Research Article Open Access
Differential Proteomic and Phospho-proteomic Analysis of Normal versus Failed Spermiation in Adult Rats by Label-Free LC-MS/MS
Rahul D Upadhyay1 , Amit Kumar Yadav2 , Shobha Sonawane1 , Reshma Goankar1 , Debasis Dash2 and BalasinorNH1*
1Department of Neuroendocrinology, National Institute for Research in Reproductive Health, J.M. Street, Parel, Mumbai, India
2CSIR-Institute of Genomics and Integrative Biology, South Campus, Mathura Road, SukhdevVihar, New Delhi, India
- *Corresponding Author:
- Balasinor NH, Ph.D
Scientist ‘E’, Departmrnt of Neuroendocrinology
National Institute for Research in Reproductive Health
J. M. Street, Parel, Mumbai-400012, India
Tel: 91-22-24192025/24192140
Fax: 91-22-24139412
E-mail: balasinorn@nirrh.res.in
Received date: August 13, 2013; Accepted date: September 12, 2013; Published date: September 16, 2013
Citation: Upadhyay RD, Yadav AK, Sonawane S, Goankar R, Dash D (2013) Differential Proteomic and Phospho-proteomic Analysis of Normal versus Failed Spermiation in Adult Rats by Label-Free LC-MS/MS. J Anal Bioanal Tech 4:172. doi: 10.4172/2155-9872.1000172
Copyright: © 2013 Upadhyay RD, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Spermiation is the final step of spermatogenesis and involves release of mature spermatids from the Sertoli cells. It is a complex process involving displacement and removal of spermatid cytoplasm, formation and degradation of tubulobulbar complexes and progressive loss of adhesive junctions, including ectoplasmic specializations and subsequent phagocytosis of residual bodies by the Sertoli cell. Spermiation occurs in stage VIII of the rat seminiferous epithelium cycle. In spite of the important role played by spermiation process in sperm output to the epididymis, very little is known about this process. To enhance our knowledge of the sperm release biology, we sought to understand the molecular events occurring at the time of spermiation. Towards this aim, we induced spermiation failure condition by 17β estradiol treatment to adult male rats and compared with the normal spermiation. On comparing both the groups, we identified a total of 104 differentially expressed proteins and 23 differentially expressed phosphoproteins by LC-MS/MS analysis and quantitation. Localization and expression of some of the validated differential proteins in the testes highlight their importance during the sperm release. The present study represents an initial step to understand the molecular basis of the process of spermiation.
Keywords
Spermiation; Cytoskeletal; Endoplasmic reticulum; Phosphoproteins; Proteomics
Introduction
In mammals, the male gametes, spermatozoa are formed in the seminiferous tubule of the testis. The diploid spermatogonia undergo several mitotic and meiotic divisions, thus forming spermatocytes and lastly the haploid spermatid [1]. The spermatid after cyto-differentiation is released into the lumen of the seminiferous tubule via a process known as spermiation. In the seminiferous tubule, specific association of different types of spermatogenic cells form stages of seminiferous epithelium. In rats, there are 14 stages and spermiation occurs in stages VII-VIII [2].
The process of spermiation is an elaborate process and takes almost 72 hours in the rat [3]. The process involves movement of spermatid towards the lumen, removal of excessive spermatid cytoplasm, dissociation of testis-specific actin based atypical adheren junction known as Ectoplasmic specialization (ES), and formation of another testis-specific actin-based structure known as tubulobulbar complex (TBC). The TBCs are involved in the removal of excess of spermatid cytoplasm, recycling of junctional molecules by endocytosis, and also act as transient attachment devices for the mature spermatid, before it is released into the lumen. Both ES and TBCs are actin-based structures with cisternae of Endoplasmic reticulum (ER) [4].
Newer advances in molecular biology and genomics have improved our knowledge of spermatogenesis by allowing the identification of a large no. of genes essential for the development of functional male gametes [5,6]. However, although proteomics has led to major breakthroughs in this era, the investigation of spermatogenesis by proteomics based strategies remains at an infant stage. Recent reports on proteomics driven studies on testis and its germ cell types suggests regulatory mechanism and heterogeneity underlying the function of testicular proteins [7,8]. All these proteomics studies serve as useful resource for comparison. Nevertheless, unprecedented efforts are still required to improve our understanding of the highly complex and sophisticated communication networks. Many studies in proteomics have been done to understand spermatogenesis and its regulation [9,10], but none of them have studied the process of sperm release, i.e. spermiation. None of the protein identified per se could be directly correlated to the process of spermiation. Also, various phosphorylated proteins and phosphorylation of various adhesion molecules have been shown to be present at the site of spermiation, but not much focus have been given to phosphoproteome profile in sperm release [11-13].
Hence, we initiated a study to identify the molecular players of sperm release. Towards this aim, comparative study of normal and failed spermiation condition was performed. Our earlier study following 17β-estradiol treatment to adult male rats at a dose of 100 μg/kg for 10 days led to spermiation failure [14,15]. Therefore, this treatment model was used for differential proteomics and phospho-proteomics by label free nano-LC-MS/MS sequencing to elucidate the importance of proteins involved during sperm release.
Materials and Methods
Extraction of proteins and phosphoproteins from control and 17β-estradiol treated adult male rats (normal vs failed spermiation)
Animal treatment: Randomly bred Holtzman strain adult male rats (75-90 days old) maintained at a temperature of 22°C-23°C, humidity 50-55% and lighting cycle of 14 h light: 10 h dark were administered 100 μg/kg bodyweight/day of 17-β Estradiol (Sigma, USA) for 10 days. The study was approved by institutional animal ethics committee for the use of animals. All experiments were done at least in triplicate using tissues from different animals.
Separation of stages VII-VIII from seminiferous tubule: 24 hrs after the administration of the last dose, testes were collected and detunicated in 0.01 M PBS. Stages of seminiferous epithelium cycle were separated by a transillumination method [16,17]. Briefly, the testis was teased apart and seminiferous tubules viewed under a stereomicroscope at 10-40 X magnification. The stages follow each other in the seminiferous tubules in a wavelike fashion and specific pattern can be observed for different stages by trans illumination in a freshly isolated rat seminiferous tubule. The method is based on the fact that condensation of the elongating/elongated spermatid nuclear chromatin is associated with increase in light absorption. Stages IX-XI has pale weak absorption with pale zone, whereas Stages VII to VIII, having homogenously dark centered absorption pattern, since the spermatids are luminally arranged in bundles. The dark zone representing Stages VII-VIII were dissected. The separated tubules were transferred to cryovials, snap frozen in liquid nitrogen and stored in -80°C until protein extraction.
Protein and phosphoprotein extraction: Protein extraction from control and treated stage VII-VIII tubules: Protein was extracted using 2D lysis buffer consisting of 50 mM Tris pH 8.5, 4% CHAPS, 9M Urea, protease inhibitor cocktail and Phospho-Stop by incubating at 4°C for at least 1 hr,and centrifuging at 12000 g for 10 min. The extracted protein from both control and estradiol treated animals was estimated by Bradford’s method, and then subjected to LC-MS/MS analysis.
Phosphoproteins enrichment by Immobilized Metal Affinity Chromatography (IMAC): A part of proteins (300 μg) extracted were processed for phosphoproteins enrichment by IMAC method. The protocol involves use of Pro-Q® Diamond Phosphoproteins Enrichment Kit (Molecular Probes, USA), which enables efficient, non-radioactive isolation of phosphoproteins from complex cellular extracts. The enrichment was performed as per the manufacturer’s protocol with no notable modifications.
