Research Article Open Access
Enzyme-Mediated Bioremediation of Organophosphates Using Stable Yeast Biocatalysts
| Randhir S. Makkar1, Augustine A. DiNovo1, Caroline Westwater2 and David A. Schofield1* | |
| 1Guild Associates, Inc., Charleston, South Carolina, 29407, USA | |
| 2Department of Craniofacial Biology, Medical University of South Carolina, South Carolina, 29425, USA | |
| Corresponding Author : | David A Schofield Guild Associates Inc. 1313B Ashley River Road Charleston, SC 29407, USA Tel: +(843) 573 0095 Fax: +(843) 573 0707 E-mail: dschofield@guildassociates.com |
| Received: February 08, 2013; Accepted: February 21, 2013; Published: February 23, 2013 | |
| Citation: Makkar RS, DiNovo AA, Westwater C, Schofield DA (2013) Enzyme- Mediated Bioremediation of Organophosphates Using Stable Yeast Biocatalysts. J Bioremed Biodeg 4:182. doi:10.4172/2155-6199.1000182 | |
| Copyright: © 2013 Makkar RS, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Organophosphates are extremely toxic compounds, which pose a threat to the environment and public due to
their widespread use as common pesticides or due to their deliberate release as chemical weapons. The bacterial enzyme organophosphorus hydrolase (OPH, EC. 3.1.8.1) can hydrolyze, and thereby detoxify a broad range of organophosphate nerve agents. This enzyme therefore offers the opportunity for the development of naturally occurring newer bioremediation strategies. The aim of this research was to generate a stable yeast biocatalyst that was capable of hydrolyzing the poorly hydrolyzed P-S class of organophosphates. The genes encoding the wild-type OPH, or the enhanced variant enzyme S308L-OPH, were integrated into the ribosomal operon of the Saccharomyces cerevisiae genome to create a stable yeast biocatalyst.
| Keywords |
| Organophosphate hydrolysis; Detoxification; Enzyme mediated |
| Introduction |
| Organophosphates are toxic man-made compounds that are used as chemical warfare agents (nerve agents), and more commonly as pesticides. Worldwide, five billion pounds of pesticides are used every year of which, organophosphate pesticides (mostly insecticides) constitute 20-38% of the total pesticides used; the most commonly used organophosphate insecticide in the U.S. is malathion [1]. As a result, low levels of organophosphate contamination are commonly found in natural habitats, in the food chain, and in our water supplies. The number of pesticide poisonings caused by inadvertent exposure has been estimated at between 1 and 3 million every year [2-4]. Organophosphate compounds exert their toxicity by inhibiting the enzyme acetylcholinesterase (AChE) which can cause a variety of ailments, including respiratory failure and death. |
| Organophosphate exposure may also be caused by the accidental or deliberate release of chemical warfare nerve agents such as VX (O-ethyl S-[2-diisopropylaminoethyl] methylphosphonothiolate), sarin, soman or tabun. The threat is compounded by the fact that some countries have massive chemical weapon stockpiles. World stockpiles are reported to exceed 200 kilotons, with U.S. reserves alone amounting to 30 kilotons. The Chemical Weapons Convention of 1997 required the destruction of all chemical weapons by the year 2007; in the U.S., this deadline has passed and is now extended until 2023. This extension has been brought about by delays in the development of disposal facilities, mostly incinerators, and the environmentally regulatory, safety, and security concerns associated with these facilities. The Pentagon has estimated that the cost of destroying U.S. stockpiles at $32 billion. This delay raises the risk of an accident or theft by terrorists. Consequently, the development of effective and environmentally-friendly methods to dispose of these man-made toxic compounds would be of value. |
| The bacterial Organo Phosphorus Hydrolase (OPH) enzyme can hydrolyze, and thereby reduce the toxicity of organophosphate pesticides and nerve agents by cleaving the various phosphorus-ester bonds (P-O, P-CN, P-F, P-S). The enzyme mediated cleavage of these bonds, however, occurs with different efficiencies [5]. For example, OPH catalyzes the P-O bond of paraoxon with a kcat of 2280 s-1, but catalyses the P-S bonds of demeton-S, malathion, phosalone and acephate with kcats of 0.63-13.16 s-1, which is approximately 1000-fold slower. Therefore, while it is very efficient at bioremediating some organophosphates, the hydrolytic efficiency of OPH towards the P-S bond class of organophosphates such as VX is very poor. |
