Research Article Open Access
ECOMICS:Ecosystem Trans-OMICS Tools and Methods for Complex Environmental Samples and Datasets
Jun Kikuchi1,2,3,4*, Yoshiyuki Ogata1 and Kazuo Shinozaki1,21RIKEN Plant Science Center, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa 230-0045, Japan
2Biomass Engineering Research Program, RIKEN Research Cluster for Innovation, 2-1, Hirosawa, Wako, 351-0198, Japan
3Graduate School of Nanobioscience, Yokohama City University, 1-7-29 Suehirocho, Tsurumi-ku, Yokohama, Kanagawa 230-0045, Japan
4Graduate School of Bioagricultural Sciences, Nagoya University, 1 Furo-cho, Chikusa-ku, Nagoya, Aichi 464-0810, Japan
- *Corresponding Author:
- Jun Kikuchi
RIKEN Plant Science Center
1-7-22 Suehiro-cho
Tsurumi-ku, Yokohama
Kanagawa 230-0045, Japan
E-mail:kikuchi@psc.riken.jp
Received date: August 02, 2010; Accepted date: August 22, 2011; Published date: August 25, 2011
Citation: Kikuchi J, Ogata Y, Shinozaki K (2011) ECOMICS: Ecosystem Trans-OMICS Tools and Methods for Complex Environmental Samples and Datasets. J Ecosys Ecograph S2:001. doi:10.4172/2157-7625.S2-001
Copyright: © 2011 Kikuchi J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and and source are credited.
Visit for more related articles at Journal of Ecosystem & Ecography
Abstract
Despite the apparent detachment of modern industrial societies from natural ecosystems, such systems continue to provide enormous and essential benefits in the form of ecosystem services, including potable water, clean air, and forest products and fisheries. Further, engineered ecosystems, but ecosystems nevertheless, provide the majority of agricultural and additional forestry products. Relationships between ecosystem services and human needs can be identified by connecting genomic information of component organisms with enzyme functions, metabolic pathways and resulting product chemicals such as biomass. Tools that advance the understanding of ecosystem function from the perspective of both environmental and metabolic systems are an important aspect of the emerging systems approach to biological science. We introduce here a web service designated “ECOMICS” to provide an omics approach to clarify such relationships. ECOMICS comprises the E-class web tool for taxonomic (metagenomic) classification based on prokaryotic and eukaryotic ribosomal sequences and selected functional (enzymatic) classification based on sequential domains, FT2DB for the digitization of NMR spectra for chemicals from metabolic to macromolecular phenotyping, Bm-Char for the chemical (macromolecular biomass) assignment of lignocellulose components, and HetMap for identifying and viewing correlations between heterogeneous trans-omics data sets that are produced by such web tools. This website is open to the public domain: https://database.riken.jp/ecomics/.
Keywords
Ecosystem; Database; Environmental samples; NMR; DNA sequences
Conversion from an underground resource-dependent to resource-recycling humanosphere
A majority of the rapid industrial development in the previous century has been dependent upon intensive exploitation of underground resources [1]. To avoid future resource depletion at a time when many developing countries are exploiting available resources, we need to rather consider the conversion from "oil-refinery" to "bio-refinery" as an innovative approach for sustainable industrial production based on primary industries [2]. Biorefinery aims to extricate us from a society dependent upon underground resources, such as ore and crude oil, to develop successful skills to recycle materials from a variety of organisms, and to utilize such skills on a grand scale. It has been studied in many countries, especially in the United States, where biorefinery factory complexes have been constructed. In a biorefinery, i) photosynthetic organisms, such as trees, grasses, and algae produce biomass by carbon metabolism through photosynthesis; ii) carbon dioxide, released from the soil since the industrial revolution, is fixed as biomass; and iii) biochar residue, after utilization of the biomass, is returned to the soil to achieve carbon-negative balance and exchange carbon dioxide, which may cause the greenhouse effect, into a usable renewed resource [3].
Photosynthetic organisms are effective resource crops in terms of untapped biomass utilization. For the utilization, it is critically important to understand a methodology to cultivate and make use of resource crops in production processes. In monocultural transformation, such as perennial grasslands transforming into cornfields, and tropical rainforest into oil palm plantations, although the transformation can be achieved in several years to decades, carbon originating from the soil is continuously released for a tremendously longer timeframe, e.g., over 840 years, as estimated by Fargiome et al. [4]. In biomass utilization, Campbell et al. [5] mentioned that the biogeneration of electricity by biomass combustion is more effective than bioethanol production by saccharification of lignocellulose. However, previous reports that estimated the influence of aerosol released by biomass combustion on cloud core production and global cooling [6] must also be considered, and also economic effects, including health costs [7].
Biomass recycling is achieved by industrial and metabolic processes. Through industrial processes, a variety of biochar components, whose chemical structure and composition depend on the burning temperature, are produced as residue [8]. In particular, well-selected biochar components may lead to the promotion of plant growth. Such application of biochar has allowed us to enter an era of advanced technology for analyzing complex organisms and chemical compounds, such as those found in soil and aerosol [9]. In grasslands, perennial weeds as resource crops are grazed by herbivores, and the excretory products are biodegraded by insects and microorganisms in the soil. Such metabolic circulating systems have existed in nature since prehistoric times. If we apply such effective and functional information to industrial and metabolic processes related to resource crops, and if we regulate the processes efficiently, then we will succeed at creating a sustainable humanosphere.