LC-MS/MS sequencing for differential protein and phosphoprotein analysis
Precipitation/on-pellet digestion procedure: Proteins extracted from control and treated samples were precipitated with Chloroformmethanol precipitation method [18]. The mixture was incubated overnight at -20°C, and then centrifuged at 12,000 g for 20 min at 4°C. The supernatant was then removed and the pellet was allowed to air dry for 3 min. The on-pellet-digestion procedure consisted of 0.05% of cleavable detergent (ProteasMAX™) in the solution to expedite the digestion and leads to the complete digestion. The peptide sample was reduced with 1 mM TCEP at 95°C for 5 min, and then alkylated with 100 mM iodoacetamide at 37°C for 30 min in the dark. A second aliquot of trypsin was added at an enzyme: substrate ratio of 1:25 (w/w). The mixture was incubated at 37°C overnight to achieve complete digestion. The final volume of the digestion mixture was approximately 100 μL. A similar protocol was followed for phosphoproteins samples.
Estimation of on-pellet-digested proteins and phosphoproteins from control and treated animals: A modified BCA method was devised to permit comparison of the peptide recoveries for different sample [18]. All analyses were performed in triplicate.
2 sets of samples were used for the sequencing as indicated below
Set 1: IMAC enriched phosphoproteins (3controls+3 treated)
Set 2: Non-enriched proteins (total proteins) (3controls+3 treated).
Each of these samples were run in duplicate
In-solution digestion protocol: The samples, obtained following on-pellet digestion, were dried using a stream of nitrogen. A trypsin solution (prepared as 40 ng/μl in 10 mM ammonium bicarbonate, containing 10% v/v acetonitrile) was added in the ratio of 1: 30. The tubes were centrifuged at 200 rpm and incubated at 37°C for 4 hrs. 50 μl of the 1 mM DTT solution was dispensed to completely cover the pellet, then vortexed, mixed and incubated at 95°C for 5 min. 50 μL (100 mM) of the iodoacetamide solution was added, vortexed and incubated at 37°C for 30 min in dark. The mixture was then dried in a vacuum concentrator. A ratio of 1:25 of 40 ng/μl trypsin was added and incubated at 37°C overnight. Next day, after a short spin to the tubes, 5 μl of 5% formic acid was added till the pH reached 3. The tubes were mixed well and centrifuged at 10000 rpm for 5 min. The supernatant was then transferred into auto-sampler vials for LC-MS/MS analysis.
Sequencing and analysis by nano LC-MS/MS
LC-MS/MS analysis of isolated peptides and phosphopeptides: 150 minute RP-LC gradient was run for optimum separation of peptides from normal and failed spermiation samples (estradiol treated). A label free quantification strategy was followed by adding peptide standards in a specific range. Prepared samples were spiked with 25 fmol/μl standard tryptic BSA digest. Samples were subjected to Nano-LC-MS/MS.
Isolated phospho-peptide from normal and failed spermiation samples were analyzed on an Agilent 1100 nano-flow system (Agilent Technologies, Palo Alto, CA), LC connected to a LTQ FT mass spectrometer (Thermo Electron, San Jose, CA), equipped with a nanoelectrospray ion source. Phosphorylation screen method was established over Nano-LC and LTQ-Orbitrap-MS. This includes standardization of Data Dependent Neutral Loss MS3 (DDNLMS3) strategy, involving screening of (S,T,Y) phosphorylation. The process of selection of phosphorylation neutral loss for MS3 was standardized for optimum gain. As a result, following m/z were chosen for MS3 neutral loss scanning: 24.5000, 32.6700, and 49.00.
Identification of proteins and phosphoproteins from normal and failed spermiation: Label free peak area based quantification was done for all the samples by using standard BSA-tryptic digest. Area of all the samples are normalized with area of BSA found in respective sample (values taken from separate analysis performed using carbamidomethylation as static modification).
Target-Decoy database search strategy was customized for maximum coverage of proteome and phospho-peptides applying a workflow merging MASCOT and SEQUEST. A decoy database search was included in the Proteome Discoverer 1.3 (PD) enabled. High confident peptides (≤ 1 %FDR), with prerequisite of minimum two peptides, leading into identification of proteins were selected and list was generated.
Spectral counts of the identified proteins were used as semiquantative measure to study the differential regulation of protein in normal and failed spermiation condition. Ratios of spectral count of Treated/control for total proteins and Phospho-treated/Phosphocontrol for phosphoproteins were calculated to study differentials. Ratios of ≥ 2.0 were considered significantly over-expressed and ratios ≤ 0.5 were considered significantly under-expressed.
Validation of differentially expressed proteins and phosphoproteins
Localization of differentially expressed proteins during normal and failed spermiation condition in the testis: Ten micron cryosections were taken on poly-L-lysine coated slides and dried at 37°C for 60 min. The sections were post-fixed in chilled acetone kept at -20°C for 10 min and once again dried at 37°C for 60 min. This was followed by three washes with PBS and blocking with iTTM FX signal enhancer (Molecular Probes) for 30 min. The blocking reagent was drained out and sections were incubated with protein-specific antibody (Calreticulin, T-Complex protein 1, Calnexin, all the antibodies were polyclonal from Abcam, Cambridge, UK) prepared in PBST (0.01 M PBS containing 0.05% Tween 20) overnight at 4°C. The following day, sections were washed in PBS and incubated with appropriate dilutions of secondary antibodies for 60 min at room temperature. 4α-6-Diamidino-2-phenylindole (DAPI) was used as a nuclear stain. The sections were subsequently washed and mounted in Prolong Gold Antifade (Molecular Probes). Co-localization of above primary antibodies with F-actin (F-actin was used as a marker for testis-specific actin-based structures, namely TBC and ES was done using phallodin labeled with BODIPY 558/568 (red); all the other primary antibodies were detected using goat anti-rabbit labeled with Alexa Flour 488. For the negative control, an equivalent concentration of rabbit IgG was used, instead of the primary antibody. Fluorescent images were captured using Carl Zeiss-LSM510-Meta Confocal system (Oberkochen, Germany). Z stacks images were generated.
Western blotting of differentially expressed proteins and phosphoproteins during normal and failed spermiation condition: The extracted proteins were run on 12% SDS-PAGE, followed by 1 hr transfer on nitrocellulose membrane. The membranes were blocked and then kept overnight at 4°C in the respective primary antibodies (Calreticulin, T-Complex protein 1, Calnexin, 14-3-3 beta). Next day, the blots then washed in PBS with 0.1% Tween-20 buffer on a rocking platform for 30 mins. Secondary antibody (goat anti-rabbit, Sigma, USA) was added to the blots and incubated for an hour at room temperature. The blots were then washed in PBS with 0.1% Tween-20 buffer on a rocking platform for an hour, and then developed on an X-ray Film. GAPDH antibody was used as a loading control to determine the expression levels of proteins and phosphoproteins.
