Research Article Open Access
Exploited Application of Bacillus spp. ETL-1979 for Degradation and Decolorization of Methyl Orange, Malachite Green and Congo Red
| Maulin P Shah1*, Kavita A Patel1, Sunu S Nair1, A M Darji1 and Shaktisinh Maharaul2 | |
| 1Industrial Waste Water Research Laboratory, Applied and Environmental Microbiology Lab, Enviro Technology Limited (CETP), Gujarat, India | |
| 2Laboratory of Environmental Bioremediation, Narmada Clean Tech Limited (FETP), Ankleshwar, Gujarat, India | |
| Corresponding Author : | Maulin P Shah Industrial Waste Water Research Laboratory Applied and Environmental Microbiology Lab, Enviro Technology Limited (CETP) Plot No: 2413/2414, GIDC, Ankleshwar- 393 002, Gujarat, India Tel: +91-90 999 65504 Fax: +91-2646-250707 E-mail: shahmp@uniphos.com |
| Received: June 15, 2013; Accepted: July 29, 2013; Published: July 31, 2013 | |
| Citation: Shah MP, Patel KA, Nair SS, Darji AM, Maharaul S (2013) Exploited Application of Bacillus spp. ETL-1979 for Degradation and Decolorization of Methyl Orange, Malachite Green and Congo Red. J Bioremed Biodeg 4:196. doi:10.4172/2155-6199.1000196 | |
| Copyright: © 2013 Shah MP, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
The local industrial dye waste water sample was collected from Final Effleunt Treatment Plant (FETP) of Ankelshwar, Gujarat, India for the isolation of dye degrading bacteria. The isolate ETL-1979 was selected based on its maximum decolorization of dyes Methyl orange, Congo red and Malachite green (each 150 mg/L). Phenotypic characterization and phylogenetic analysis based on 16S rDNA sequence confirmed ETL-1979 as Bacillus sp. On physiochemical parameters optimization, Bacillus sp. ETL-1979 degraded 10.13% (0.15 mg/mL), 11.76% (0.11 mg/ mL) and 9.32% (0.8 mg/mL) of Methyl orange, Congo red and Malachite green, respectively within 2 h, whilst 100% degradation (2.97 mg/mL) at 18 h for Methyl orange, 100% (2.43 mg/mL) at 16 h for Congo red and 100% (3.18 mg/mL) at 20 h for Malachite green were recorded. The degraded products were found non-toxic in nature, while bacterial cells showed elongation and membrane rupturing under SEM taken from dye degraded media.
| Keywords |
| Methyl orange; Malachite green; Congo red; Bacillus; SEM |
| Introduction |
| Rapid industrialization has necessitated the manufacture and use of different chemicals in day to day life [1]. Reactive dyes, including many structurally different dyes, are extensively used in the textile industry because of their wide variety of color shades, high wet fastness profiles, ease of application, brilliant colors and minimal energy consumption. The three most common groups are azo, anthraquinone and phthalocyanine [2]. The synthetic dyes, especially azo dyestuff, have been produced and widely used in paper, textile and coating industries, which caused serious environmental pollution, especially in developing countries. Synthetic azo dyestuff is very difficult to remove and biodegrade due to its complex aromatic molecular structure, they are also highly colored, toxic and can heavily contaminate water source [3,4]. The dyeing industry has been referred as one of the four major polluting industries in India. The Indian textile industry is the world’s second largest after China. There are about 1, 200 