Information resources as a blueprint for biomass utilization
The number of different species of organisms is over 100,000, even by underestimation, and it is estimated that we are only fully aware of approximately 1% of these. Genomes in such diverse organisms include the blueprints to acquire nutrients in various environments and survive using their metabolic systems. The blueprint, i.e., genomic DNA, contains the information required for synthesizing amino acids and then incorporating them into proteins. By specifying correct functional groups, such as amino acid side chains at different sites in a protein structure, a variety of molecular functions, which are carried out by the proteins and necessary for survival, is expressed.
Information science has had marked influences in protein science. Along with structural genomics [10-12], nuclear magnetic resonance (NMR) technology [13], and instrument development in the field [14], global application of technological innovations in DNA sequencers has extended the grasp of our analysis into entirely new areas such as transcriptomics and metagenomics, and the present sophistication of retrieval techniques from mass data resources have had a tremendous impact in the area. In recent years, much of protein science tends to focus on interaction and binding affinity of candidate medicines with receptor proteins, in studies of drug design, and these types of experiments are crucial in applications discovery. However, that constitutes only a small component of the total biological information resources at our disposal.
To give an example of a much more encompassing framework for visualizing this knowledge, we have focused on the metabolome and biomass. The metabolome is based on the proposition that the adjustment of balance between metabolites contributes to homeostasis [15,16]. Biomass is based on the physical principle that the conversion of energy into polysaccharides, lignins, lipids, etc. leads to the production of complex structures. However, much of the chemistry that would be necessary for effective utilization of unused biomass has not yet been described. Although woody biomass has been actively utilized because of its reasonable strength and refractoriness, exploitation has led to a decrease in soil fertility, deforestation, and groundwater supply. To avoid depletion of such resources, the improvement of monitoring and analysis technologies that are applicable to complex systems including plants, insects, and microorganisms in the soil is required. For such technologies, integrated applications and quantitative information retrieval (profiling) based on bioinformatics, stable isotopic labeling [17-19], NMR [20-24], and giga-sequencers are effective approaches that we now have at hand.
Profiling the forms and chemical composition of material resources
If we assume that strings of nucleotides (building blocks of genetic information) are primary biopolymers, and proteins (which carry out molecular mechanisms based on the translation of genetic information) are secondary biopolymers, then by an extension of this logic, biomass is composed additionally of polysaccharides as tertiary biopolymers (i.e., the product of proteins), and lignin as quaternary biopolymers (higher order biopolymers). Biomass has low solubility and degradability and thus has limited development of molecular biological protocols for biomass research, when compared with what has been attained for proteins and nucleotides. To measure solid samples, scanning electron microscope is used for analysis of surface structure, and solid-state NMR and Fourier transformation infrared spectrometry are used to gain information about internal chemical structure. For example, to utilize trees and grasses in biorefinery processes, it is important to optimize mechanical or ionic liquid-pretreated decay of their crystalline structure. To evaluate such decay, chemical shift values of C4 or C1 sites in cellulose, which may be measured by solid-state NMR, have been utilized as a marker of structural change.
Although physical properties that are characteristic of biomass are based on its macromolecular structure, higher resolution information acquired on its chemical composition leads to association of its physical properties with coordinates on a higher-order genetic blueprint. For example, genetic modification (GM) of the syringaldehyde:vanillin ratio, or compositional unit, of biomass has been attempted, especially in the United States, in order to increase its degradability, and there are extensive projects on strategic GM of biomass grasses and phenotypic profiling of their cell walls that have been described [25-29]. For such phenotypic profiling, it was shown that imidazole is effective at breaking hydrogen bonds in cellulose. However, it is still difficult to demarcate the chemical signals of hemicellulose, which shows broader chemical signals, and therefore, higher resolution will be necessary.
A similar strategy has been applied to the analysis of biochar and humic acids, which are utilized to return biorefinery residues to farmlands. However, chemical information on these macromolecules is insufficient to promote the utilization of unused biomass, and thus, technical developments for measurement, and informational developments based on NGS output, are required.
In analyses of small molecules, technical developments in metabolic profiling are continuously advancing, in areas such as mass spectrometry and NMR data processing. These developments have been reviewed by Kikuchi and Hirayama [30]. In this paper, we will introduce approaches we have considered to elicit information from extraction residue and unassigned chemical signals. NMR can be utilized for evaluating intact living tissue, low-solubility residue, as well as soluble extracts. Thus, we have developed a novel protocol of metabolite annotation to more effectively utilize plant resources. For this purpose, a systematic methodology to evaluate components of extracts and their residues and to detect changes in the components was required. We established this methodology by binning and matrix operation of two-dimensional (2-D) NMR data using 13C-labeled plants [31].
Information on chemical structure is also as remarkable as the material resources themselves. Therefore, we devised a quantitative index named "p-value" to evaluate metabolic profiling data, similar to the E-value of the BLAST program, which is the prevalent homology search engine, and we developed the SpinAssign program, which adopts a method using p-value for metabolite profiling [32]. By applying the program to 13C-labeled plants, we were able simultaneously to acquire 221 metabolite candidates. This methodology is a useful approach for databases of reference metabolite chemical shift, and it is available at the SpinAssign website [33] on the PRIMe server [34].