| The goal of our research is to develop environmentally-friendly enzymes and biocatalysts for the bioremediation of organophosphates. The P-S class of organophosphates such as malathion, demeton-S, and VX was the focus of this study because: (i) these organophosphates pose a serious decontamination challenge since they are hydrolyzed very inefficiently by OPH; (ii) malathion is the most commonly used organophosphate insecticide, and (iii) VX is extremely toxic, and the most stable of the warfare nerve agents. Towards this goal, a variant OPH enzyme (termed S308L-OPH) was generated which displayed a 25-fold increase in hydrolytic efficiency against P-S organophosphates [6]. In this report, the Generally Regarded As Safe (GRAS) yeast Saccharomyces cerevisiae was utilized for the generation of a stable recombinant yeast biocatalyst expressing OPH or the S308L-OPH. The biocatalyst was able to hydrolyze and thereby detoxify organophosphate pesticides (malathion, demeton-S) and warfare agents (VX). |
| Materials and Methods |
| Microorganisms and propagation |
| The Escherichia coli and S. cerevisiae strains used and generated in this study are described in table 1. E. coli was propagated in Luria Bertani (LB) broth or LB agar at 37°C supplemented with 100 μg/mL ampicillin when needed. S. cerevisiae was grown and maintained on 1% yeast extract, 2% peptone, and 2% dextrose (YPD, Difco) broth or YPD agar at 30°C, where appropriate, YPD was supplemented with 200 μg/ mL G418. |
| Construction of the yeast Oph and S308l-Oph integration cassettes |
| An integration/expression cassette containing preferential yeast expression signals and the necessary sequences to direct targeted genome integration into the yeast rDNA unit was constructed. The wild-type gene encoding OPH, but lacking the signal peptide sequence, was kindly provided by Mulbry and Karns [7]. The variant S308L-OPH was generated from the wild-type gene and harbors the following amino acid substitutions; A80V, I106V, F132D, K185R, D208G, H257W, I274N, S308L, R319S [6]. The wild-type and variant S308L-OPH, originally derived from the bacterium Flavobacterium species, were yeast codon optimized (Bio S&T Inc., Montreal, Canada) to ensure efficient expression in yeast (GenBank numbers GU947025 and GU947026, respectively). Both coding sequences were designed to incorporate a preferred yeast translation initiation sequence (5’-AAAAGTATG) immediately preceding the ATG (bold) start codon [8] and cloned into the HindIII/XhoI sites of pBluescript SKII- to generate pOPH-SK- and pS308L-OPH-SK (Table 2). |
| A 635 bp TaqI fragment of the yeast GAPDH 5’-untranslated region has been shown to contain all the sequences necessary for promoter function in vivo [9]. Therefore, the GAPDH promoter (653 bp) was generated by PCR using a high fidelity DNA polymerase and S. cerevisiae genomic DNA as template. The 5’ and 3’ PCR primers were designed to incorporate PstI and HindIII sites, respectively (Table 3). The resulting PCR product was cloned into the corresponding sites of pOPH-SK- and pS308L-OPH-SK-, thereby placing the GAPDH promoter upstream of the target genes. |
| The gene encoding neomycin resistance (NEOr) was used as the marker for the selection of recombinant clones. A 916 bp fragment containing the NEOr gene and an abbreviated (120 bp) 5’ SV40 flanking promoter sequence was PCR-amplified using pEGFP-N1 (Clontech, Mountain View CA, USA) as template. The 5’ and 3’ PCR primers contained SpeI and PstI restriction endonuclease sites for cloning into the pGAPDH-OPH-SK- and pGAPDH-S308L-OPH-SK- vectors (Table 2). This placed the NEOr gene 5’ of the GAPDH promoter. |
| The integration/expression cassette was targeted to the nontranscribed spacer in the rDNA between the 5S and 18S subunits (located on chromosome XII) by homologous recombination. Two rDNA fragments comprising of 592 bp and 663 bp were PCR amplified using S. cerevisiae genomic DNA as template, and primers R4F/R5R and R6F/R7R, respectively (Table 3). The R4F/R5R and R6F/R7R primers were designed to contain SacI/SpeI and XhoI/KpnI sites respectively, for cloning into the 5’ or 3’ ends of the integration cassette, thereby providing flanking sequences for targeted homologous recombination into the yeast genome. Lastly, the TcycI transcriptional terminator was PCR-amplified using pESC-URA (Agilent Technologies, Inc, Santa Clara, CA, USA) as template and cloned 3’ of the target genes in the XhoI site. All cloning steps were performed in E. coli ER2738 using standard methodology [10]. The sequences of the cloned fragments were verified by deoxy dye terminator sequencing. The wild-type OPH and S308L-OPH integration cassettes were designed to allow the cassette to be released from the plasmid backbone using the unique enzymes SacI and KpnI. |