Immuno-precipitation to study phosphorylation status of 14-3- 3beta protein: Immuno-precipitation was performed, as described by Upadhyay et al. [17], except for the primary antibody used here was polyclonal 14-3-3 beta (Epitomics, Germany). Normal rabbit serum, instead of primary antibody, was used as negative control. Briefly, 150- 200 μg of protein extract from stages VII–VIII micro-dissected tubules was incubated with 7.5 μg of Pan-phospho antibody (polyclonal) for 1 h at 4°C. As much as 50 μl of washed protein G slurry pre-chilled at 4°C, was added to the immune complex and further incubated for 1 h at 4°C on a rocking platform. The immunoprecipitated complex was then centrifuged at 10,000 g for 30 s at 4°C and the supernatant was discarded. Beads containing bound immuno-complex were then washed thrice with 500 μl of lysis buffer, each time centrifuging for 30 s at 10,000 g. After the last wash, the supernatant was removed and 50 μl of 1×Laemmli buffer was added to the bead pellet. The bead pellet was boiled to 90-100°C for 5 min and centrifuged at 10,000 g for 5 min, and the supernatant was loaded on to 10% SDS-PAGE gel, followed by blotting on nitrocellulose membrane, and then probing with 14-3-3 beta antibody. Normal rabbit serum and mouse IgG, instead of primary antibody, were used as negative control.
Results
Database search for identification of proteins and phosphoproteins
LC MS/MS coupled to Mascot and Sequest database search was used to identify the proteins and phosphoproteins. 2078 total proteins were identified, of which 427 were of Rattus origin (Supplementary file 1). A total of 121 phospho-peptides with 201 phosphorylation sites, and of these 23 showed Rattus assigned phospho-peptides, as shown in Table 1.
| Samples set considered for analysis | Search engines | Total proteins | Rattus proteins | Total phospho-peptides | Total phosphorylation sites | Assigned Phospho peptides | Rattus assigned Phospho-peptides |
|---|---|---|---|---|---|---|---|
| PC1, PC2, PC3, C1, C2, C3, PT1, PT2, PT3, T1, T2, T3 | SequestMascot | 2078 | 427 | 121 | 201 | 29 | 23 |
Table 1: Summary of results after LC-MS run of both normal and failed spermiation sample.
Differentials expression analysis of proteins and phosphoproteins
A total of 25 proteins were found to be up-regulated and 79 proteins to be down regulated in the differential protein analysis data. A total of 11 phospho-proteins were found to be up-regulated and 12 phosphoproteins to be down regulated in the differential phosphoprotein analysis data.
Detailed of all the differentials are listed in Table 2 (up-regulated protein), Table 3 (down-regulated protein), Table 4 (up-regulated phosphoprotein) and Table 5 (down-regulated phosphoprotein).
| Protein ID | Protein Name | Fold Change |
|---|---|---|
| Q3KRD5 | Mitochondrial import receptor subunit TOM34 | 3.71 |
| P34058 | Heat shock protein HSP 90-beta | 17.53 |
| Q66HD0 | Endoplasmin | 2.14 |
| Q7TP40 | PEST proteolytic signal-containing nuclear protein | 2.36 |
| P34058 | Heat shock protein HSP 90-beta | 17.53 |
| Q66HD0 | Endoplasmin | 2.14 |
| O08629 | Transcription intermediary factor 1-beta | 2.53 |
| P11980 | Pyruvate kinase isozymes M1/M2 | 2.70 |
| Q5RK28 | Normal mucosa of esophagus-specific gene 1 protein | 3.37 |
| Q68FR9 | Elongation factor 1-delta | 4.50 |
| P15865 | Histone H1.2 | 4.79 |
| P07943 | Aldose reductase | 2.77 |
| P02770 | Serum albumin | 2.90 |
| Q3KRD5 | Mitochondrial import receptor subunit TOM34 | 3.71 |
| Q63525 | Nuclear migration protein nudC | 2.08 |
| Q60587 | Trifunctional enzyme subunit beta, mitochondrial | 6.40 |
| Q498U4 | SAP domain-containing ribonucleoprotein | 2.15 |
| Q64060 | Probable ATP-dependent RNA helicase DDX4 | 2.14 |
| P38983 | 40S ribosomal protein SA | 3.56 |
| Q6P6R2 | Dihydrolipoyl dehydrogenase, mitochondrial | 2.33 |
| P07483 | Fatty acid-binding protein, heart | 2.29 |
| B0K020 | CDGSH iron-sulfur domain-containing protein 1 | 2.20 |
| Q9ER34 | Aconitate hydratase, mitochondrial | 3.52 |
| P54921 | Alpha-soluble NSF attachment protein | 8.80 |
| P18418 | Calnexin | 3.31 |
Table 2: List of up-regulated proteins during normal and failed spermiation condition.
| Protein ID | Protein Name | Fold change |
|---|---|---|
| P15999 | ATP synthase subunit alpha, mitochondrial | 0.32 |
| P11598 | Protein disulfide-isomerase A3 | 0.23 |
| Q6AYX5 | Outer dense fiber protein 2 | 0.47 |
| Q68FX6 | Calcium-binding and spermatid-specific protein 1 | 0.44 |
| P02401 | 60S acidic ribosomal protein P2 | 0.14 |
| P35565 | Calreticulin | 0.36 |
| Q63429 | Polyubiquitin-C | 0.48 |
| P97536 | Cullin-associated NEDD8-dissociated protein 1 | 0.47 |
| P10719 | ATP synthase subunit beta, mitochondrial | 0.45 |
| Q6PEC1 | Tubulin-specific chaperone A | 0.22 |
| Q9EPH8 | Polyadenylate-binding protein 1 | 0.47 |
| Q63081 | Protein disulfide-isomerase A6 | 0.30 |
| P47245 | Nardilysin | 0.20 |
| Q6P502 | T-complex protein 1 subunit gamma | 0.43 |
| Q6AY33 | Acrosin-binding protein | 0.39 |
| P13084 | Nucleophosmin | 0.22 |
| P04797 | Glyceraldehyde-3-phosphate dehydrogenase | 0.30 |
| P02793 | Ferritin light chain 1 | 0.16 |
| Q68FQ0 | T-complex protein 1 subunit epsilon | 0.20 |
| Q68FS2 | COP9 signalosome complex subunit 4 | 0.20 |
| P35571 | Glycerol-3-phosphate dehydrogenase, mitochondrial | 0.38 |
| Q63945 | Protein SET | 0.07 |
| P19945 | 60S acidic ribosomal protein P0 | 0.38 |
| P45592 | Cofilin-1 | 0.28 |
| Q63617 | Hypoxia up-regulated protein 1 | 0.213572 |
| Q64428 | Trifunctional enzyme subunit alpha, mitochondrial | 0.30 |
| P10111 | Peptidyl-prolyl cis-trans isomerase A | 0.25 |
| P16036 | Phosphate carrier protein, mitochondrial | 0.19 |