Technical development to retrieve information on genome resources using next-generation DNA sequencers (NGSs)
Recent advances in sequencing techniques to decode genomic and transcribed sequences have enabled us to acquire a vast amount of sequence information. The acquisition of genomic information leads to the regulation of biomass synthesis and degradation in order to utilize unused biomass and develop strategies for GM. Several equipment makers have developed NGSs, such as the Genome Sequence FLX system of Roche (http://www.454.com/), the 5500xl SOLiD system of Applied Biosystems (http://www.appliedbiosystems.com/), and the HiSeq 2000 system of Illumina (http://www.illumina.com/). Although the length of readable sequence obtained from such NGSs is still shorter than routinely obtained by Sanger's method, the amount of available sequence information is rapidly increasing. A transition from decoding genomes of model organisms to decoding genomes of various organisms, at the level of individual variation, may be observed in research in the field. Since 2005, NGS data for genome decoding have been continuously and increasingly published [35-50], and this trend will continue and intensify.
Sequencing using NGSs is also applicable to transcriptomics [51- 58], which evaluates quantitative gene expression, to metagenomics [59-63], which obtains genomic sequences of whole microorganisms in a specific environment, as well as to genomic sequencing. Especially, gene expression analysis of practical organisms whose genomes are not yet decoded and metagenomic analysis of microorganism flora will provide the genetic information needed for utilization of unused biomass. In general, the biological phenomena involved in biomass synthesis and degradation include various elements, such as elements in the metagenome, metatranscriptome, meta-metabolome, etc. [64- 69]. To reveal and understand such phenomena, a nonlinear approach, such as network analysis, will be useful.
EcoNOmics without the "No": an omics approach for social planning around sustainable bioresource utilization, with less regard for economic activity
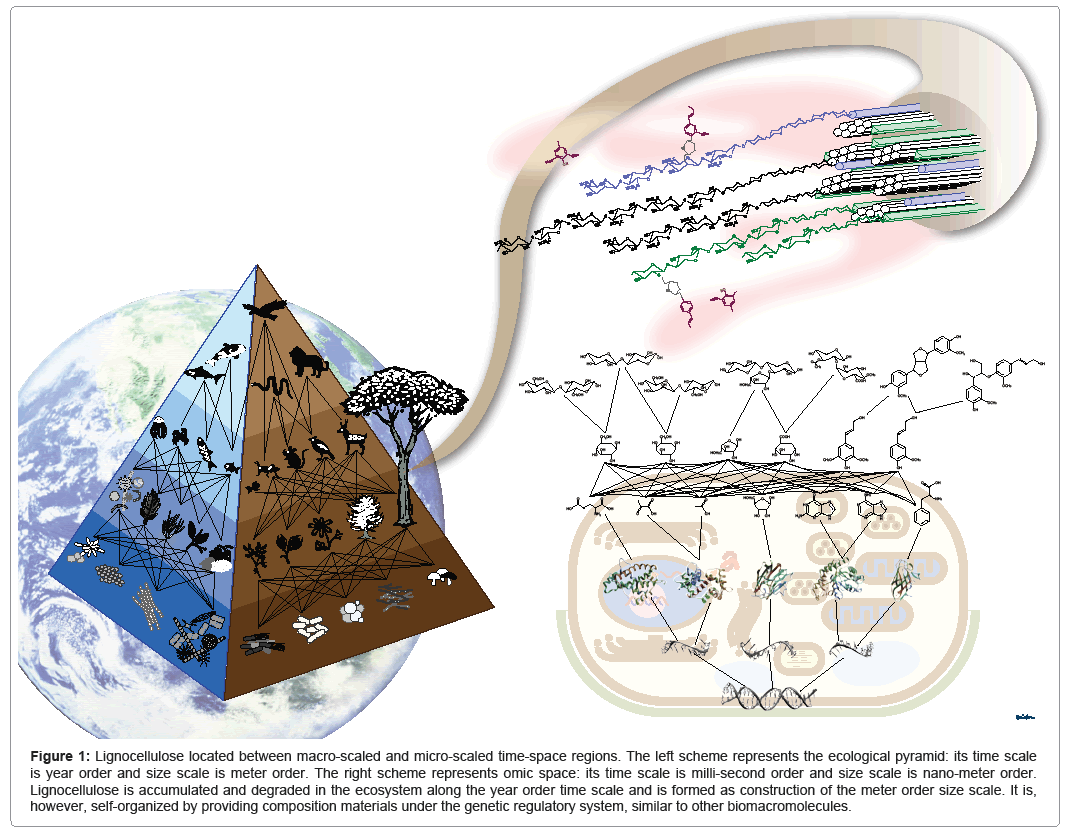
As mentioned in the previous sections, rapid development of information science is revolutionizing the life sciences. However, omics research in model organisms, such as yeast and rice, represents the tendency of "linear" analyses, in which each omics output is collected and analyzed without integrating with other omics information. Recent advances in sequencing technology has promoted the shift of decoding genomic sequences of model organisms to profiling genomic information included in environmental and ecological samples, i.e., metagenomics [70]. Figure 1 represents a hierarchical pyramid of various organisms including plants, animals, and microorganisms. Organisms interact not on the basis of "linear" but rather "nonlinear" relationships, as in a network or web, through a food chain and symbiotic relationships. Similarly, there is a hierarchy of molecules in a cell; it start with genomic DNA, followed by RNA, protein, metabolites, and, finally, biomass supramolecules, such as lignocellulose. Organisms are also related to each other through their metabolic systems.
Figure 1: Lignocellulose located between macro-scaled and micro-scaled time-space regions. The left scheme represents the ecological pyramid: its time scale is year order and size scale is meter order. The right scheme represents omic space: its time scale is milli-second order and size scale is nano-meter order. Lignocellulose is accumulated and degraded in the ecosystem along the year order time scale and is formed as construction of the meter order size scale. It is, however, self-organized by providing composition materials under the genetic regulatory system, similar to other biomacromolecules.