medium to largescale textile mills in India. Sources also claim about 3, 000 garment manufacturing units employing about 3 million people. All together, India exports to 162 countries, which accounts for 38% of India’s export ($ 35 billion in 2000) [5]. Chemical degradation technologies, such as catalytic ozonation, photocatalysis, ultrasonic irradiation and electrochemical oxidation, have been studied for the degradation of azo dyes in wastewater [6]. Biological degradation technology used bacteria or fungi to decolor, which is kind to human and environment, nontoxic, and no secondary contamination due to its source (natural environment) [7,8]. Many bacteria and fungi can efficiently biodegrade azo dyes and decolorized them, such as Aeromonas, Pseudomonas, Bacillus, Rhodococcus, Shigella, Klebsiella, Rhizopus oryzae, Penicillium oxalicum and Phanerochaete chrysosporium [9-13]. Bacteria could produce azo reductase, showing maximum decolori-zation activity, while fungi could absorb the dying stuff [14,15]. Azo reductase has low substrate specificity, and can break the dye molecules in the highaffinity electron azo bond, producing colorless and aromatic amines [16,17]. Bioremediation offers a cost effective and environmentally friendly method for decolorization and degradation of dyes in industrial effluents and contaminated soil [9,18]. The present study emphasized on the degradation of three dyes belonging to monoazo, diazo and triphenylmethane group by the bacteria isolated from local industrial waste, which can be useful in providing an alternate method to accomplish dye degradation of a wide range of dyes in an ecofriendly manner. |
| Materials and Methods |
| Dyes and chemicals |
| The dyes, Methyl orange, Congo red and Malachite green used in the study were of analytical grade procured from Himedia, Mumbai, India. The Methyl orange is a mono azo dye of the 4-dimethylaminoazobenzene-4'-sulfonic acid sodium salt, and Congo red is the sodium salt of benzidinediazo-bis-1-naphtylamine- 4-sulfonic acid, a secondary diazo dye, whereas Malachite green is the triphenylmethane dye of {4-[(4-dimethylaminophenyl)- phenylmethylidene]-dimethyl-azanium chloride}. The Methyl orange, Congo red and Malachite green has maximum absorbance (λmax) of light at 444 nm [19], 530 nm [20] and 620 nm [21], respectively. |
| Isolation, identification and genomic DNA extraction |
| The wastewater sample was collected from final effluent treatment plant (FETP) of Ankleshwar; Gujarat, India, was screened for the dye degrading bacteria. The screening and routine sub culturing of dye decolorizing bacterial isolate was carried out on nutrient media (in g/L: Peptone, 10; Sodium chloride, 5; Yeast extract, 5) at pH 7 on 35°C. The wastewater sample was tenfold serially diluted and 100 μL of 10-5 dilution was spread plated on the nutrient agar plates. After 24 h incubation, morphologically distinct colonies obtained were picked, purified by streak plate method and inoculated into the separate liquid medium amended with individual 150 mg/L Methyl orange, Congo red and Malachite green dyes to analyze their decolorizing ability. Following 10 h interval, aliquot of 1 mL was withdrawn and centrifuged at 8000 rpm for 10 min. The clear supernatant obtained was used to measure the dye decolorization