Research Article Open Access
Differential Expression of miR-130a in Postmortem Prefrontal Cortex ofSubjects with Alcohol Use Disorders
Fan Wang1,4, Joel Gelernter1-4 and Huiping Zhang1,4*1Departments of Psychiatry, Yale University School of Medicine, New Haven, CT, USA
2Departments of Genetics, Yale University School of Medicine, New Haven, CT, USA
3Departments of Neurobiology, Yale University School of Medicine, New Haven, CT, USA
4VA Medical Center, VA Connecticut Healthcare System, West Haven, CT, USA
- *Corresponding Author:
- Huiping Zhang, Ph.D.
Department of Psychiatry, Yale University School of Medicine
VA Medical Center/116A2, 950 Campbell Avenue, West Haven, CT 06516, USA
Tel: (203) 932-5711 ext. 5245
Fax: (203) 937-4741
E-mail: huiping.zhang@yale.edu
Received date: June 26, 2013; Accepted date: July 18, 2013; Published date: July 23, 2013
Citation: Wang F, Gelernter J, Zhang H (2013) Differential Expression of miR- 130a in Postmortem Prefrontal Cortex of Subjects with Alcohol Use Disorders. J Addict Res Ther 4:155. doi:10.4172/2155-6105.1000155
Copyright: © 2013 Wang F, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Addiction Research & Therapy
Abstract
Background: Emerging evidence suggests that neuroadaptations to alcohol may result from chronic alcohol consumption-induced expression changes of microRNAs (miRNAs) and their target genes. Studies with animal or cell culture models have demonstrated that ethanol exposure leads to miRNA expression alterations. However, there is limited information on miRNA expression in the brains of subjects with alcohol use disorders (AUDs). The present study aimed to analyze expression changes of miRNAs and their target genes in postmortem prefrontal cortex (PFC) of AUD subjects.
Methods: Genome-wide miRNA and mRNA expression was examined in postmortem PFC of 23 European Australia AUD cases and 23 matched controls using the Illumina HumanHT-12 v4 Expression Bead Chip array, which targets 43,270 coding transcripts and 3,961 non-coding transcripts (including 574 miRNA transcripts). Multiple linear regression analysis and permutation test were performed to identify differentially expressed miRNAs and their target mRNAs. Target gene prediction, Gene Set Enrichment Analysis (GESA), and DAVID functional annotation clustering analysis were applied to identify AUD-associated gene sets and biological modules.
Results: Two miRNAs and 787 coding genes were differentially expressed in the PFC of AUD cases [miR-130a (downregulated): Ppermutation=0.023, miR-604 (upregulated): Ppermutation=0.019, coding genes: 1.6×10-5≤Ppermutation≤0.05; but all P values did not survive multiple-testing correction]. GESA showed that the 202 predicted target genes of miR-130a were highly enriched in differentially expressed genes (Pnominal<0.001), but not the 116 predicted target genes of miR- 604 (Pnominal=0.404). DAVID functional clustering further revealed that the hub target genes (e.g., ITPR2 and ATP1A2) of miRNA130a were mainly responsible for regulating ion channel function.
Conclusion: This study provides evidence that downregulation of miR-130a may lead to altered expression of a number of genes in the PFC of AUD subjects. Further studies are warranted to confirm these findings in replication samples and other reward-related brain regions.
Keywords
MicroRNA; Alcohol use disorders; Postmortem prefrontal cortex; Expression microarray
Introduction
Alcohol use disorders (AUDs), including abuse or dependence, are prevalent health and social problems. Despite the high prevalence (over 8%) of AUDs [1], their molecular mechanism is not well understood; it is known that AUDs are influenced by multiple genes and geneenvironment interactions [2]. Environmental factors alone (e.g., chronic alcohol consumption) may also lead to alcohol tolerance or dependence through neuroadaptations. Emerging evidence suggests that alcohol-induced neuroadaptations are at least partially linked to altered expression of microRNAs (miRNAs) and their target genes that are involved in AUD-relevant biological pathways [3].