The era of "planar" or integrative approaches is dawning in omics analysis. For example, the RIKEN Omics Research Platform, including NMR and Life Science Accelerator, is applicable to assembling information on lives existing in the global environment such as "population-omics" to extensively collect polymorphism of sequences and phenotypes, and "eco-omics" to systematically collect information on interactions between environmentally-related organisms. Research based on techniques to utilize biomass require high-throughput analyses of variability of non-model plants and of interactions between microorganisms, and, thus, datasets obtained from such high-throughput analyses are accumulating at an increasing rate. A database to provide such integrated information will be a model for next-generation information resources.
The ECOMICS web suite to perform trans-omics analysis of environmental samples
To apply an eco-omics approach to environmental samples, we developed the ECOMICS web services as a source of information and tools useful for trans-omics approaches in ecosystem and biomass research. ECOMICS is composed of i) the E-class web tool for taxonomical and functional classification of nucleotide and peptide sequence data (e.g., metagenomic datasets obtained from an NGS); ii) the FT2DB tool to digitize NMR spectra for downstream chemical phenotyping in quantitative analysis of chemical signals; iii) the Bm-Char webtool to assign biomass-related components (especially components of lignocelluloses, such as hemicellulose and lignin); and iv) the HetMap tool to integrate and view associations between heterogeneous trans-omics datasets (such as metagenomic or enzymatic data obtained from E-class, chemical signal data from FT2DB, and lignocellulosic data from Bm-Char).
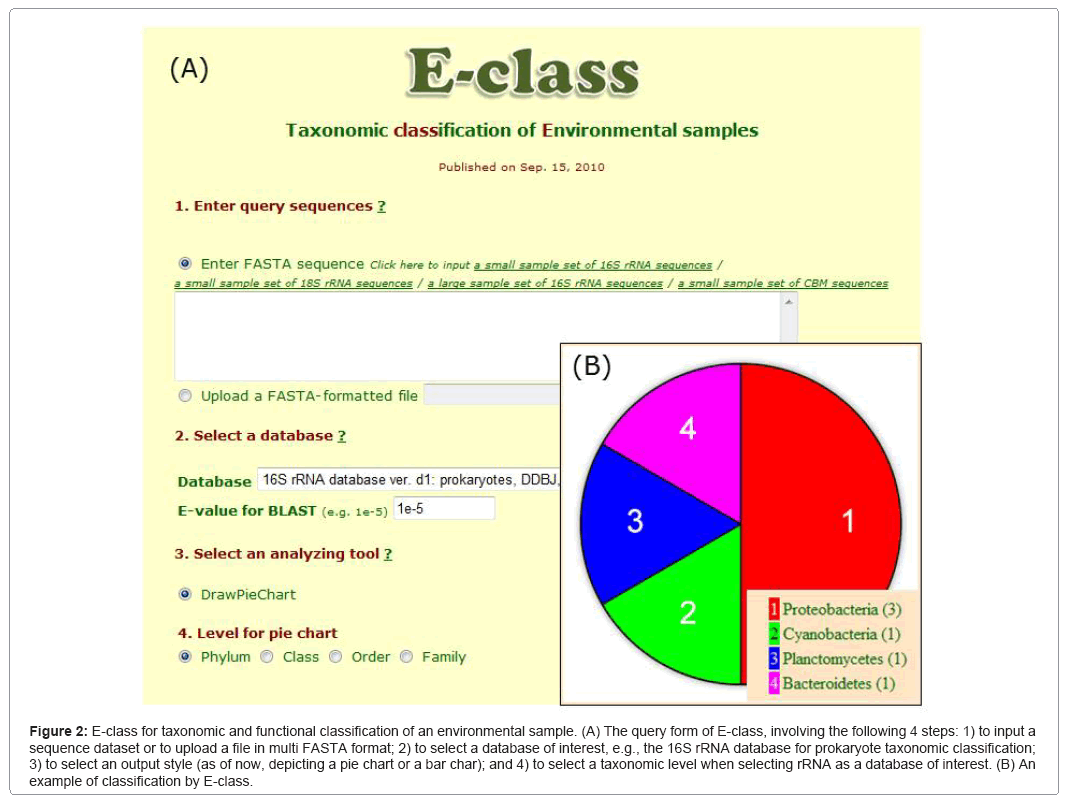
E-class (Figure 2) implements a BLAST search for a large number of sequences (e.g., a metagenomic dataset) in useful sequence databases, using the search for taxonomic and protein domain classification (i.e., small subunit ribosomal RNA or ssrRNA) of both prokaryotes and eukaryotes and enzymatic domains of carbohydrate-binding modules (CBM) and cytochrome P450 (CYP). The categorization of ssrRNA, CBM, and CYP are based on public websites, i.e., SILVA [71], Cazy [72], and Nelson [73], respectively. When a user submits a sequence dataset in multi-FASTA format, E-class provides a pie chart that represents the categorized distribution of query sequences, the BLAST output data, and the pie chart legend with the numbers of categorized sequences. We are planning to add databases for various protein functional analyses.
Figure 2: E-class for taxonomic and functional classification of an environmental sample. (A) The query form of E-class, involving the following 4 steps: 1) to input a sequence dataset or to upload a file in multi FASTA format; 2) to select a database of interest, e.g., the 16S rRNA database for prokaryote taxonomic classification; 3) to select an output style (as of now, depicting a pie chart or a bar char); and 4) to select a taxonomic level when selecting rRNA as a database of interest. (B) An example of classification by E-class.