| Integration into the yeast genome by homologous recombination |
| The wild-type and S308L-OPH cassettes were excised from the pBluescript-SKII- backbone using SacI and KpnI, resolved using agarose gel electrophoresis, and gel purified using the QIAquick gel extraction kit (Qiagen, Valencia, CA, USA). S. cerevisiae BY4741 was transformed with approximately 0.5 μg of DNA using a high-efficiency LiAc/PEG yeast transformation method [11]. The transformation mixes were plated onto YPD plates supplemented with 200 μg/mL G418 to select for NEOr colonies. After incubation at 30°C for 72 to 96 h, NEOr colonies were restreaked onto master plates (YPD/200 μg/ mL G418). |
| Quantitative real time PCR |
| The number of integrated S308L-OPH copies in the yeast genome was measured using quantitative real-time PCR. Quantification was assessed using iQ SYBR Green qPCR Super Mix (BioRad, Hercules, CA, USA) and a BioRad iCycler iQ real-time PCR detection system. Real time PCR primers were designed for the target (S308L-OPH) and internal reference control (YLR346C) genes (Table 3). Both primer pairs produced a single amplicon with a uniform melting curve as determined by the dissociation profile of the product. The absolute copy number of the target gene (S308L-OPH) and reference gene (YLR346C; single-copy) were determined from a corresponding standard curve, using the CT values. For each genomic preparation, the target gene copy number was divided by the copy number of YLR346C. Therefore, the target gene copy number was determined as the ratio of S308L to YLR346C. Real time qPCR amplifications of genomic DNA (from two independent cultures; duplicate reactions) were performed simultaneously with standards representing 1×103 to 1 x 107 copies of S308L or YLR346C (triplicate reactions). Both standard curves were linear in the range tested (correlation coefficient= 0.999). The slopes of the standard curves for S308L and YLR346C were -3.379 and -3.365, respectively. A high amplification efficiency was obtained for both primer pairs (S308L, 97.7% and YLR346C, 98.2%). |
| Demeton-S methyl and malathion hydrolysis assays |
| S. cerevisiae BY4741 (control cells), wild-type OPH, and S308LOPH were grown in YPD media (± G418 when appropriate) supplemented with 0.1 mM CoCl2 at 30°C. At an A600 of 0.6 to 0.8, the cells were harvested by centrifugation (10,000×g, 2 min), washed with 35 mM HEPES pH 8.0, and the cell pellets were incubated with 0.5 mL Y-MER dialyzable lysis buffer (Thermo Scientific) for 20 min at room temperature (RT), followed by centrifugation at 24,000×g for 10 min at 4°C. The ability of yeast lysates to hydrolyze demeton-S methyl (Chem Service, West Chester, PA, USA) or malathion (Cerilliant, Round Rock, Texas, USA) were measured in reactions consisting of 50 mM HEPES pH 7.5, 1.0 mM DTNB (Ellman’s reagent), 0.1 mM CoCl2 and either 1 mM demeton-S methyl or 0.4 mM malathion. The rate of hydrolysis was measured by following the appearance of 2-nitro-5-thiobenzoate at 412 nm at RT (∼25°C) using a BioTek Synergy 2 microplate reader. Control rates of hydrolysis from lysates lacking OPH were measured and subtracted from enzymatic hydrolysis rates. Each clone was measured using triplicate lysates prepared from triplicate cultures and the specific activity of yeast lysates to hydrolyze demeton-S methyl or malathion was measured as μmoles hydrolyzed/min/mg of total protein. The ability of yeast cells to hydrolyze demeton-S methyl and malathion was measured by resuspending washed intact yeast cells directly in the reaction buffer used for the enzyme lysates. After various incubation periods, cells were collected by centrifugation and the resulting cell-free supernatant was measured at A412. Results were normalized to the A600 and presented as μ moles hydrolyzed/min/OD600 of 1. Paraoxon assays were performed as described above and consisted of 50 mM HEPES pH 7.5, 0.1 mM CoCl2, and 0.1 mM paraoxon. |
| Acetylcholinesterase (AChE) assays |