miRNAs are a group of small non-coding RNAs (~19 - 24 nucleotides) that regulate the expression of protein-coding genes. The regulatory function of miRNAs is accomplished through the RNAinduced silencing complex (RISC). miRNAs guide the RISC to the 3’ untranslated regions (3’ UTRs) of their target mRNAs via base pairing, leading to translation inhibition or mRNA degradation [4,5]. miRNAs are abundant in the brain and participate in many biological processes such as neuronal differentiation [6], synapse formation and plasticity [7,8], and neurodegeneration [9]. Chronic alcohol consumption may alter miRNA expression in the brain, and dysregulation of miRNAs may lead to diseases such as AUDs. Studies with cell culture models demonstrated that alcohol-induced miRNA alterations were associated with alcohol tolerance (in rat striatal primary neurons) [10], gut leakiness (in the human intestinal epithelial cell line Caco-2) [11], and neural proliferation and differentiation (in fetal mouse cerebral cortexderived neurosphere) [12]. Moreover, Guo et al. [13] reported that chronic intermittent ethanol exposure or removal induced different miRNA expression patterns in mouse cortical primary neuronal cultures. Studies with a mouse model also showed that ethanol exposure caused dysregulation of specific miRNAs, which target a number of genes that are crucial in the context of fetal alcohol spectrum disorders (FASDs) [14].
To date, there is limited information on miRNA expression alterations in the brains of human AUD subjects; to our knowledge, only one study examined AUD-associated miRNAs in postmortem prefrontal cortex (PFC) of human AUD subjects [15]. This study identified 35 miRNAs that were upregulated in the PFC of AUD subjects, and those genes potentially targeted by the upregulated miRNAs were overrepresented by biological processes such as apoptosis, cell cycles, cell adhesion, and nervous system development. Given the relatively small size of the PFC tissue sample (14 cases vs. 13 controls) in that study, additional studies with larger samples are warranted. In the present study, we examined genome-wide gene expression in postmortem PFC of 23 AUD cases and 23 matched controls using the microarray approach, and the differentially expressed miRNAs and mRNAs in AUD cases were identified. In addition, the enrichment of the putative target genes of AUD-associated miRNAs in differentially expressed genes was also analyzed.
Materials and Methods
Study subjects
Autopsy brain tissue samples were obtained from the New South Wales Tissue Resource Centre (NSW TRC) at the University of Sydney. The NSW TRC is sponsored by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) for collecting human brain tissues for alcohol-related research. It has ethics approval from the Sydney Local Health Network and The University of Sydney. Fresh-frozen sections of Brodmann area 9 (BA9, mainly the dorsolateral PFC of the brain) were obtained from 23 European Australian AUD cases and 23 matched controls. Case and control samples were matched by race, sex, age, brain pH, brain weight, and postmortem interval (PMI). Cases were affected with alcohol abuse or dependence (with no comorbid drug abuse or dependence or major psychotic disorders such as schizophrenia and bipolar disorder) according to the criteria in the Diagnostic and Statistical Manual of Mental Disorders 4th Edition (DSM-IV) [16]. Controls were assessed to be free of substance (alcohol or drug) abuse or dependence or major psychotic disorders. The case group included 23 AUD subjects and 16 (69.6%) were males. The average age of AUD cases was 56 ± 9 years (mean ± S.D.) and the average amount of daily use of alcohol in AUD cases was 165 ± 81 gram. The control group included 23 matched healthy subjects and 16 (69.6%) were males. The average age of controls was 56 ± 9 years and the average amount of daily use of alcohol in control subjects was 11 ± 9 gram.
Transcriptome analysis by microarray-based assay
Total RNA was extracted from postmortem PFC tissue using the miRNeasy Mini Kit (QIAGEN, Valencia, CA, USA). Total RNA was quantified by NanoDrop 8000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The RNA integrity number (RIN) was determined using the RNA 6000 Nano Lab-on-a-Chip kit and the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). The RIN ranged from 5 to 7 for the 46 RNA samples. Genome-wide gene expression levels were quantified using the Illumina HumanHT-12 v4 Expression BeadChip (Illumina Inc, San Diego, CA, USA). To avoid potential batch effects in expression array-based assays, biotinlabeled cDNAs generated from total RNA samples of six AUD cases and six matched controls were loaded onto the same BeadChip (i.e., 12 samples per Chips; in total, four chips were used for the 46 RNA samples). Each array targets 43,270 coding transcripts and 3,961 noncoding transcripts (including 574 miRNA transcripts). Probe intensity and gene expression data were analyzed using Illumina GenomeStudio software V2011.1 (Gene Expression Module V1.9.0).