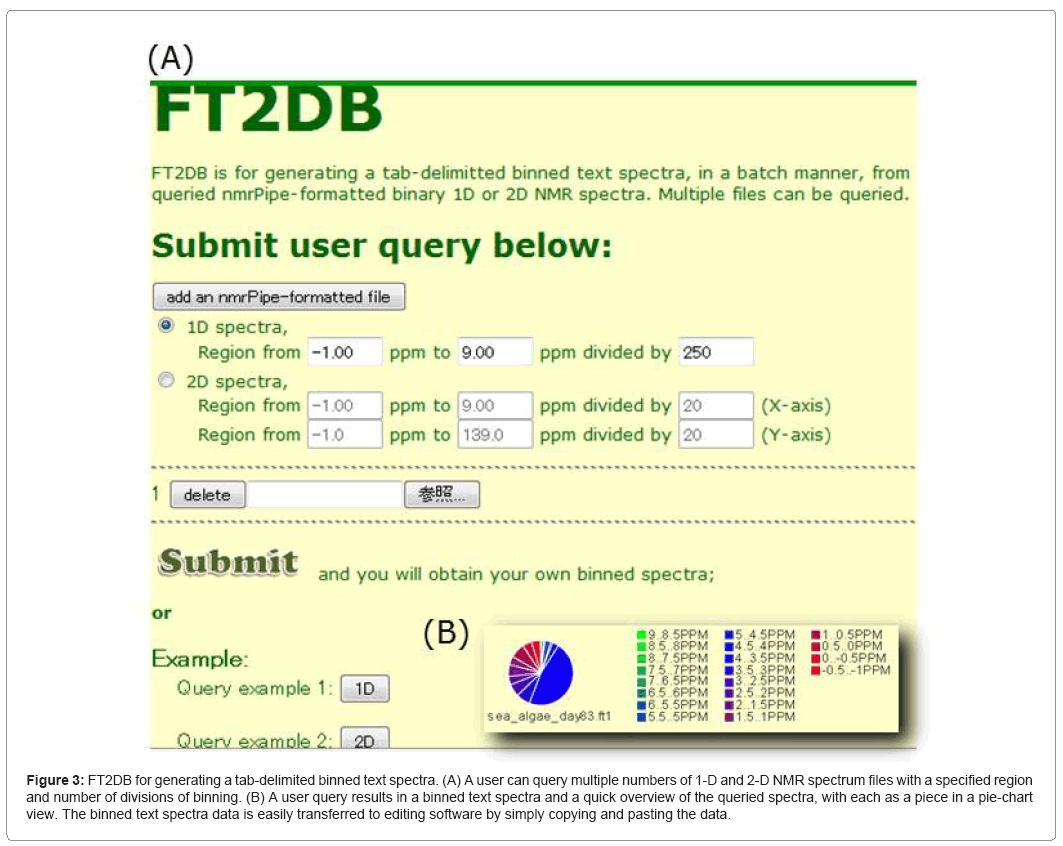
FT2DB (Figure 3) provides a web-based service and a downloadable suite of programs to produce a digitized data format of NMR spectra. FT2DB accepts "nmrPipe"-formatted data of 1- and 2-D NMR spectra 81 in the ECOMICS MS. It results in binning data of an NMR spectrum, which allows a user to specify a region of chemical shift of interest. In the web service, it provides a pie chart representation of the overall distribution of chemical shift regions, which alleviates the need for a comparison of different NMR spectra. Output data from FT2DB may serve as a query data set for HetMap.
Figure 3: FT2DB for generating a tab-delimited binned text spectra. (A) A user can query multiple numbers of 1-D and 2-D NMR spectrum files with a specified region and number of divisions of binning. (B) A user query results in a binned text spectra and a quick overview of the queried spectra, with each as a piece in a pie-chart view. The binned text spectra data is easily transferred to editing software by simply copying and pasting the data.
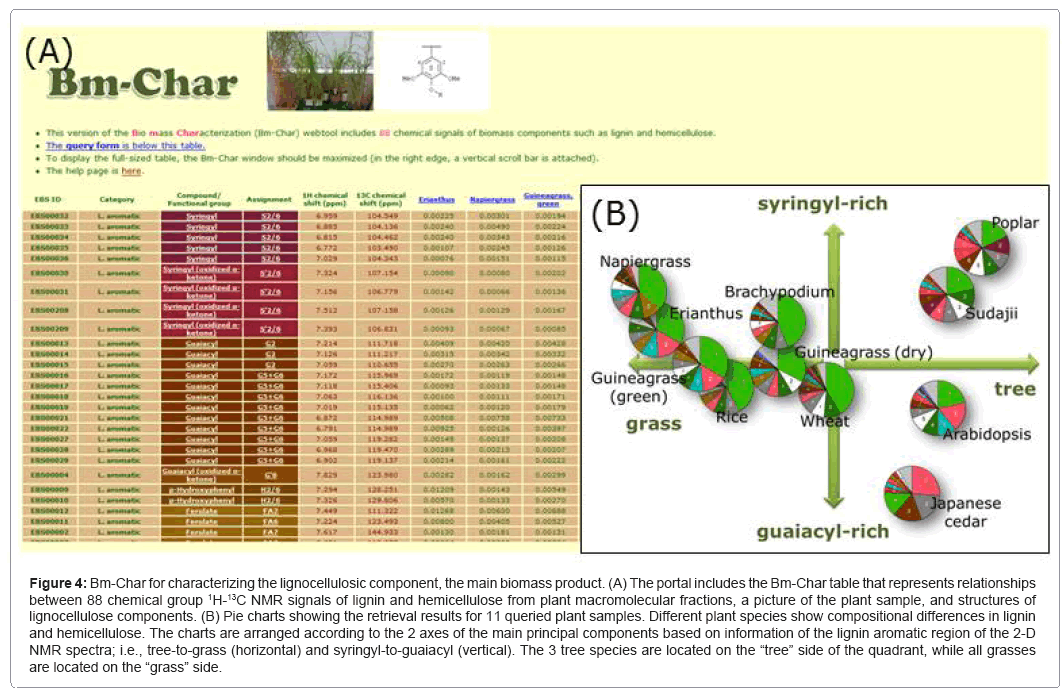
Bm-Char (Figure 4) is able to assign query chemical shifts to 88 known chemical signals of lignin and hemicellulose, including signals of 42 aromatic and 17 aliphatic sites in lignin, 26 hemicellulosic sites, and 3 uncategorized sites, as previously reported [74]. Results are represented as a pie chart of categorized chemical signals, according to the assignment of known signals, with the numbers of the categorized signals included in the legend. As our assignment of chemical shifts related to lignocellulose improves, we plan to adopt the newly assigned signals into Bm-Char.
