, Tyldesley S, Gelmon K, Nielsen TO, et al. (2012) Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol 28:1684-1691.
- Robertson FM, ChuK, Circo R, Wulfkuhle J, Krishanmurthy S, et al. (2011) Genomic and proteomic pathway mapping reveals signatures of mesenchymal epithelial plasticity in Inflammatory Breast Cancer. Breast Cancer: Recent Advances in Biology, Imaging and Therapeutics: 978-953-307-730-7.
- Hurteau GJ, Carlson JA, Roos E, Brock GJ (2009) Stable expression of miR-200c alone is sufficient to regulate TCF8 (ZEB1) and restore E-cadherin expression. Cell Cycle 8: 2064-2069.
- Charafe-Jauffret E, Ginestier C, Iovino F,Tarpin C, Diebel M, et al. (2010) Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res 16:45-55.
- Van Laere S, Limame R, Van Marck EA, Vermeulen PB, Dirix LY. (2010) Is there a role for mammary stem cells in inflammatory breast carcinoma? a review of evidence from cell line, animal model, and human tissue sample experiments. Cancer 116 : 2794-2805.
- Giampieri S, Manning C, Hooper S, Jones L, Hill CS, et al. (2009) Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol 11: 1287-1296.
- Talmadge JE, Fidler IJ (2010) AACR centennial series: The biology of cancer metastasis: Historical perspective. Cancer Res 70: 5649-5669.
- Chao YL, Shepard CR, Wells A (2010) Breast carcinoma cells re-express E-cadherin during mesenchymal to epithelial reverting transition. Mol Cancer 9: 179.
- Rodriguez FJ, Lewis-Tuffin LJ, Anastasiadis PZ (2012) E-cadherin's dark side: Possible role in tumor progression. Biochim Biophys Acta 1826: 23-31.
Citation: Robertson FM, Chu K, Fernandez SV, Mu Z, Zhang X, et al. (2012) Genomic Profiling of Pre-Clinical Models of Inflammatory Breast Cancer Identifies a Signature of Epithelial Plasticity and Suppression of TGFß Signaling. J Clin Exp Pathol 2:119. DOI: 10.4172/2161-0681.1000119
Copyright: © 2012 Robertson FM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 17644
- [From(publication date): 7-2012 - Apr 08, 2025]
- Breakdown by view type
- HTML page views: 12634
- PDF downloads: 5010