Low-level analysis of microarray data was performed in R 2.15.1 (http://www.r-project.org/) using the Bioconductor package lumi. The variance stabilizing transformation (VST) method and the robust spline normalization (RSN) method were applied to all arrays. After normalization, annotated genes with intensities indistinguishable from background noise (detection p value > 0.05) in more than half of the RNA samples were removed. The R package sva was applied to remove batch effects directly.
Differential expression analysis of miRNAs and mRNAs
Genome-wide gene expression alterations in postmortem PFC of AUD cases were examined using multiple linear regression analysis with adjustment for covariates such as sex, age, and PMI. To exclude false positive signals by chance, a permutation test with a million iterations was employed by use of the R package lmPerm for multiple linear regression analyses, in which the AUD status of each subject was relabeled randomly in each test. The q-value was computed for each nominal P value by controlling the false discovery rate (FDR) at 0.05 using the R package qvalue [17]. All statistical analyses were implemented using the statistical package R (version 2.15.1) (http:// www.r-project.org/).
miRNA-mRNA expression correlation analysis and miRNA target prediction
Given that miRNAs potentially suppress the expression of their target genes, Pearson correlation analysis was applied to identify mRNAs that were negatively correlated with differentially expressed miRNAs in their expression levels using gene expression data from the control subjects. The obtained miRNA-mRNA pairs were subsequently validated by two integrative bioinformatics programs: miRWalk [18] and miRecords [19]. For each differentially expressed miRNA, we obtained a list of putative target genes that were predicted by at least two independent programs under default settings.
Gene set enrichment analysis
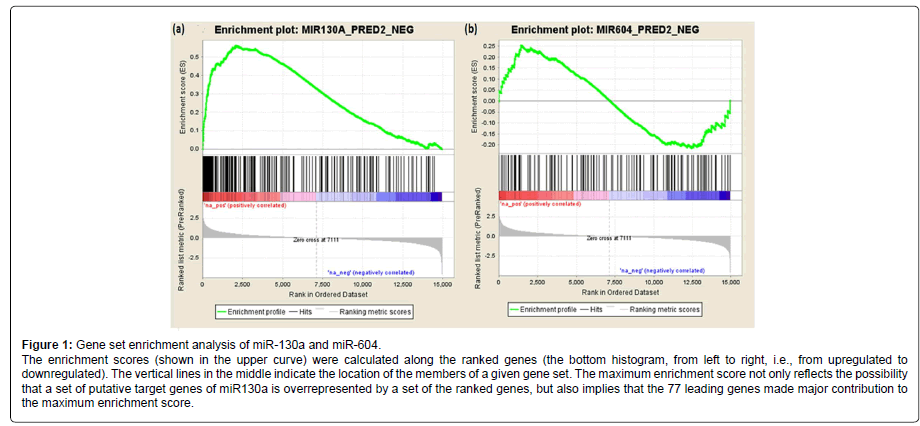
Gene set enrichment analysis (GSEA) was performed as described previously [20,21]. 14,950 genes with expression levels detectable by the microarray approach were ranked by the direction of expression changes (up- or downregulated) and -log10 (p values) that reflected gene expression differences between AUD cases and matched controls. Then the list of pre-ranked genes and the list of predicted target genes of differentially expressed miRNAs were used to run GSEA. A set of miRNA target genes that were enriched in differentially expressed genes was obtained.
Results
Differentially expressed miRNAs and mRNAs in AUD cases
Two miRNAs showed differential expression in AUD cases [miR- 130a (downregulated): regression coefficients = -0.05, Ppermutation= 0.023; miR-604 (upregulated): regression coefficients = 0.05, Ppermutation = 0.019], but neither P values survived multiple-testing correction (q > 0.05). Additionally, we found that 787 coding genes were differentially expressed (351 were upregulated and 436 were downregulated) in AUD cases (1.6×10-5 ≤ Ppermutation ≤ 0.05), but none of these P values survived multiple-testing correction (q > 0.05).