| O35180 | Endophilin-A3 | 0.41 |
| Q80U96 | Exportin-1 | 0.19 |
| Q5HZV9 | Protein phosphatase 1 regulatory subunit 7 | 0.13 |
| P13383 | Nucleolin | 0.08 |
| P11442 | Clathrin heavy chain 1 | 0.21 |
| Q6AY30 | Probable saccharopine dehydrogenase | 0.10 |
| Q5XI62 | Uncharacterized protein C1orf56 homolog | 0.20 |
| Q62703 | Reticulocalbin-2 | 0.32 |
| Q6T393 | Spermatogenesis-associated protein 20 | 0.04 |
| P04904 | Glutathione S-transferase alpha-3 | 0.38 |
| P04785 | Protein disulfide-isomerase | 0.35 |
| Q9R063 | Peroxiredoxin-5, mitochondrial | 0.05 |
| Q91Y78 | Ubiquitin carboxyl-terminal hydrolase isozyme L3 | 0.16 |
| Q63610 | Tropomyosin alpha-3 chain | 0.37 |
| O35244 | Peroxiredoxin-6 | 0.49 |
| P05197 | Elongation factor 2 | 0.18 |
| Q62764 | DNA-binding protein A | 0.23 |
| P11884 | Aldehyde dehydrogenase, mitochondrial | 0.26 |
| Q08163 | Adenylyl cyclase-associated protein 1 | 0.22 |
| P10888 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial | 0.47 |
| Q06647 | ATP synthase subunit O, mitochondrial | 0.39 |
| Q9Z2L0 | Voltage-dependent anion-selective channel protein1 | 0.14 |
| P38656 | Lupus La protein homolog | 0.11 |
| P52555 | Endoplasmic reticulum resident protein 29 | 0.17 |
| P37996 | ADP-ribosylation factor-like protein 3 | 0.37 |
| P17164 | Tissue alpha-L-fucosidase | 0.16 |
| P80254 | D-dopachrome decarboxylase | 0.34 |
| O88600 | Heat shock 70 kDa protein 4 | 0.21 |
| P50554 | 4-aminobutyrate aminotransferase, mitochondrial | 0.18 |
| P85972 | Vinculin | 0.24 |
| O08651 | D-3-phosphoglycerate dehydrogenase | 0.23 |
| P45479 | Palmitoyl-protein thioesterase 1 | 0.34 |
| Q5XHZ2 | Synaptonemal complex central element protein 1 | 0.07 |
| Q5XI60 | Receptor expression-enhancing protein 6 | 0.28 |
| Q07936 | Annexin A2 | 0.26 |
| O88767 | Protein DJ-1 | 0.24 |
| Q6P6V0 | Glucose-6-phosphate isomerase | 0.26 |
| P04642 | L-lactate dehydrogenase A chain | 0.46 |
| P21533 | 60S ribosomal protein L6 | 0.35 |
| O35774 | A-kinase anchor protein 4 | 0.34 |
| P11762 | Galectin-1 | 0.13 |
| P11030 | Acyl-CoA-binding protein | 0.46 |
| B0BN93 | 26S proteasome non-ATPase regulatory subunit 13 | 0.24 |
| Q64560 | Tripeptidyl-peptidase 2 | 0.16 |
| P24155 | Thimet oligopeptidase | 0.45 |
| P14668 | Annexin A5 | 0.28 |
| P30009 | Myristoylated alanine-rich C-kinase substrate | 0.46 |
| Q7TP40 | PEST proteolytic signal-containing nuclear protein | 0.36 |
| Q68FY0 | Cytochrome b-c1 complex subunit 1, mitochondrial | 0.16 |
| P14841 | Cystatin-C | 0.37 |
| Q9ER24 | Ataxin-10 | 0.30 |
Table 3: List of down-regulated proteins during normal and failed spermiation condition.
| Protein ID | Protein Name | Fold Change |
|---|---|---|
| P35213 | 14-3-3 protein beta/alpha | 3.24 |
| O08629 | Transcription intermediary factor 1-beta | 2.77 |
| O35180 | Endophilin-A3 | 3.39 |
| Q4V8E4 | Coiled-coil domain-containing protein 104 | 13.96 |
| P31000 | Vimentin | 3.36 |
| Q62703 | Reticulocalbin-2 | 22.00 |
| Q4QR76 | Actin-like protein 7B | 3.64 |
| Q6P6S3 | B box and SPRY domain-containing protein | 2.09 |
| O35987 | NSFL1 cofactor p47 | 3.44 |
| Q3KR86 | Mitochondrial inner membrane protein (Fragment) | 3.11 |
| P05371 | Clusterin | 4.59 |
Table 4: List of up-regulated differential Phosphoproteins during normal and failed spermiation condition.
| Protein ID | Protein Name | Fold change |
|---|---|---|
| Q05962 | ADP/ATP translocase 1 | 0.22 |
| Q6P4Z9 | COP9 signalosome complex subunit 8 | 0.42 |
| Q5U216 | ATP-dependent RNA helicase DDX39A | 0.42 |
| Q07647 | Solute carrier family 2, facilitated glucose transporter member 3 | 0.12 |
| P21670 | Proteasome subunit alpha type-4 | 0.07 |
| Q6AYG7 | NFATC2-interacting protein | 0.33 |
| P38983 | 40S ribosomal protein SA | 0.26 |
| Q9ESW0 | DNA damage-binding protein 1 | 0.29 |
| Q10728 | Protein phosphatase 1 regulatory subunit 12A | 0.19 |
| Q5XIP1 | Protein pelota homolog | 0.46 |
| Q10728 | Protein phosphatase 1 regulatory subunit 12A | 0.19 |
| P21670 | Proteasome subunit alpha type-4 | 0.07 |
Table 5: List of down-regulated differential Phosphoproteins during normal and failed spermiation condition.
Expression analysis of differentially regulated proteins and phosphoproteins
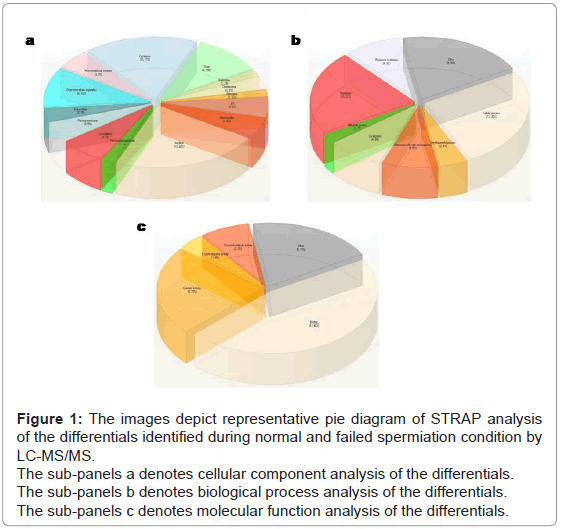
Gene ontology analysis by STRAP (Software Tool for Researching Annotation of Proteins): Analysis of the differential proteins in terms of cellular component revealed 22% of proteins belong to the nucleus, 17% to the cytoplasm, 7% to the cytoskeleton and 5% each to endoplasmic reticulum and mitochondria [19] (Figure 1a). Analysis in-terms of biological component showed 26% of proteins involved in cellular process, 22% in regulation (Figure 1b), while molecular function analysis demonstrated 46% of proteins involved in binding and 23% in catalytic activity (Figure 1c).
Figure 1: The images depict representative pie diagram of STRAP analysis of the differentials identified during normal and failed spermiation condition by LC-MS/MS.
The sub-panels a denotes cellular component analysis of the differentials.
The sub-panels b denotes biological process analysis of the differentials.
The sub-panels c denotes molecular function analysis of the differentials.
Interestingly, gene ontology analysis by STRAP showed a number of proteins of endoplasmic reticulum (ER) to be differentially expressed, using cellular component as a determinant, suggesting involvement of ER. Since, in the testis, endoplasmic reticulum (ER) forms an essential component of ES and TBCs [4], we studied the expression of two ER related proteins, i.e. calreticulin and calnexin, in stages where spermiation occurs.