Figure 4: Bm-Char for characterizing the lignocellulosic component, the main biomass product. (A) The portal includes the Bm-Char table that represents relationships between 88 chemical group 1H-13C NMR signals of lignin and hemicellulose from plant macromolecular fractions, a picture of the plant sample, and structures of lignocellulose components. (B) Pie charts showing the retrieval results for 11 queried plant samples. Different plant species show compositional differences in lignin and hemicellulose. The charts are arranged according to the 2 axes of the main principal components based on information of the lignin aromatic region of the 2-D NMR spectra; i.e., tree-to-grass (horizontal) and syringyl-to-guaiacyl (vertical). The 3 tree species are located on the “tree” side of the quadrant, while all grasses are located on the “grass” side.
HetMap is an integrated service that calculates a correlation matrix of heterogeneous datasets, including the common multiple variables (such as datasets that originate from a common environmental sample), that are obtained through the E-class, FT2DB, and Bm-Char analyses. It depicts a correlation matrix based on query datasets as a heat map, in which each cell represents a correlation of a pair of elements. The colors in the heat map correspond to the correlation coefficient (i.e., red and blue cells represent positive and negative correlation, respectively). HetMap enables the user to color cells according to statistically significant correlation coefficients.
A suite of the ECOMICS web services is applicable to datasets acquired from an environmental sample, in which various organisms, proteins, metabolites, and biomass coexist and interrelate in terms of particular processes, many of which are still unknown or only poorly described. We believe that these tools will provide a useful approach to understand environmental processes at a higher level. All software and web services are freely available at the ECOMICS web server (https://database.riken.jp/ecomics/).
Acknowledgements
This research was supported in part by Grants-in-Aid for Scientific Research for challenging exploratory research (J.K.) and Scientific Research (A) (J.K.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. This work was also supported, in part, by grants from Research and Development Program for New Bio-industry Initiatives of the Bio-oriented Technology Research Advancement Institution (BRAIN) (J.K.), and the New Energy and Industrial Technology Development Organization (NEDO) (J.K.).
References
- Lopes Ferreira N (2008) Industrial exploitation of renewable resources: from ethanol production to bioproducts development. J Soc Biol 202: 191-199.
- Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, et al. (2006) The path forward for biofuels and biomaterials. Science 311: 484-489.
- Lehmann J (2007) A handful of carbon. Nature 447: 143-144.
- Fargione J, Hill J, Tilman D, Polasky S, Hawthorne P (2008) Land clearing and the biofuel carbon debt. Science 319: 1235-1238.
- Campbell JE, Lobell DB, Field CB (2009) Greater transportation energy and GHG offsets from bioelectricity than ethanol. Science 324: 1055-1057.
- Normile D (2009) Round and round: a guide to the carbon cycle. Science 325: 1642-1643.
- Hill J, Polasky S, Nelson E, Tilman D, Huo H, et al. (2009) Climate change and health costs of air emissions from biofuels and gasoline. Proc Natl Acad Sci U S A 106: 2077-2082.
- Keiluweit M, Nico PS, Johnson MG, Kleber M (2010) Dynamic molecular structure of plant biomass-derived black carbon (biochar). Environ Sci Technol 44: 1247-1253.
- Sannigrahi P, Sullivan AP, Weber RJ, Ingall ED (2006) Characterization of water-soluble organic carbon in urban atmospheric aerosols using solid-state 13C NMR spectroscopy. Environ Sci Technol 40: 666-672.
- Kikuchi J, Iwahara J, Kigawa T, Murakami Y, Okazaki T, et al. (2002) Solution structure determination of the two DNA-binding domains in the Schizosaccharomyces pombe Abp1 protein by a combination of dipolar coupling and diffusion anisotropy restraints. J Biomol NMR 22: 333-347.
- Fukushima K, Kikuchi J, Koshiba S, Kigawa T, Kuroda Y, et al. (2002) Solution structure of the DFF-C domain of DFF45/ICAD. A structural basis for the regulation of apoptotic DNA fragmentation. J Mol Biol 321: 317-327.
- Sakata E, Yamaguchi Y, Kurimoto E, Kikuchi J, Yokoyama S, et al. (2003) Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO Rep 4: 301-306.
- Horiuchi T, Takahashi M, Kikuchi J, Yokoyama S, Maeda H (2005) Effect of dielectric properties of solvents on the quality factor for a beyond 900 MHz cryogenic probe model. J Magn Reson 174: 34-42.