Expression correlation of miR-130a (or miR-604) and its target mRNAs
Among all the genes (n = 14,950) that were detectable by the microarray approach, 467 genes showed negative correlations with miR-130a in their expression levels (Pcorrelation < 0.05), and 327 genes showed negative correlations with miR-604 in their expression levels (Pcorrelation < 0.05). With the use of two prediction programs miRWalk and miRecords, out of the 467 genes showing negative correlation with miR-130a in gene expression, 202 genes were predicted to be putative targets of miR-130a; out of the 327 genes showing negative correlation with miR-130a in gene expression, 116 genes were predicted to be putative targets of miR-604.
| Genes | Expression (controls)a | Expression (AUD cases)a | Effect sizeb | Pnominalc | Ppermutationd |
|---|---|---|---|---|---|
| FBXO21 | 9.47 | 9.59 | 0.12 | 0.001 | 0.002 |
| ITPR2e | 6.72 | 6.82 | 0.12 | 0.002 | 0.002 |
| FAM65B | 7.21 | 7.37 | 0.15 | 0.005 | 0.004 |
| ZFYVE21 | 10.06 | 10.26 | 0.23 | 0.006 | 0.006 |
| BBS2 | 9.17 | 9.31 | 0.18 | 0.005 | 0.007 |
| GPR177 | 7.78 | 7.99 | 0.23 | 0.007 | 0.008 |
| JARID2e | 8.27 | 8.41 | 0.17 | 0.011 | 0.010 |
| RAB34 | 7.05 | 7.34 | 0.29 | 0.012 | 0.011 |
| C10orf54e | 6.89 | 7.25 | 0.31 | 0.012 | 0.012 |
| SPON1 | 8.02 | 8.43 | 0.43 | 0.034 | 0.022 |
| ATP1A2 | 9.85 | 10.17 | 0.44 | 0.027 | 0.024 |
| ATP1B2 | 9.41 | 9.83 | 0.44 | 0.056 | 0.029 |
| WIF1 | 7.18 | 7.44 | 0.39 | 0.037 | 0.032 |
| AFF4 | 7.66 | 7.80 | 0.17 | 0.022 | 0.033 |
| BAG3e | 7.74 | 8.25 | 0.45 | 0.035 | 0.033 |
| TUBB2Be | 12.31 | 12.66 | 0.32 | 0.029 | 0.033 |
| SDC4 | 8.01 | 8.53 | 0.49 | 0.040 | 0.034 |
| CPT2 | 7.43 | 7.59 | 0.15 | 0.029 | 0.035 |
| SLC39A12 | 7.25 | 7.54 | 0.35 | 0.060 | 0.038 |
| PCDHB5 | 6.65 | 6.69 | 0.05 | 0.073 | 0.040 |
| SLC1A2 | 10.29 | 10.74 | 0.54 | 0.040 | 0.041 |
| GJA1 | 8.78 | 9.23 | 0.54 | 0.054 | 0.047 |
| TMEM192 | 6.71 | 6.75 | 0.04 | 0.050 | 0.048 |
aMean gene expression levels in controls (or AUD cases) measured by Illumina's
HT-12 v4 Gene Expression BeadChip.
bEffect size (or regression coefficients) obtained by multiple linear regression
analysis with adjustment for sex, age, and PMI.
cObserved P values obtained by multiple linear regression analysis with adjustment
for sex, age, and PMI.
dPermutation P values obtained by R package lmPerm analysis.
eGenes with expression levels negatively correlated with that of miR-130a.
Table 1: 23 differentially expressed genes predicted to be targets of miR-130a.
Gene set enrichment analysis of miR-130a and miR-604 target genes
We further analyzed the association of the 202 predicted target genes of miR-130a (or 116 predicted target genes of miR-604) with AUDs using gene set enrichment analysis (GESA). The pre-ranked [by -log10(p-value)] list of all detectable genes (i.e., 14,950 genes) and the list of the 202 predicted target genes of miR-130a (or the list of the 116 predicted target genes of miR-604) were uploaded into the program GSEA to analyze whether the predicted target genes of miR-130a or miR-604 were overrepresented by the differentially expressed genes. As shown in Figure 1, the predicted target genes of miR-130a were overrepresented by those genes that were differentially expressed in AUD cases (Pnominal < 0.001), and 77 of the 202 predicted target genes of miR130a were found to be the leading genes for obtaining the maximum enrichment score. However, the predicted target genes of miR-604 were not enriched by the differentially expressed genes (Pnominal > 0.05).