Biological process analysis showed the maximum no. of differential proteins to be involved in cellular activity and its regulation. Moreover, molecular function analysis revealed that lot of differentials to be involved in binding of proteins. Based on these indications, we selected 14-3-3 beta from differential phosphoprotein list, which have been shown to have a large number of binding partners, and is involved in various cellular activities like protein-protein interaction, cell adhesion, cellular trafficking, etc. [20].
Collectively, from the differential total protein list, we selected two down-regulated proteins (Calreticulin and T-Complex protein 1), and one up-regulated protein (Calnexin), for validation. From the differential phosphoprotein list, we selected up-regulated phosphoprotein 14-3-3 beta, to validate LC-MS/MS results. Validation of the above proteins was performed by immunofluroscence, western blotting and immunoprecipitation experiments.
Localization of differential proteins during normal and failed spermiation condition: Calnexin and calreticulinare components of the quality control system that promotes correct folding of proteins that enter the secretory pathway and targets mis-folded proteins for degradation. Both calnexin and calreticulin reside in the ER [21]. Hence, their localization was studied during normal and failed spermiation in the testis.
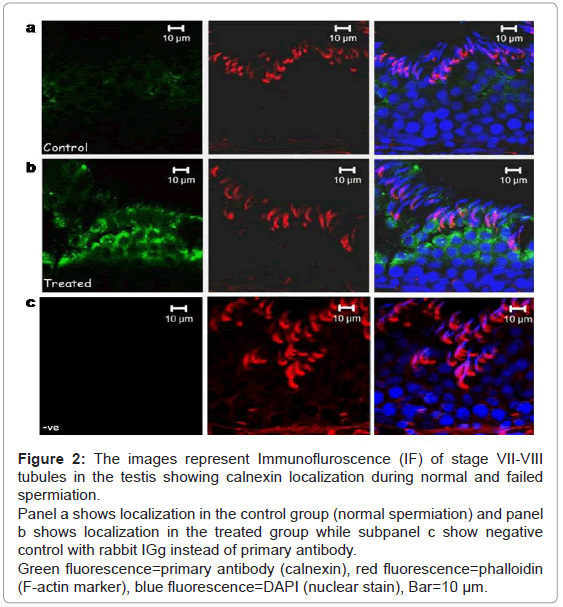
Calnexin was seen to be localized in the adluminal compartment of the seminiferous epithelium of the testis during normal and failed spermiation condition (Figure 2). The signal intensity (expression level) of calnexin seemed to be increased during failed spermiation, i.e. estradiol treated group (Figure 2b).
Figure 2: The images represent Immunofluroscence (IF) of stage VII-VIII tubules in the testis showing calnexin localization during normal and failed spermiation.
Panel a shows localization in the control group (normal spermiation) and panel b shows localization in the treated group while subpanel c show negative control with rabbit IGg instead of primary antibody.
Panel a shows localization in the control group (normal spermiation) and panel b shows localization in the treated group while subpanel c show negative control with rabbit IGg instead of primary antibody.
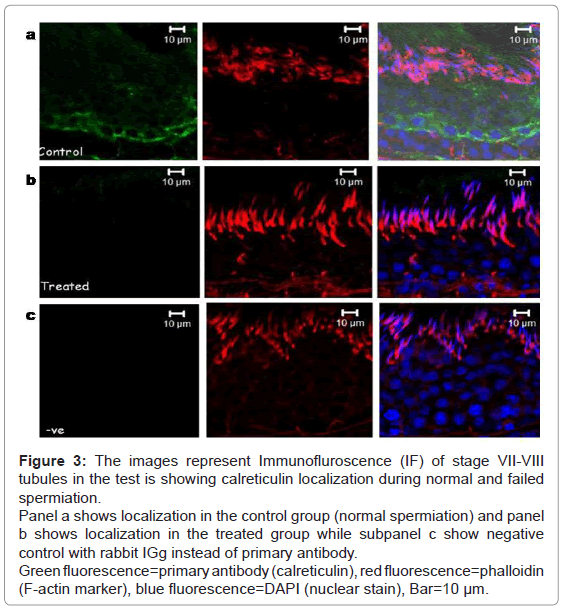
Calreticulin was seen to be localized in the adluminal compartment surrounding round spermatids of the seminiferous epithelium of the testis during normal and failed spermiation condition (Figure 3). The signal intensity (expression level) of calreticulin seemed to be decreased during failed spermiation, i.e. estradiol treated group (Figure 3b).
Figure 3: The images represent Immunofluroscence (IF) of stage VII-VIII tubules in the test is showing calreticulin localization during normal and failed spermiation.
Panel a shows localization in the control group (normal spermiation) and panel b shows localization in the treated group while subpanel c show negative control with rabbit IGg instead of primary antibody.
Green fluorescence=primary antibody (calreticulin), red fluorescence=phalloidin (F-actin marker), blue fluorescence=DAPI (nuclear stain), Bar=10 μm.
The expression of F-actin (marker for TBC and ES) was localized in the TBCs and ES in control group, i.e. normal spermiation (both Figures 2a and 3a-red panels), whereas in failed spermiation, it was only observed in ES as TBC were absent (both Figures 2b and 3b-red panels).
The T-complex protein 1 (TCP-1) of the mouse has been shown to play an important role in male germ cell development. In the yeast, TCP-1 is involved in actin and tubulin cytoskeletal protein folding [22-24].
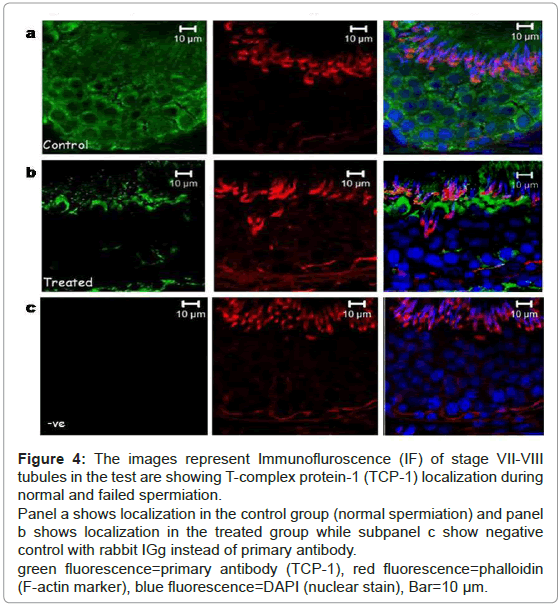
Hence, testicular localization of TCP-1 was performed during normal and failed spermiation condition (Figure 4). Localization of T-complex protein-1 during spermiation revealed that it was mainly localized in the seminiferous epithelium. The signal intensity was seen to localize around all the germ cell types, both in the basal and in the adluminal compartment during normal spermiation (Figure 4a). In the failed spermiation condition, i.e. in the treated group the expression of TCP-1 protein was observed only at the luminal edge of the epithelium (Figure 4b).
Figure 4: The images represent Immunofluroscence (IF) of stage VII-VIII tubules in the test are showing T-complex protein-1 (TCP-1) localization during normal and failed spermiation.
Panel a shows localization in the control group (normal spermiation) and panel b shows localization in the treated group while subpanel c show negative control with rabbit IGg instead of primary antibody.
green fluorescence=primary antibody (TCP-1), red fluorescence=phalloidin (F-actin marker), blue fluorescence=DAPI (nuclear stain), Bar=10 μm.