| All organophosphate compounds act by inhibiting the enzyme acetylcholinesterase (AChE). To confirm the yeast lysates and intact cells hydrolyzed demeton-S methyl and thereby detoxified the agent, samples from the demeton-S methyl hydrolysis assays were analyzed for a reduction in the ability of the agent to inhibit AChE. Aliquots of the hydrolysis assays (after varying incubation periods) were mixed with purified human erythrocyte AChE (8.56 nM) for 30 min at RT. Demeton-S methyl controls (in reaction buffer) lacking OPH enzyme and AChE positive controls were run in parallel. The AChE assay mix was then diluted and mixed with the substrate, acetylcholine iodide (ACh, 0.5 mM), and DTNB (1.0 mM) in 100 mM Na2HPO4 buffer (pH 7.4). Absorbance changes due to ACh hydrolysis were monitored at 412 nm every 2 min for 30 min and the slope of the regression line of the reaction was used for calculating percentage AChE inhibition. |
| Yeast VX hydrolysis assays |
| VX hydrolysis assays were performed in a chemical surety materiel laboratory at the Southwest Research Institute (San Antonio, Texas). The laboratory meets safety, physical security, and personnel selection requirements established by the U.S Army Materiel Command and by the Army Standard Surety Clauses for use of chemical agents. VX P-S bond cleavage was detected using Ellman’s reagent essentially as described previously [12]. Assay conditions consisted of 0.5 mM VX, yeast cells (control and S308L-OPH), 50 mM HEPES (pH 7.5), 100 μM CoCl2, and 1 mM DTNB. Thiol release was measured at 415 nm at 37°C. Hydrolytic activity from recombinant S308L-OPH yeast were measured and subtracted from control rates. |
| Results |
| Rationale for the design of the integration cassette |
| The genes encoding the wild-type OPH and variant S308LOPH were targeted for yeast genome integration rather than relying on heterologous expression from an episomal plasmid. The major drawback of using 2 μ episomal plasmids in yeast is that they suffer from low segregational stability, i.e. they are not stably maintained in the yeast population [13]. Therefore, the target genes were integrated into the yeast genome in order to generate a biocatalyst that can stably maintain and express the heterologous gene for many generations even in the absence of selective pressure [14,15]. A drawback of integration is that in general, integrated sequences are maintained at a lower copy number (1-5) compared to episomal plasmids (30-50) [16]; however, since the number of integrated copies is proportional to the number of target sites in the yeast genome [17], yeast cells carrying multiple copies of the integrated DNA can be generated when the insertion sequence is present in multiple copies. Consequently, the integration cassette was targeted to the non-transcribed spacer in the rDNA between the 5S and 18S subunits (located on chromosome XII). Since there are approximately 140 copies of the rDNA unit, integration at this site has yielded recombinant strains with a similar number of integrated sequences [14]. |
| OPH can hydrolyze and thereby reduce the toxicity of a wide range of organophosphate pesticides and nerve agents; however, the catalytic efficiency of the enzyme against P-S substrates such as demeton-S, malathion, phosalone and VX is approximately 103 lower than the preferred substrate paraoxon [18,19]. The variant S308LOPH, which harbors 9 amino acid changes compared to the wildtype OPH, exhibits an approximately 25-fold improvement in the ability to hydrolyze demeton-S, malathion and VX [6]. Analysis of the coding sequences for the bacterial OPH and S308L-OPH using the graphical codon usage analyzer indicated that there were a number of codons that are rarely used in S. cerevisiae. Therefore, both genes were codon-optimized for expression in yeast and integrated into the yeast genome. To drive expression of OPH and S308L-OPH, the GAPDH promoter was placed immediately 5’ of the genes of interest. The GAPDH promoter is routinely used to achieve high level, foreign gene expression in S. cerevisiae since it is both strongly and constitutively expressed [14,15,20,21]. The TcycI terminator was placed 3’ of the genes of interest to ensure efficient processing of the gene transcripts. |
| Verification of yeast genome integration |