Functional annotation of putative target genes of miR-130a
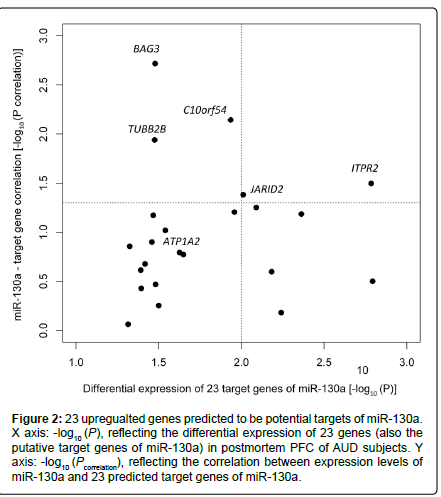
Among the 202 putative target genes of miR-130a, 24 (11.9 %) showed differential expression in AUD cases, and 23 of the 24 genes were upregulated in AUD cases at Ppermutation ≤ 0.05 (Table 1). The proportion (23/202) of upregulated putative target genes of miR-130a was significantly higher than that of the set of all upregulated genes among all genes detected (351/14,950) (hypergeometric test: p = 4.0×10-10). We further examined the correlation between the expression levels of these 23 upregulated genes and the downregulated miR-130a in all 46 samples (23 cases + 23 matched controls) with adjustment for covariates (AUD status, sex, age, and PMI). Five (ITPR2, JARID2, C10orf54, BAG3, and TUBB2B) of the 23 upregulated genes showed significant negative correlations with miR-130a (downregulated) in their expression levels. Moreover, ITPR2 and JARID2 were among the top genes (Ppermutation < 0.010) showing differential expression in postmortem PFC of AUD cases (ITPR2: Ppermutation = 1.6×10-3; JARID2: Ppermutation = 9.8×10-3) (Figure 2). To explore the biological function of the above 23 upregulated genes, we uploaded the list of these 23 genes into the DAVID web server. Highly enriched gene ontology (GO) terms were assigned into eight clusters. As shown in Figure 3, the top cluster (with a fold enrichment score of 1.25) consisted of 14 GO terms, and 11 genes in this top cluster mainly participate in cell projection, ion transport, and neuron projection. Two genes (ITPR2 and ATP1A2) were implicated in all 14 GO terms.
Figure 2:23 upregualted genes predicted to be potential targets of miR-130a. X axis: -log10 (P), reflecting the differential expression of 23 genes (also the putative target genes of miR-130a) in postmortem PFC of AUD subjects. Y axis: -log10 (Pcorrelation), reflecting the correlation between expression levels of miR-130a and 23 predicted target genes of miR-130a.
Figure 3: DAVID functional clustering of 23 upregulated genes predicted to be the targets of miR-130a. 23 upregulated genes (also the putative targets of miR-130a) were uploaded into the DAVID server to highlight those genes that were highly enriched by functionally related GO terms. 11 of the 23 upregulated genes were associated with specific GO terms. Each column indicates a particular GO term, and each row indicates a particular gene. Colors in the 2-D map indicate whether a gene is reported in a corresponding GO term (green) or not (dark). ITPR2, inositol 1,4,5-triphosphate receptor, type 2; ATP1A2, ATPase, Na+/K+ transporting, alpha 2 (+) polypeptide; SLC39A12, solute carrier family 39 (zinc transporter), member 12; ATP1B2, ATPase, Na+/ K+ transporting, beta 2 polypeptide; SLC1A2, solute carrier family 1 (glial high affinity glutamate transporter), member 2; CPT2, arnitine palmitoyltransferase 2; PCDHB5, protocadherin beta 5; ZFYVE21, zinc finger, FYVE domain containing 21; RAB34, member RAS oncogene family; FAM65B, family with sequence similarity 65, member B; BBS2, Bardet-Biedl syndrome 2.
Discussion
The chronic use of alcohol provokes gene expression changes, allowing the brain to adapt to the environment through homeostatic mechanisms in distinct signaling pathways. In the present study, genome-wide differential expression analysis revealed that two miRNAs (miR-130a and miR-604) and 787 coding genes were dysregulated in postmortem PFC of AUD subjects. Since miRNAs are able to inhibit the expression of their target mRNAs, the significantly negatively correlated miRNAs and mRNAs potentially constitute the functional miRNAmRNA pairs. We used an integrative analysis approach to identify the putative target genes (or mRNAs) of miR-130a and miR-604. Gene set enrichment analysis indicated that the putative target genes of miR-130a (but not miR-604) were over-represented by differentially expressed genes.