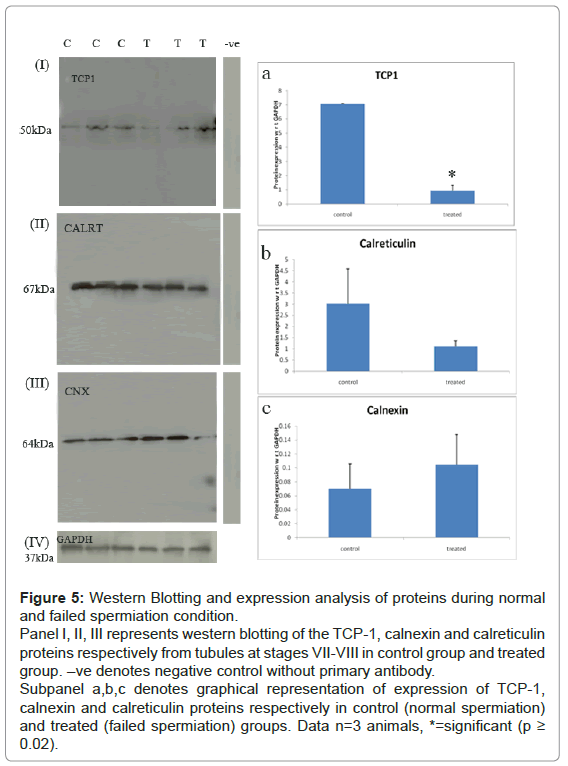
Expression levels of all the three proteins during normal and failed spermiation condition: Western Blotting of calnexin, calreticulin and TCP-1: Western blotting experiments showed significant down regulation of Tcp-1with p-value ≤ 0.05, and trend toward increase and decrease for calnexin and calreticulin, respectively, as compared with GAPDH expression levels (loading control). The results were seen to be in concordance with the LC-MS differential data (Figure 5).
Figure 5: Western Blotting and expression analysis of proteins during normal and failed spermiation condition.
Panel I, II, III represents western blotting of the TCP-1, calnexin and calreticulin proteins respectively from tubules at stages VII-VIII in control group and treated group. –ve denotes negative control without primary antibody.
Subpanel a,b,c denotes graphical representation of expression of TCP-1, calnexin and calreticulin proteins respectively in control (normal spermiation) and treated (failed spermiation) groups. Data n=3 animals, *=significant (p ≥ 0.02).
Detection of phosphorylation status of 14-3-3 beta
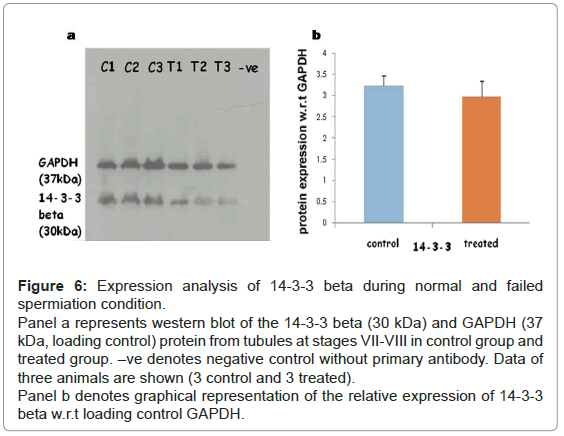
Expression of 14-3-3 protein in testis during normal and failed spermiation: As we observed an increase in the phosphorylation of 14- 3-3 beta in the differential phosphoprotein list, we sought to determine the expression of 14-3-3 beta at protein level during normal and failed spermiation condition, i.e. in stages VII-VIII of the seminiferous tubule. Western blotting showed no change in the expression levels of 14-3-3 beta (30 kDa) when compared with GAPDH (37 kDa), as a loading control in normal and failed spermiation condition (Figures 6a and 6b).
Figure 6: Expression analysis of 14-3-3 beta during normal and failed spermiation condition.
Panel a represents western blot of the 14-3-3 beta (30 kDa) and GAPDH (37 kDa, loading control) protein from tubules at stages VII-VIII in control group and treated group. –ve denotes negative control without primary antibody. Data of three animals are shown (3 control and 3 treated).
Panel b denotes graphical representation of the relative expression of 14-3-3 beta w.r.t loading control GAPDH.
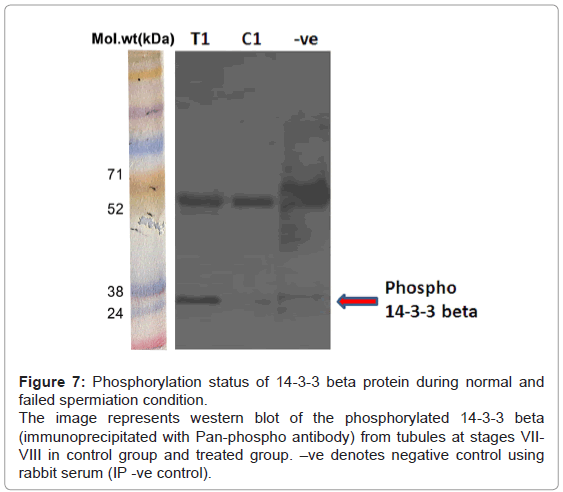
Phosphorylation status of 14-3-3 beta by immuno-precipitation: The phosphorylation status of 14-3-3 beta by immuno-precipitation experiment revealed increased phosphorylation of 14-3-3 beta in the treated group as compared to controls (Figure 7), indicating upregulation of phosphorylation of 14-3-3 beta, as seen in the differentials of phosphoproteins.
Figure 7: Phosphorylation status of 14-3-3 beta protein during normal and failed spermiation condition.
The image represents western blot of the phosphorylated 14-3-3 beta (immunoprecipitated with Pan-phospho antibody) from tubules at stages VIIVIII in control group and treated group. –ve denotes negative control using rabbit serum (IP -ve control).
Discussion
Our study suggests that there is a change in the expression of proteins and phosphoproteins during normal and failed spermiation condition in stages VII-VIII of rat spermatogenic cycle. A total of 427 proteins and phosphoproteins belong to rattus species (rat) of proteins identified. Differential protein and phosphoprotein profile revealed 24 up-regulated and 80 down-regulated proteins, and 11 up-regulated and 12 down-regulated phosphoproteins.
Gene Ontology study by using STRAP software [19] revealed that a number of proteins were related to endoplasmic reticulum (ER), as determined using cellular component as a determinant. Since ES and TBCs have ER as a part of their structure, we studied the expression of two ER related proteins. Biological process analysis showed the maximum number of differential proteins to be involved in cellular activity and its regulation. In addition, molecular function analysis revealed that maximum number of differentials proteins to be involved in binding.
Amongst the other proteins in the differential list, a number of proteins important for the process of spermatogenesis were demonstrated, For example, Aldose reductase was seen to up-regulated in the proteomic profile; it is known to be involved in reduction of cytotoxic metabolites produced during spermatogenesis in addition to its metabolic function [25]. Cofilin-1, which is down-regulated, is known to plays a role in actin turn-over by depolymerization of actin filament, suggesting a defect in actin remodeling during stage VII-VII in normal versus treated animals could have led to absence of TBCs reported in our earlier study and spermiation failure [15,26]. A number of the differentials belong to mitochondrial component of the cell, as demonstrated by STRAP software analysis, suggesting mitochondrial dysfunction. One such protein, phosphorylated adenine nuclease translocase-1, which was seen to be down-regulated in the present study, has been reported to be involved in mitochondrial DNA maintenance [27]. MARCKS (myristoylated alanine-rich C-kinase substrate) is known to be calcium regulated, and its phosphorylation is important for acrosomal exocytosis [28]. Down-regulation of MARCKS in the present study suggests the importance of calcium homeostasis during spermatogenesis and acrosomal formation. The role of the above mentioned proteins in the process of spermiation is unclear and further studies in this direction needs to be done.