| S. cerevisiae NEOr colonies were analyzed by PCR to confirm the presence of the genes encoding OPH and S308L-OPH. PCR analysis of the genomic DNA from the NEOr clones using the OPH or S308LOPH primers (Table 3) generated PCR products of the correct predicted sizes (190 and 137 bp, respectively). This indicated that the recombinant yeast clones harbored the genes encoding OPH and S308L-OPH as expected. To analyze whether the genes were integrated into the yeast genome, and at the predicted chromosomal location, primers were designed such that the forward primer was internal to the gene of interest while the reverse primer was located out with the integration cassette (in the ribosomal DNA). Consequently, PCR products should only be obtained if the gene of interest is integrated into the genome at the correct genomic location. PCR analysis using primers designed to span the 3’ site of integration generated PCR products for OPH and S308L-OPH of the correct predicted sizes (1236 and 1250 bp, respectively). This strongly suggested that both genes were integrated into the S. cerevisiae genome at the correct predicted genomic location. Quantitative real time PCR (qPCR) was used to determine the number of integrated copies of the gene encoding S308LOPH. Using absolute qPCR, the copy number from two independent yeast genomic preparations was 25.16 ± 3.60 and 34.03 ± 7.46 (mean copy number ± SD). Although not empirically determined, a similar copy number of integrants for the wild-type OPH is expected. Of note, growth curves for both wild-type OPH and S308L-OPH recombinant yeast were similar when grown in the presence of 200, 400 and 600 μg/ mL G418. |
| Hydrolytic activity of recombinant yeast lysates against Paraoxon, Demeton-S Methyl and Malathion |
| To investigate whether the recombinant yeast produced functional protein that was capable of hydrolyzing organophosphates, lysates from S308L-OPH recombinant yeast, and from E. coli BL21(DE3) pLysS harboring pS308L-ET30a (bacterial S308L protein expression strain and plasmid) were assessed for the ability to hydrolyze paraoxon (Table 4). Lysates prepared from both yeast and E. coli were able to efficiently hydrolyze paraoxon. Surprisingly however, yeast lysates had specific activities that were 6.7-fold higher than E. coli lysates. This indicates that the yeast lysates harbored functional S308L-OPH protein, and the production of protein (expression signals, solubility) was significantly higher in yeast compared to a bacterial protein over expression system. |
| To investigate whether the yeast paraoxonase activity translated to the hydrolysis of the P-S organophosphate pesticides, lysates prepared from OPH (wild-type) and S308L-OPH recombinant yeast were assessed for the ability to hydrolyze demeton-S methyl and malathion. Lysates from the recombinant yeast were able to hydrolyze demeton-S methyl and malathion (Table 5). The specific activities of lysates against both substrates were relatively similar. As expected, the specific activities of the variant S308L-OPH lysates were significantly higher than wild-type OPH lysates; the fold increase for demeton-S methyl and malathion were 106- and 37-fold, respectively. Collectively, the data indicated that the recombinant yeast expressed functional protein that could hydrolyze P-S organophosphates. |
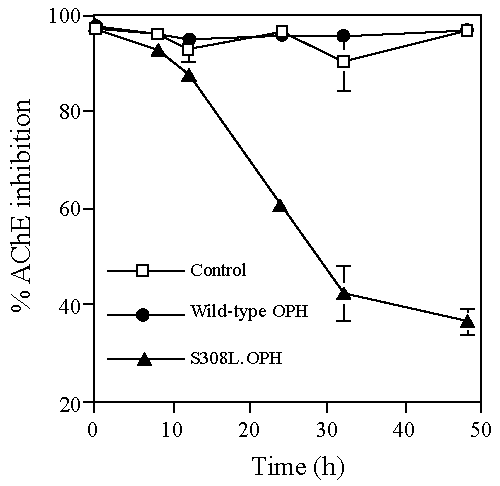
| All organophosphate compounds act by inhibiting the enzyme acetylcholinesterase (AChE). To confirm that the recombinant yeast lysates hydrolyzed demeton-S methyl and thereby detoxified the agent, samples from the demeton-S methyl hydrolysis assays (after 0, 8, 12, 24, 32 and 48 h incubation) were analyzed for a reduction in the ability of the agent to inhibit AChE (Figure 1). AChE inhibition was approximately 95% for lysates prepared from control (no OPH) and wild-type OPH yeast over the time period analyzed (Figure 1). Therefore, these lysates were unable to prevent demeton-S methyl from inhibiting AChE activity. In contrast, yeast lysates prepared from S308L-OPH yeast resulted in a significant reduction in AChE inhibition. Therefore, the results demonstrated the ability of the recombinant yeast S308L-OPH lysates to hydrolyze and thereby detoxify demeton-S methyl. |
| Yeast biocatalytic activity |
| To investigate whether the recombinant yeast could function as a biocatalyst and hydrolyze organophosphate agents directly, the ability of intact yeast cells to hydrolyze demeton-S methyl and malathion was determined (Table 6). The specific activity of recombinant yeast cells harboring the wild-type OPH was not detectable above background controls (wild-type yeast) for demeton-S methyl, but exhibited moderate activity against malathion. In contrast, the recombinant yeast harboring S308L-OPH exhibited good activity against both demeton-S methyl and malathion. In agreement with the lysate data, the S308LOPH yeast displayed a large increase in activity against both substrates compared to yeast harboring the wild-type OPH. |