- Kiyoshi T, Maeda H, Kikuchi J, Ito Y, Hirota H, et al. (2004) Present status of 920 MHz high-resolution NMR spectrometers. Ieee Transactions on Applied Superconductivity 14: 1608-1612.
- Okamoto M, Tsuboi Y, Chikayama E, Kikuchi J, Hirayama T (2009) Metabolic movement upon abscisic acid and salicylic acid combined treatments. Plant Biotechnology 26: 551-560.
- Mochida K, Furuta T, Ebana K, Shinozaki K, Kikuchi J (2009) Correlation exploration of metabolic and genomic diversities in rice. BMC Genomics 10: 568.
- Kikuchi J, Shinozaki K, Hirayama T (2004) Stable isotope labeling of Arabidopsis thaliana for an NMR-based metabolomics approach. Plant Cell Physiol 45: 1099-1104.
- Ohyama K, Suzuki M, Kikuchi J, Saito K, Muranaka T (2009) Dual biosynthetic pathways to phytosterol via cycloartenol and lanosterol in Arabidopsis. Proc Natl Acad Sci U S A 106: 725-730.
- Date Y, Nakanishi Y, Fukuda S, Kato T, Tsuneda S, et al. (2010) New monitoring approach for metabolic dynamics in microbial ecosystems using stable-isotope-labeling technologies. J Biosci Bioeng 110: 87-93.
- Fukuda S, Nakanishi Y, Chikayama E, Ohno H, Hino T, et al. (2009) Evaluation and characterization of bacterial metabolic dynamics with a novel profiling technique, real-time metabolotyping. PLoS One 4: e4893.
- Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, et al. (2011) Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469: 543-547.
- Nakanishi Y, Fukuda S, Chikayama E, Kimura Y, Ohno H, et al. (2011) Dynamic omics approach identifies nutrition-mediated microbial interactions. J Proteome Res 10: 824-836.
- Sekiyama Y, Kikuchi J (2007) Towards dynamic metabolic network measurements by multi-dimensional NMR-based fluxomics. Phytochemistry 68: 2320-2329.
- Tian C, Chikayama E, Tsuboi Y, Kuromori T, Shinozaki K, et al. (2007) Topdown phenomics of Arabidopsis thaliana: metabolic profiling by one- and twodimensional nuclear magnetic resonance spectroscopy and transcriptome analysis of albino mutants. J Biol Chem 282: 18532-18541.
- Cheng G, Varanasi P, Li C, Liu H, Melnichenko YB, et al. (2011) Transition of cellulose crystalline structure and surface morphology of biomass as a function of ionic liquid pretreatment and its relation to enzymatic hydrolysis. Biomacromolecules 12: 933-941.
- Feng D, Li L, Yang F, Tan W, Zhao G, et al. (2011) Separation of ionic liquid [Mmim][DMP] and glucose from enzymatic hydrolysis mixture of cellulose using alumina column chromatography. Appl Microbiol Biotechnol 91: 399-405.
- Gremos S, Zarafeta D, Kekos D, Kolisis F (2011) Direct enzymatic acylation of cellulose pretreated in BMIMCl ionic liquid. Bioresour Technol 102: 1378-1382.
- Ha SH, Mai NL, An G, Koo YM (2011) Microwave-assisted pretreatment of cellulose in ionic liquid for accelerated enzymatic hydrolysis. Bioresour Technol 102: 1214-1219.
- Kilpelainen I, Xie H, King A, Granstrom M, Heikkinen S, et al. (2007) Dissolution of wood in ionic liquids. J Agric Food Chem 55: 9142-9148.
- Kikuchi J, Hirayama T (2007) Practical aspects of uniform stable isotope labeling of higher plants for heteronuclear NMR-based metabolomics. Methods Mol Biol 358: 273-286.
- Sekiyama Y, Chikayama E, Kikuchi J (2010) Profiling polar and semi-polar plant metabolites throughout extraction processes using a combined solutionstate and HR-MAS NMR approach. Anal Chem 82: 1643-1652.
- Chikayama E, Sekiyama Y, Okamoto Y, Nakanishi Y, Tsuboi Y, et al. (2010) Statistical indices for simultaneous large-scale metabolite detections for a single NMR spectrum. Anal Chem 82: 1653-1658.
- Chikayama E, Suto M, Nishihara T, Shinozaki K, Kikuchi J (2008) Systematic NMR analysis of stable isotope labeled metabolite mixtures in plant and animal systems: coarse grained views of metabolic pathways. PloS one 3: e3805.
- Akiyama K, Chikayama E, Yuasa H, Shimada Y, Tohge T, et al. (2008) PRIMe: a Web site that assembles tools for metabolomics and transcriptomics. In Silico Biol 8: 339-345.
- Cannon SB, Sterck L, Rombauts S, Sato S, Cheung F, et al. (2006) Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc Natl Acad Sci U S A 103: 14959-14964.
- Chan AP, Crabtree J, Zhao Q, Lorenzi H, Orvis J, et al. (2010) Draft genome sequence of the oilseed species Ricinus communis. Nat Biotechnol 28: 951- 956.
- Huang S, Li R, Zhang Z, Li L, Gu X, et al. (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41: 1275-1281.
- International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763-768.
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463-467.
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245-250.
- Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, et al. (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452: 991-996.
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551-556.
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64-69.
- Sato S, Hirakawa H, Isobe S, Fukai E, Watanabe A, et al. (2011) Sequence analysis of the genome of an oil-bearing tree, Jatropha curcas L. DNA Res 18: 65-76.
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, et al. (2008) Genome structure of the legume, Lotus japonicus. DNA Res 15: 227-239.