During spermiation, endoplasmic reticulum is known to show dynamic changes. Flattened ER (f-ER, narrow cisternae of ER), is associated with ES and arranged in concentric layers in the Sertoli cell cytoplasm covering the spermiated head. The f-ER disappeared just prior to spermiation and another ER structure, i.e. tubular ER appears during late stage-VII surrounding both dorsal and ventral surface of the spermatid head, which also disappeared at the time of spermiation [29]. In addition, during spermatogenesis, ER proteins are known to be important in regulating sperm motility and acrosomes reaction [30]. In the present study, two ER resident proteins, calnexin and calreticulin, are shown to be up and down regulated, respectively during failed spermiation condition, suggesting their involvement in the dynamic changes in ER form during spermiation, but the mechanism remains to be elucidated.
Immuno-localization studies with calnexin showed its expression in the adluminal compartment of the seminiferous epithelium and the intensity of staining showed an increase in treated group. In addition, western blotting also showed the protein to be up regulated. It has been established that it plays a role in protein folding and its quality control. Knockout of a testis-specific variant of calnexin, calmegin, showed mice were infertile, highlighting the importance of calnexin in male fertility [31]. Furthermore, localization of calmegin was reported to be in the meiotic and post meiotic germ cell in the test is suggesting its role in spermatid differentiation [32]. In yeast cells, calnexin have been shown to regulate the process of apoptosis during their fission [33]. In the present study, an increase of calnexin in spermiation failure group may be involved in similar function of regulating apoptosis, as our earlier study showed increase in tunnel positive cell in the treatment group [14].
Calreticulin, a multi-functional protein resides mainly in ER is a chaperone, and is involved inthe regulation of cell adhesion, gene expression and intracellular Ca+2 homeostasis. It has been shown to be present in the acrosomes of rat sperm [30,34]. A decrease of calreticulin in the treated group could have affected the process of acrosomes formation. Over-expression of calreticulin has shown to regulate vinculin assembly at focal contacts and decreases phosphorylation status of β-catenin [35]. Calreticulin affects β-catenin-associated pathway, and hence cell adhesion [36]. Moreover, β-catenin along-with cadherins forms an actin-based adherens junctions associated with ES during spermatogenesis [37], suggesting the involvement of calreticulin in spermatogenesis.
Cytoskeleton reorganization is shown to be important in maintaining the cytoarchitecture of seminiferous epithelium [38]. Our earlier studies demonstrated disruption in the actin and microtubular organization and spermiation failure, following 17β-estradiol treatment [15]. The present study shows significant down regulation of T-complex protein-1, a molecular chaperone, activated when bound to c-AMP. Mutational studies with TCP-1 genes in the yeast have shown disorganization of actin and microtubular network [23,24]. The disruption in actin and microtubules observed earlier could be due to the down regulation. Futhermore, TCP-1 protein has been shown to be present in chromatin of highly condensed spermatid, suggesting its involvement in sperm head formation [39]. We hypothesize that down regulation of TCP-1 affected actin/microtubule organization, affecting spermatid differentiation and sperm release.
Moreover, we observed phosphorylated 14-3-3 beta protein to be over expressed in the differential phosphorylation data. Studies in humans and rats have shown binding of 14-3-3 beta to vimentin and tubulin in the seminiferous epithelium [17,40]. The upregulation of phosphorylated 14-3-3 beta observed in the present study may have affected the phosphorylation status of vimentin, leading to reorganization of vimentin and observed apical projection retention in stages VIII in the testis, as described earlier [17]. In addition, binding of 14-3-3 to Protein Phosphatase 1 gamma isoform have been shown to be important during sperm function and its motility [41]. Thus, increased phosphorylation of 14-3-3 beta may have affected vimentin and tubulin reorganization leading to spermiation failure.
In summary, the present study demonstrates differential protein and phosphoprotein expression during spermiation and spermiation failed condition by LC-mass spectrometry. The differential proteins identified suggest an effect on endoplasmic reticulum and cytoskeleton reorganization, which forms an important component of ES and TBCs, indicating that the function of these two structures maybe affected, leading to spermiation failure. Some of the differentials that may serve as potential biomarker for sperm release, however, more studies need to be done to achieve this. Alternatively, since a number of proteins involved in calcium homeostasis and acrosome biogenesis is affected, suggesting that estrogen treatment could have affected the spermatid maturation. The affected spermatids are then not released but retained and phagocytosed by the Sertoli cell. This may prevent defective spermatid from fertilizing the oocyte. Since, spermiation is the final stage of spermatogenesis molecules regulating this event offer a great potential for reversible contraceptive development without affecting the other germ cell population.
Acknowledgements
We would like to thank Department of Science and Technology, New Delhi, India, for partially funding the study. We would acknowledge ICMR for SRF fellowship to Rahul D Upadhyay. We thank the staff of the C-CAMP facility, Mass Spectrometry Lab, National Centre for Biological Sciences, Bangalore for the LCMS/ MS sequencing of samples, and we are grateful to Dr. Dominik Schwudke, Mr. Karthik Kamath for their valuable assistance in the sequencing, data generation of proteins and phosphoproteins of our samples. The assistance of Dr. Carmen Tekwe is also acknowledged in analyzing the spectrometry data. We would also like to acknowledge the efforts of Mr. Suryakant Mandavkar in the animal experiment and Mr. Deepak Shelar for his technical assistance. Authors would like to thank Ms Annette Fonseca for her help in correcting grammatical mistakes of the manuscript.
Conflict of Interest
Authors declare no conflict of interest.
References
- Hess RA (1998) Spermatogenesis: Overview. In: Encyclopedia of Reproduction. Knobil E, Neill JD (Eds.), Academic Press, San Diego, USA 539-545.
- O’Donnell L, Nicholls PK, O’Bryan MK, McLachlan RI, Stanton PG (2011) Spermiation: The process of sperm release. Spermatogenesis 1: 1-22.
- Leblond CP, Clermont Y (1952) Definition of the stages of the seminiferous epithelium in the rat testes. Ann NY Acad Sci 5: 548-573
- Upadhyay RD, Kumar AV, Ganeshan M, Balasinor NH (2012) Tubulobulbar complex: Cytoskeletal remodeling to release spermatozoa. Reprod Biol Endocrinol 10: 27.
- deRooij DG (2001) Proliferation and differentiation of spermatogonial stem cells. Reproduction 121: 347-354.
- Matzuk MM, Lamb DJ (2002) Genetic dissection of mammalian fertility pathways. Nat Cell Biol 4: 41-49.
- Rolland AD, Evrard B, Guitton N, Lavigne R, Calvel P, et al. (2007) Two-dimensional fluorescence difference gel electrophoresis analysis of spermatogenesis in the rat. J Proteome Res 6: 683-697.
- Chocu S, Calvel P, Rolland A, Pineau C (2012) Spermatogenesis in mammals: proteomic insights. Syst Biol Reprod Med 58: 179-190.