| To investigate whether the biocatalytic activity of the recombinant yeast against P-S organophosphate pesticides, translated to P-S chemical warfare agents, the ability of the S308L-OPH yeast to hydrolyze VX was determined. Assays were performed in a chemical materiel surety laboratory under the strict regulatory guidelines governing the use and testing of chemical warfare agents. VX P-S bond cleavage was detected using Ellman’s reagent essentially as described previously [12]. The results demonstrated that the yeast biocatalyst was able to hydrolyze VX (Table 6). The specific activity of the biocatalyst against VX was similar, if not greater, than the activity against the pesticides. Therefore, the biocatalyst displayed broad-substrate activity against various P-S bond containing organophosphate pesticides and nerve agents. |
| Substrate concentration dependent activity |
| The efficiency of the biocatalyst is likely to be dependent on the rate of substrate entry into the yeast cell, which in turn may be dependent on the concentration of the agent. Therefore, the specific activity of the S308L-OPH biocatalyst in the presence of different demeton-S methyl concentrations was analyzed (Figure 2A). The yeast S308L-OPH biocatalyst was incubated in the presence of 0.33, 0.66, 1.31, or 2.63 mM demeton-S methyl and the specific activity was determined. As the demeton-S methyl concentration increased, the specific activity increased; the specific activity of the S308L-OPH yeast was approximately 6-fold higher at the highest demeton-S methyl concentration analyzed (2.63 mM) compared to the lowest concentration (0.33 mM). The data therefore indicated that the activity of the biocatalyst was dependent on the substrate concentration. |
| To confirm that the S308L-OPH yeast biocatalyst hydrolyzed demeton-S methyl and thereby detoxified the agent, samples from the demeton-S methyl hydrolysis assays were analyzed for a reduction in the ability of the agent to inhibit AChE. The ability of the yeast biocatalyst to detoxify demeton-S methyl was analyzed using 50 or 250 μM demeton-S methyl (Figure 2B). At both demeton-S methyl concentrations, the recombinant yeast mediated a complete reduction in AChE inhibition. As expected, the time required to alleviate AChE inhibition was longer for the assays containing the higher concentration of demeton-S methyl. In contrast, yeast harboring wildtype OPH did not cause a reduction in AChE inhibition over the time period analyzed. Therefore, the results confirm the ability of the yeast S308L biocatalyst to hydrolyze and thereby detoxify demeton-S methyl. |
| Temperature dependent activity |
| Most biological-based assays are influenced by the assay temperature. Since variation in temperature is likely to affect enzyme activity, and hence whole cell biocatalytic activity, the specific activity of the S308L-OPH yeast to hydrolyze demeton-S methyl was measured at 4, 22, 30 and 37°C (Table 7). The results demonstrated that the specific activity of the OPH expressing yeast cells was temperature dependent; as the temperature increased, the specific activity increased resulting in a 43-fold difference between assays performed at 37°C compared to 4°C. The temperature-dependent specific activity of the yeast cultures directly correlated with the ability to detoxify demeton-S methyl at the varying temperatures. |
| Discussion |
| Organophosphates are extremely toxic compounds which exert their mammalian toxicity by inhibiting AChE activity. The overuse of these compounds has resulted in a number of environmental problems such as contamination of waterways, air, and terrestrial ecosystems. With this overuse, there are legitimate concerns over organophosphate exposure to the public and our natural wildlife. Unintentional pesticide exposure (water, soil, food, handling) is thought to cause 1 to 3 million poisonings annually worldwide and 200,000 deaths [2-4]. This number is considered to be an underestimation and does not fully reflect the full extent of the problem. Therefore, there is a need for newer bioremediation strategies that can safely dispose of these toxic manmade compounds. |