at their respective absorbance maxima (λmax), using spectrophotometer, where a medium without dye and inoculum was used as blank, while medium with dye but without inoculum was taken as control. The absorbance for all the isolates was taken similarly up to 50 h. All the experiments were carried out in triplicates and the mean value was taken. The decolorization of dyes was calculated by the given formula [22], as Decolorization efficiency (%) = I − F / I ×100 , Where I=Absorbance of media prior to incubation, F=Absorbance of decolorized media. After calculating the percentage decolorization of dyes up to 50 h, the bacterial isolate with the strongest decolorizing ability, designated as ETL-1979, was selected and preserved on slant and broth as stock. The isolate ETL-1979 was initially studied for the phenotypic characteristics on the nutrient agar media, followed by Gram staining 16S rDNAtechnique [23]. Genomic DNA of the isolate was extracted with a GenElute DNA extraction kit from Sigma. The 16S rRNA gene of isolate was amplified using the universal primers 8F (5′-AGAGTTTGATCCTGGCTCAG) and 1541R (50-AAGGAGGTGATCCAGCCGCA-3′). The amplification was done by initial denaturation at 95°C for 5 min followed by 10 cycles of 93°C for 1 min, 63°C for 1 min, 71°C for 1.5 min; 20 cycles of 93°C for 1 min, 67°C for 1 min, 71°C for 2 min and final extension at 71°C for 5 min. The purified PCR product was sequenced in both directions using an automated sequencer by Bangalore Genei (India). The phylogenic relationship of the isolate was determined by comparing the sequencing data with sequences of some members of the genus Bacillus, available through the GenBank database of the National Center for Biotechnology Information. The gene sequences of each isolate obtained in this study were compared with known 16s rRNA gene sequences in the GenBank database. |
| Physiochemical parameters analysis |
| The isolated strain ETL-1979 was further studied for the physiochemical parameters to check their effect on the dyes decolorization taking different carbon sources (1%), nitrogen sources (1%), pH (4, 5, 6, 7, 8, 9 and 10), temperature (25, 30, 35 and 40°C), various dye concentrations (150, 200, 250, 300 and 350 mg/L), different volumes of inoculum (100, 150, 200, 250 and 300 μL) and static/shaker condition. The dyes decolorization was monitored as described in the earlier section. The decolorization analysis of the dyes was then further observed using all the optimized parameters with respect to time after every 2 h interval, while the cell pellets obtained were dried at 85°C and weighed mass was then expressed in mg/mL. |
| Decolorization study of ETL-1979 on dyes and scanning electron microscope |
| Decolorization of dyes may take place by adsorption [24], or degradation [25]. Dye adsorption can be also easily judged by an evidently colored cell pellet, whereas those retaining their original colors are accompanied by the occurrence of biodegradation [16]. The three sets of optimized nutrient broth (25 mL) inoculated (1% v/v) with pure bacterial isolate ETL-1979 were incubated overnight and before addition of dyes (individually150 mg/L in separate flask), one flask from each set were autoclaved at 121°C for half an hour, while the unautoclaved served as control. After 24 h incubation, samples from the flasks were withdrawn and centrifuged at 8000 rpm for 10 min. The supernatant was used for monitoring the dye decolorization, as described earlier. To determine the effect of dyes on the isolate ETL- 1979 cell morphology, visualization of the 24 h old cultures grown in normal and individual dye (150 mg/L) amended optimized nutrient broth were performed under Scanning Electron Microscope (SEM) [26]. |