- Stanton PG, Sluka P, Foo CF, Stephens AN, Smith AI, et al. (2012) Proteomic changes in rat spermatogenesis in response to in vivoandrogen manipulation; Impact on meiotic cells. PLoS ONE 7: e41718.
- Chalmel F, Lardenois A, Evrard B, Mathieu R, Feig C, et al. (2012) Global human tissue profiling and protein network analysis reveals distinct levels of transcriptional germline-specificity and identifies target genes for male infertility. Hum Reprod 27: 3233-3248.
- Chapin R, Wine RN, Harris MW, Borchers CH, Haseman JK (2001) Structure and control of a cell-cell adhesion complex associated with spermiation in rat seminiferous epithelium. J Androl 22: 1030-1052.
- Mruk DD, Cheng CY (2004) Sertoli-Sertoli and sertoli-germ cell interactions and their significance in germ cell movement in the seminiferous epithelium during spermatogenesis.Endocr Rev 25: 747-806.
- Beardsley A, Robertson DM, O’Donnell L (2006) A complex containing α6ß1-integrin and phosphorylated focal adhesion kinase between Sertoli cells and elongated spermatids during spermatid release from the seminiferous epithelium. J Endocrinol 190: 759-770.
- D'Souza R, Gill-Sharma MK, Pathak S, Kedia N, Kumar R, et al. (2005) Effect of high intratesticular estrogen on the seminiferous epithelium in adult male rats. Mol Cell Endocrinol 241: 41-48.
- D'Souza R, Pathak S, Upadhyay R, Gaonkar R, D'Souza S, et al. (2009) Disruption of tubulobulbar complex by high intratesticular estrogens leading to failed spermiation. Endocrinology 150: 1861-1869.
- Parvinen M, Toppari J, Lahdetie J (1993) Transillumination-phasecontrast microscopy techniques for evaluation of male germ cell toxicity and mutagenicity. In: Chapin RE, Heindel J (Eds), Methods in toxicology, Academic Press, Orlando, USA 3A: 142-165.
- Upadhyay R, D'Souza R, Sonawane S, Gaonkar R, Pathak S, et al. (2011) Altered phosphorylation and distribution status of vimentin in rat seminiferous epithelium following 17β-estradiol treatment. Histochem Cell Biol 136: 543-555.
- Duan X, Young R, Straubinger RM, Page B, Cao J, et al. (2009) A straightforward and highly efficient precipitation/on-pellet digestion procedure coupled with a long gradient nano-LC separation and Orbitrap mass spectrometry for label-free expression profiling of the swine heart mitochondrial proteome. J Proteome Res 8: 2838-2850.
- Bhatia VN, Perlman DH, Costello CE, McComb ME (2009) Software tool for researching annotations of proteins: open-source protein annotation software with data visualization. Anal Chem 81: 9819-9823
- Haian Fu, Subramanian RR, Masters SC (2000) 14-3-3 proteins: Structure, function, and regulation. Annu Rev Pharmacol Toxicol 40: 617-647.
- Taubenberger AM, Lupas AN, Li H, Ecke M, Simmeth E, et al. (2001) Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis. EMBO 20: 6772-6782.
- Lader E, Ha H, O'Neill M, Artzt K, Bennett D (1989) Tctex-1: A candidate gene family for a mouse t complex sterility locus. Cell 58: 969-979
- Horwich AL, Fenton WA, Chapman E, Farr GW (2007) Two families of chaperonin: Physiology and mechanism. Annu Rev Cell Dev Biol 23: 115-145.
- Altschuler GM, Willison KR (2008) Development of free-energy-based models for chaperonin containing TCP-1 mediated folding of actin. J R Soc Interface 5: 1391-1408.
- Kobayashi T, Kaneko T, Iuchi Y, Matsuki S, Takahashi M, et al. (2002) Localization and physiological implication of aldose reductase and sorbitol dehydrogenase in reproductive tracts and spermatozoa of male rats. J Androl 23: 674-683.
- Svitkina TM, Borisy GG (1999) Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol 145: 1009-1026.
- Kaukonen J, Juselius JK, Tiranti V, Kyttälä A, Zeviani M, et al. (2000) Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science 289: 782-785.
- Rodriguez Pena MJ, Castillo Bennett JV, Soler OM, Mayorga LS, Michaut MA (2013) MARCKS protein is phosphorylated and regulates calcium mobilization during human acrosomal exocytosis. PLoS ONE 8: 64551.
- Morales C, Clermont Y (1993) Structural changes of the Sertoli cell during the cycle of the seminiferous epithelium. In: Russell LD, Griswold MD (Eds), The Sertoli Cell, Clearwater FL: Cache River Press, USA 305-329.
- Nakamura M, Moriya M, Baba T, Michikawa Y, Yamanobe T, et al. (1993) An endoplasmic reticulum protein, calreticulin, is transported into the acrosome of rat sperm. Exp Cell Res 205: 101-110.
- Ohsako S, Janulis L, Hayashi Y, Bunick D (1998) Characterization of domains in mice of calnexin-t,a putative molecular chaperone required in sperm fertility, with use of glutathione S-transferase fusion proteins. Biol Reprod 59: 1214-1223.
- Yoshinaga K, Tanii I, Toshimori K (1999) Molecular chaperone calmegin localization to the endoplasmic reticulum of meiotic and post-meiotic germ cells in the mouse testis. Arch Histol Cytol 62: 283-293.
- Guerin R, Beauregard PB, Leroux A, Rokeach LA (2009) Calnexin regulates apoptosis induced by inositol starvation in fission yeast. PLoS ONE 4: 6244.
- Nakamura M, Michikawa Y, Baba T, Okinaga S, Arai K (1992) Calreticulin is present in the acrosome of spermatids of rat testis. Biochem Biophys Res Commun 186: 668-673.
- Opas M, Pawlikowski SM, Jass GK, Meseali N, Michalak M (1996) Calreticulin modulates cell adhesiveness via regulation of vinculin expression. J Cell Biol 12: 294-307.
- Fadel MP, Szewczenko-Pawlikowski P, Dziak LE, Symonds JM, Blaschuk O, et al. (2001) Calreticulin affects beta-catenin associated pathways. J Biol Chem 276: 27083-27089.
- Lee NP, Mruk DD, Conway AM, Cheng CY (2004) Zyxin, axin and Wiskott-Aldrich syndrome protein are adaptors that link the cadherin/catenin protein complex to the cytoskeleton at adherens junctions in the seminiferous epithelium of the rat testis. J Androl 25: 200-215.
- Lie PY, Mruk DD, Lee WM, Cheng CY (2010) Cytoskeletal dynamics and spermatogenesis. Phil Trans R Soc B 365: 1581-1592.
- Souès S, Kann ML, Fouquet JP, Melki R (2003) The cytosolic chaperonin CCT associates to cytoplasmic microtubular structures during mammalian spermiogenesis and to heterochromatin in germ-line and somatic cells. Exp Cell Res 288: 363-373.
- Graf M, Brobeil A, Sturm K, Steger K, Wimmer M (2011) 14-3-3 beta in the healthy and diseased male reproductive system. Hum Reprod 26: 59-66.
- Puri P, Myers K, Kline D, Vijayaraghavan S (2008) Proteomic analysis of bovine sperm YWHA binding partners identify proteins involved in signaling and metabolism. Biol Reprod 79: 1183-1191.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 15315
- [From(publication date):
November-2013 - Apr 03, 2025] - Breakdown by view type
- HTML page views : 10662
- PDF downloads : 4653