| Recombinant yeast cells harboring integrated copies of the gene encoding the variant S308L-OPH were able to hydrolyze and detoxify the organophosphate pesticides malathion and demeton-S methyl, and the chemical warfare agent VX. Previous studies have shown that the variant S308L-OPH enzyme exhibits an increased hydrolytic efficiency to hydrolyze P-S organophosphates compared to the wild-type OPH enzyme; this ability was due to improvements in the enzymes catalytic rate (kcat) and Michaelis constant (Km) [6]. Improved enzyme characteristics are necessary against these substrates since the hydrolytic efficiency is generally very low in comparison to other organophosphate substrates. Moreover, nerve agents such as VX are particularly problematic since they are stable, not very volatile, and are likely to persist in the environment following an accidental or deliberate release [22]. Our results demonstrated that the recombinant S308LOPH yeast biocatalyst displayed significantly increased hydrolytic capability compared to recombinant yeast harboring the wild-type OPH as expected. |
| The hydrolytic ability of the biocatalyst was dependent on the efficiency of the enzyme hydrolyzing the organophosphate agent, and also the rate of substrate entry into the cell. Bacterial biocatalysts heterologously expressing the gene encoding OPH have been tested for their ability to hydrolyze organophosphates [23]. Although effective, the cellular location of the enzyme has been shown to influence reaction rates. For example, OPH expressed on the cell surface hydrolyzes organophosphates more effectively than whole cell biocatalysts where OPH resides within the cytoplasm [24,25]. This is most likely due to the fact that the bacterial cell envelope acts as a permeability barrier to molecules [26-28]. Permeabolizing the outer membrane using solvents, freeze/thaw methods, or using outer membrane permeable mutants can overcome these issues and increase the rate of passive diffusion and hydrolysis [26,27]. The mechanism of organophosphate entry into the yeast cell is unknown, but is most likely by passive diffusion. The rate of passive diffusion into the cell may be specific to the particular agent, for example, the rate of passive diffusion may be size, structure, and charge dependent. The specific activity of the biocatalyst was concentration dependent, with activity increasing as the concentration of the agent increased. This is most likely due to increased entry of the substrate into the cell since the substrate will flow down the concentration gradient in an attempt to achieve a constant equilibrium inside the cell, and in the external milieu. Therefore, the biocatalyst is likely to function optimally at high substrate concentrations, but display reduced hydrolytic efficiency at low concentrations. To overcome this possible limitation, the enzyme may be fused to a glycosylphosphatidylinositol anchor and expressed at the yeast cell surface [29], and thereby bypass substrate diffusion into the cell. Alternatively, mutations in ergosterol biosynthesis, which have been shown to change membrane fluidity and increase permeability [30-32], may be introduced into the yeast biocatalyst to increase passive diffusion rates. |
| In conclusion, strategies are needed for the safe and environmentally-friendly removal of toxic organophosphates in the environment or the decontamination of surplus chemical agent material. Naturally occurring enzymes and recombinant biocatalysts that are capable of efficiently hydrolyzing and detoxifying these compounds offer the opportunity for viable alternatives to harsher chemical decontamination methods. |
| Acknowledgements |
| Sequencing data was provided by the Medical University of South Carolina Biotechnology Resource Facility. VX hydrolysis assays were performed by Joseph Brewer and James Scott at the Southwest Research Institute in San Antonio. This work was supported by the Defense Advanced Research Project Agency (DARPA contract W31P4Q-06-C-0474, Distribution Statement A) and the Defense Threat Reduction Agency (contract # HDTRA1-10-C-0035) awarded to D.A.S of Guild Associates, Inc. The views, opinions, and/or findings contained in this article are those of the authors and should not be interpreted as representing the official views or policies, either expressed or implied, of the Defense Advanced Research Projects Agency or the Department of Defense. |
| References |
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 | Table 4 |
| Table 5 | Table 6 | Table 7 |
Figures at a glance
 |
 |
| Figure 1 | Figure 2 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 15330
- [From(publication date):
March-2013 - Dec 16, 2025] - Breakdown by view type
- HTML page views : 10495
- PDF downloads : 4835