| Toxicity study |
| Microbial toxicity test was performed in order to assess the toxicity of dyes and their degraded products formed. The degraded products of each 150 mg/L Methyl orange, Congo red and Malachite green from the 50 mL optimized nutrient media were extracted in equal volume of ethyl acetate, dried, and dissolved in 5 mL sterilized distilled water to make a final concentration of 500 ppm and 1000 ppm. These final concentrations were then used for the microbial toxicity studies carried out in relation to Staphylococcus aureus, Pseudomonas aeruginosa and Aspergillus niger, while the respective concentrations of dyes were taken as control. The growth inhibition zones of microbes (diameter in cm) were recorded after 24 h and 48 h incubation at 35°C for the bacterial and fungal strains, respectively. |
| Results |
| Screening and identification |
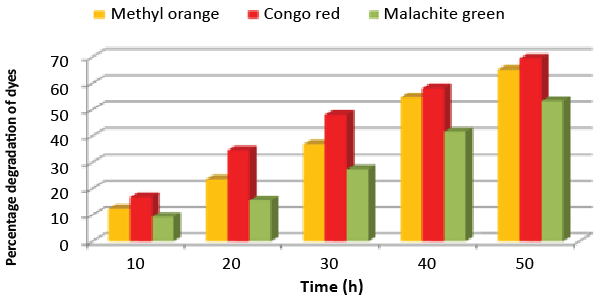
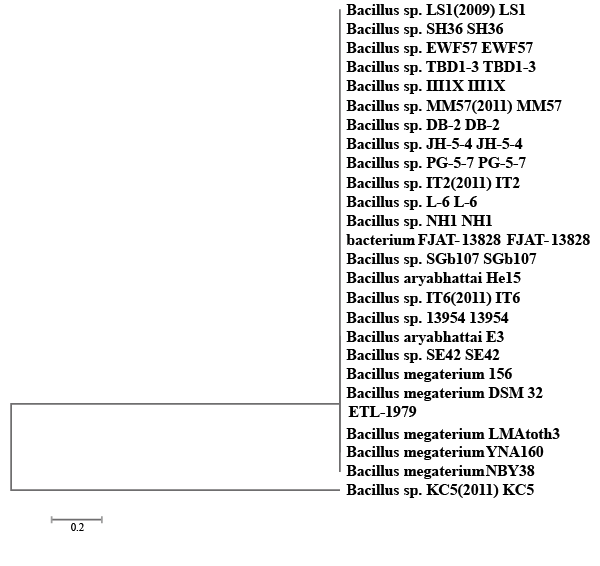
| A bacterial isolate ETL-1979 was able to degrade 65.17% of Methyl orange, 69.41% of Congo red and 53.23% of Malachite green, within 50 h incubation in individual dye amended nutrient broth (Figure 1). The isolate ETL-1979 colonies were white, glossy, round, with regular margins and 1 to 3 mm in diameter, and were observed to be Gram positive rods. Sequence analysis of 16S rDNA showed that the isolate ETL-1979 had highest similarity of 100% with Bacillus megaterium DMZ 32 (X60629). Based on the phenotypic characteristics and phylogenetic analysis, isolate ETL-1979 was identified as Bacillus sp. Phylogenetic relationship between the isolate ETL-1979 with different members of the Bacillus sp. are illustrated in the Figure 2. |
| Physiochemical parameters analysis |
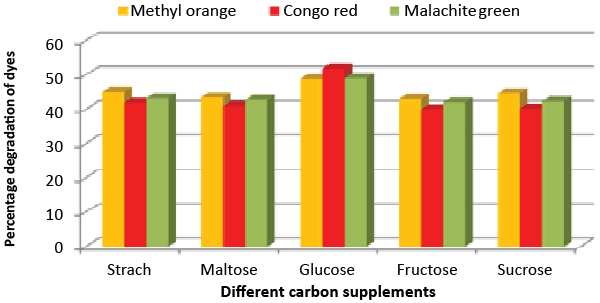
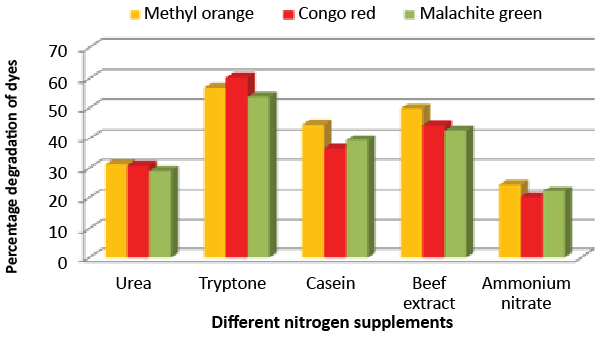
| Effect of carbon and nitrogen sources: The dye decolorization efficiency of the bacterial strain ETL-1979 was monitored with 1% different carbon and nitrogen sources as supplement in the nutrient media. Results concluded that among the different carbon sources, glucose showed maximum decolorization of 48.7, 51.8 and 49.9% for the Methyl orange, Congo red and Malachite green, respectively (Figure 3). Whilst among the different nitrogen supplements used, beef extract showed maximum decolorization of 49.7, 44.3 and 42.6% for the Methyl orange, Congo red and Malachite green, respectively (Figure 4). |
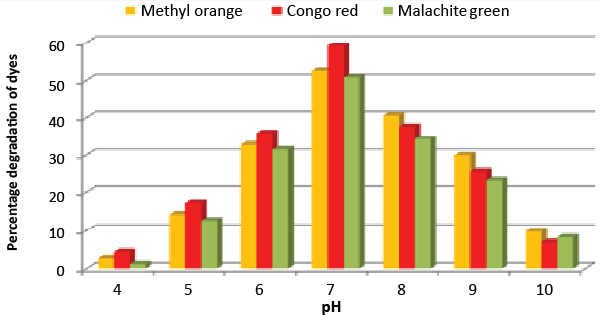
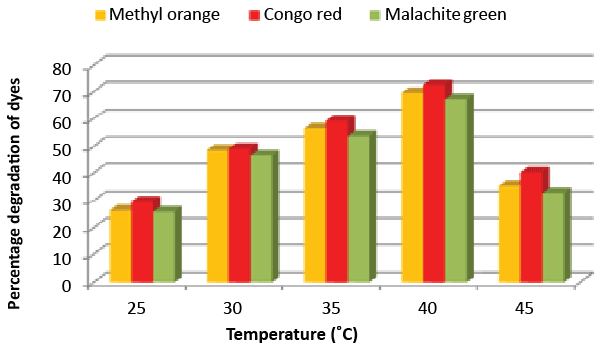
| Effect of pH and temperature: The strain ETL-1979 represented lowest dye degradation of 2.67, 4.34 and 1.21% at pH 4, while maximum degradation of 52.4, 59 and 50.7% were observed for Methyl orange, Congo red and Malachite green, respectively at pH 7 (Figure 5). The lowest dyes degradation of 26.2, 29.6 and 25.8% at temperature of 25°C, while maximum dye degradation of 69.5, 72.3 and 67.4% at 40°C were observed for Methyl orange, Congo red and Malachite green, respectively (Figure 6). |