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178-183.
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112-1115.
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, et al. (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43: 109-116.
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596-1604.
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, et al. (2010) The genome of the domesticated apple (Malus x domestica Borkh.). Nat Genet 42: 833-839.
- Bolon YT, Joseph B, Cannon SB, Graham MA, Diers BW, et al. (2010) Complementary genetic and genomic approaches help characterize the linkage group I seed protein QTL in soybean. BMC Plant Biol 10: 41.
- Buggs RJ, Chamala S, Wu W, Gao L, May GD, et al. (2010) Characterization of duplicate gene evolution in the recent natural allopolyploid Tragopogon miscellus by next-generation sequencing and Sequenom iPLEX MassARRAY genotyping. Mol Ecol 19 Suppl 1: 132-146.
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, et al. (2010) Genomewide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45-58.
- Legrand S, Valot N, Nicole F, Moja S, Baudino S, et al. (2010) One-step identification of conserved miRNAs, their targets, potential transcription factors and effector genes of complete secondary metabolism pathways after 454 pyrosequencing of calyx cDNAs from the Labiate Salvia sclarea L. Gene 450: 55-62.
- Libault M, Farmer A, Brechenmacher L, Drnevich J, Langley RJ, et al. (2010) Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol 152: 541-552.
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, et al. (2010) An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J 63: 86-99.
- Picardi E, Horner DS, Chiara M, Schiavon R, Valle G, et al. (2010) Largescale detection and analysis of RNA editing in grape mtDNA by RNA deepsequencing. Nucleic Acids Res 38: 4755-4767.
- Sun C, Li Y, Wu Q, Luo H, Sun Y, et al. (2010) De novo sequencing and analysis of the American ginseng root transcriptome using a GS FLX Titanium platform to discover putative genes involved in ginsenoside biosynthesis. BMC genomics 11: 262.
- Barriuso J, Marin S, Mellado RP (2010) Effect of the herbicide glyphosate on glyphosate-tolerant maize rhizobacterial communities: a comparison with preemergency applied herbicide consisting of a combination of acetochlor and terbuthylazine. Environ Microbiol 12: 1021-1030.
- Campbell BJ, Polson SW, Hanson TE, Mack MC, Schuur EA (2010) The effect of nutrient deposition on bacterial communities in Arctic tundra soil. Environ Microbiol 12: 1842-1854.
- Koopman MM, Fuselier DM, Hird S, Carstens BC (2010) The carnivorous pale pitcher plant harbors diverse, distinct, and time-dependent bacterial communities. Appl Environ Microbiol 76: 1851-1860.
- Manter DK, Delgado JA, Holm DG, Stong RA (2010) Pyrosequencing reveals a highly diverse and cultivar-specific bacterial endophyte community in potato roots. Microb Ecol 60: 157-166.
- Teixeira LC, Peixoto RS, Cury JC, Sul WJ, Pellizari VH, et al. (2010) Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. Isme J 4: 989-1001.
- Ogata Y, Sakurai N, Aoki K, Suzuki H, Okazaki K, et al. (2009) KAGIANA: an excel-based tool for retrieving summary information on Arabidopsis genes. Plant cell physiol 50: 173-177.
- Ogata Y, Sakurai N, Suzuki H, Aoki K, Saito K, et al. (2009) The prediction of local modular structures in a co-expression network based on gene expression datasets. Genome Inform 23: 117-127.
- Ogata Y, Suzuki H, Sakurai N, Shibata D (2010) CoP: a database for characterizing co-expressed gene modules with biological information in plants. Bioinformatics 26: 1267-1268.
- Ozaki S, Ogata Y, Suda K, Kurabayashi A, Suzuki T, et al. (2010) Coexpression analysis of tomato genes and experimental verification of coordinated expression of genes found in a functionally enriched coexpression module. DNA Res 17: 105-116.
- Sakurai N, Ara T, Ogata Y, Sano R, Ohno T, et al. (2011) KaPPA-View4: a metabolic pathway database for representation and analysis of correlation networks of gene co-expression and metabolite co-accumulation and omics data. Nucleic Acids Res 39: 677-684.
- Urano K, Maruyama K, Ogata Y, Morishita Y, Takeda M, et al. (2009) Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J 57: 1065-1078.
- Raes J, Foerstner KU, Bork P (2007) Get the most out of your metagenome: computational analysis of environmental sequence data. Curr Opin Microbiol 10: 490-498.
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, et al. (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188-7196.
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, et al. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37: 233-238.
- Nelson DR (2009) The cytochrome p450 homepage. Hum Genomics 4: 59-65.
- Kim H, Ralph J (2010) Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d(6)/pyridine-d(5). Org Biomol Chem 8: 576-591.
Relevant Topics
- Aquatic Ecosystems
- Biodiversity
- Conservation Biology
- Coral Reef Ecology
- Distribution Aggregation
- Ecology and Migration of Animal
- Ecosystem Service
- Ecosystem-Level Measuring
- Endangered Species
- Environmental Tourism
- Forest Biome
- Lake Circulation
- Leaf Morphology
- Marine Conservation
- Marine Ecosystems
- Phytoplankton Abundance
- Population Dyanamics
- Semiarid Ecosystem Soil Properties
- Spatial Distribution
- Species Composition
- Species Rarity
- Sustainability Dynamics
- Sustainable Forest Management
- Tropical Aquaculture
- Tropical Ecosystems
Recommended Journals
Article Tools
Article Usage
- Total views: 15156
- [From(publication date):
March-2013 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 10559
- PDF downloads : 4597