| Decolorization on static and shaking conditions |
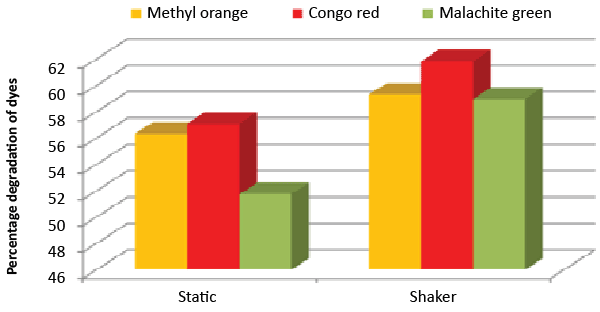
| It was monitored that under static condition, isolate ETL-1979 degraded 56.3, 57 and 51.7% of the Methyl orange, Congo red and Malachite green, respectively, whereas higher degradation of 59.3, 61.8 and 58.9% for Methyl orange, Congo red and Malachite green, respectively, were recorded on the shaker condition (Figure 7). |
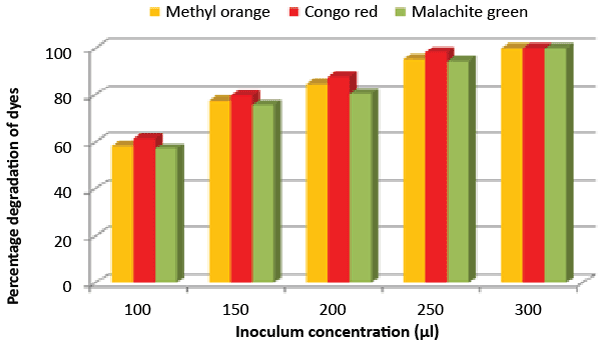
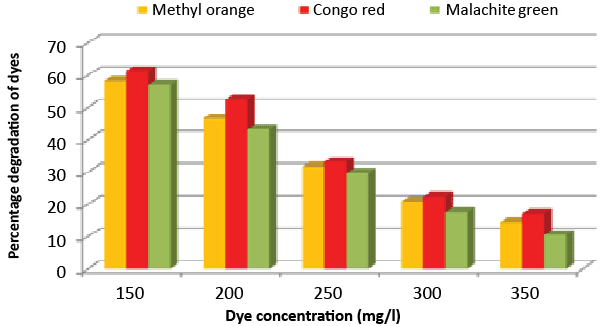
| Effect of inoculum and dye concentration: The decolorization of 58.3, 61.3 and 57.1% were recorded for the 100 μL volume of inoculums, whereas 100% decolorization were recorded with the use of 300 μL volume of inoculum for the 150 mg/L of Methyl orange, Congo red and Malachite green after 24 h incubation at 35°C (Figure 8). The study also recorded that at 150 mg/L concentration the isolate showed 58.3, 61.3 and 57.1% decolorization, which decreased to 14.2, 17.1 and 10.4% for the 350 mg/L of Methyl orange, Congo red and Malachite green, respectively (Figure 9). |
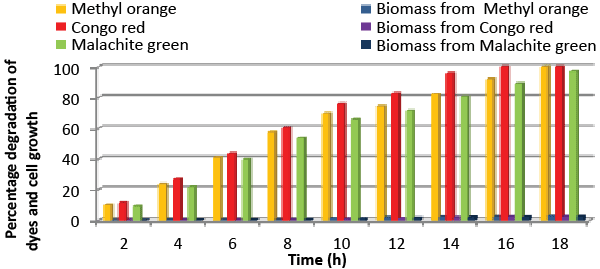
| Effect of total physiochemical parameters: The decolorization efficiency of the bacterial isolate ETL-1979 on individual 150 mg/L dyes and its biomass was calculated using all of the optimized parameters, with respect to time (Figure 10). It was deduced that bacteria degraded 10.1% (0.15 mg/mL), 11.7% (0.11 mg/mL) and 9.32% (0.08 mg/mL) of Methyl orange, Congo red and Malachite green, respectively within 2 h, whilst 100% degradation (2.97 mg/mL) at 18 h for Methyl orange, 100% (2.43 mg/mL) at 16 h for Congo red and 100% (3.18 mg/mL) at 20 h for Malachite green were recorded. This proves that the combined effect of optimized parameters attributed to the decolorization of the dyes by the isolated bacteria Bacillus sp. ETL-1979. |
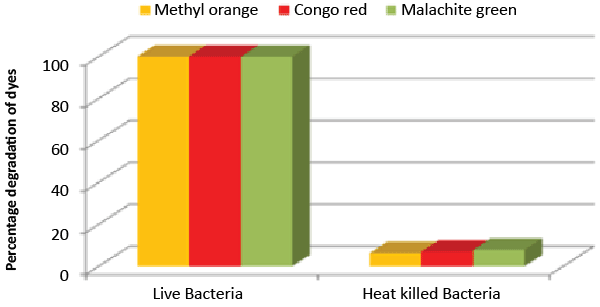
| Decolorization study of Bacillus sp. ETL-1979 and SEM analysis: In the heat-killed bacterial cultures, only 6.22, 6.91and 7.87% decolorization of Methyl orange, Congo red and Malachite green were recorded after 24 h incubation, while in the control culture, 100% decolorization of Methyl orange, Congo red and Malachite green were obtained in 24 h (Figure 11), and the cell pellets were not pigmented. Moreover, the scanning micrographs showed elongation and cell membrane rupturing for cells taken from the dye degraded media, as compared to the normal media, as depicted in Figures 12a-12d revealed the toxic effect of dyes on the bacteria. |
| Microbial toxicity study: The inhibition zones were observed with control Methyl orange, Congo red and Malachite green with all bacterial strains studied, whereas their degraded products did not show any growth inhibition. |
| Discussion |
| This study has been aimed at isolating and characterizing the potential dye degrading bacteria from the local industrial wastes. The strain Bacillus sp. ETL-1979, isolated from Final Effluent Treatment Plant (FETP) of Ankleshwar, Gujarat, India used glucose and beef extract as carbon and nitrogen supplement, respectively. Although effects of some other carbon sources on bacterial decolorization performance have been studied in former researches, where lactate, peptone, succinate, yeast extract and format were proved to enhance decolorization, but sucrose and dextrin resulted in lower decolorization activities [27]. It was also reported that addition of inorganic nutrients like nitrogen does not always enhance degradation of organic compounds, because there are many other factors which many decrease microbial activity [28]. Urea when dissolved in liquid culture causes a shift of pH more towards acidic side, which decreased the color removal, growth, as well as enzyme activity of strains. Also, presence of nitrate in culture media might slow down color removal process [29]. The strain Bacillus sp. ETL-1979 showed maximum dye decolorization at neutral pH, was in accordance with the report that optimum pH for color removal often exists between 6.0 and 10.0 [30]. Further, the isolate Bacillus sp. ETL-979 showed highest dye degradation on shaker must be because of dissolved oxygen concentration, which creates the microaerophillic environment in the glass vessel for the bacteria bringing about the decolorization of dyes at a faster rate [31]. Results also concluded that for 100 μL inoculum, 350 mg/L dye acted as inhibitor for the bacterial growth and resulted in less decolorization, whereas 300 μL inoculum showed 100% decolorization of 150 mg/L of dyes within 24 h, suggested that the concentration of dye as substrate can influence the efficiency of dye removal through a combination of factors, including the toxicity of dyes at higher concentrations and the ability of the enzyme to recognize the substrate efficiently at very low concentrations [32]. Further, the toxicity results for Methyl orange, Congo red and Malachite green coincides with the other research work reported for the respective dyes degradation using other bacteria [20,21,33-37]. The non-toxicity of dye degraded products can be suggested because of hydroxylase and oxygenase produced by the bacteria [38]. |
| Conclusion |
| It was inferred that the bacterial strain Bacillus sp. ETL-1979 shows 100% degradation with the optimized physiochemical parameters. Since, the degraded products of the dyes showed no toxic effect on the microbes and plants, implies that maximum degradation of a wide range of different dyes can be meted out by employing the potential bacterial strains from the local industrial wastes, using optimized physiochemical parameters, as it renders the ability to bio transform the toxic dyes into non-toxic products without any additional treatment. This study further recommends the identification, purification of enzymes and their kinetics involved in the degradation of dyes by the isolate, and exploitation of potential bacterial consortium from the local industrial wastes in the treatment of dye polluted waste water, which would be cost effective, and can contribute to eradication of the dye pollution from the water bodies. |
References
|
Figures at a glance
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
 |
 |
 |
 |
| Figure 5 | Figure 6 | Figure 7 | Figure 8 |
 |
 |
 |
 |
| Figure 9 | Figure 10 | Figure 11 | Figure 12 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 14478
- [From(publication date):
July-2013 - Apr 04, 2025] - Breakdown by view type
- HTML page views : 9874
- PDF downloads : 4604