Among the 23 upregulated genes that were predicted to be potential targets of miR-130a (Table 1), five (ITPR2, JARID2, C10orf54, BAG3, and TUBB2B) were negatively correlated with miR-130a in their expression levels. Two (ITPR2 and JARID2) of them are of interest. Functional clustering analysis suggests that ITPR2, which is an ion channel gene, might be involved in the development of AUDs. ITPR2 encodes the inositol 1, 4, 5-triphosphate receptor (type 2), which is a ligand-gated ion channel that can be activated by cytosolic Ca2+ and inositol trisphosphate (IP3). A growing number of studies indicate that ethanol alters nerve signaling by interacting with neurotransmitter receptors and ion channels in the central nervous system, particularly the pentameric ligand-gated ion channels [e.g., GABA(A) and nicotinic acetylcholine receptors] [22] and voltage-gated calcium channels [23]. Genetic variants (rs1049380 and rs4654) in ITPR2 haven been reported to be associated with alcohol dependence by Genome-wide association studies [24]. Moreover, ITPR2 and several other genes form a network to influence the function of neural synapses, and thus are potentially involved in traits such as substance use, stress, obesity and so on [25]. JARID2 encodes a nuclear protein that is essential for embryogenesis, and overexpression of JARID2 negatively regulates cell proliferation. Although the direct influence of JARID2 on the risk of AUDs has not been observed, variation in JARID2 has been found to be associated with AUD-relevant disorders such as schizophrenia [26,27]. To extend these findings, future studies are needed to examine whether the impact of miR-130a on the susceptibility of individuals to AUDs is through regulating the ion channel activity.
To our current knowledge, only one study conducted by Lewohl et al. [15] is known to have systematically examined AUD-associated miRNAs in postmortem brains of AUD subjects (14 AUD cases vs. 13 controls), and a total of 35 miRNAs were identified to be up-regulated in AUD cases. Compared to the study by Lewohl et al. [15], the novelty of the present study is that we used an integrative approach to screen the putative targets of miRNAs by combining miRNA target prediction and miRNA-mRNA expression correlation analysis. Nevertheless, the two AUD-associated miRNAs (miR-130a and miR-604) identified in the present study were not among the 35 miRNAs identified in the study by Lewohl et al. [15]. The inconsistent findings may be due to (1) the inclusion criteria of AUD cases were similar but may not be exact the same, thus leading to different genetic backgrounds; (2) the sample size of these two studies were different (the sample size was larger for the present study); and (3) both studies did not consider the accumulative effect of multiple miRNAs as well as the interactive effect of miRNAs and environmental factors on the risk for AUDs.
To have a better understanding of the role of miRNA-mRNA pairs in the etiology of AUDs, the present study needs to be improved in several aspects. First, although the sample size is larger than that of any published studies that used postmortem PFC tissues for AUD studies, the moderate sample size (23 cases and 23 controls) may not have sufficient statistical power to identify most differentially expressed miRNAs and mRNAs. Thus, studies with a larger sample are warranted. Second, although we applied an integrative approach to identify putative targets of miRNAs, more efforts should be made to confirm the predicted target genes of miRNAs. Functional study approaches (e.g., 3’ UTR-reporter gene assays) are necessary to validate the interaction of miRNAs and their target mRNAs. Third, the present study investigated the miRNA regulatory activity in only one brain region (PFC). The findings should be extended to other reward-related brain regions. Fourth, given the relatively low specificity and sensitivity of the microarray method, other advanced gene expression detection systems such as the next-generation sequencing technology (e.g., miRNA-Seq and mRNA-Seq) would more ideally be used.
In summary, the present study provided evidence that miR-130a and its target genes are involved in the neuroadaptations to alcohol. We thus conclude that dysfunction of miRNAs in human PFC may contribute to the etiological process of AUDs, and that miR-130a and its target genes are potential targets for pharmacotherapy of AUDs.
Acknowledgements
This study was supported by the National Institutes of Health (NIH) Grants K99/R00 DA022891 (HZ) and P50 AA12870 (JHK & JG). The authors are grateful to the Australian Brain Donor Programs New South Wales Tissue Resource Centre for providing alcoholic and control brain tissues for this study. The centre is supported by the University of Sydney, the National Health and Medical Research Council of Australia, and the National Institute on Alcohol Abuse and Alcoholism. We also thank the deceased subjects’ next of kin for providing informed written consent for the studies. The authors declare that they have no competing interests.
References
- Grant BF, Dawson DA, Stinson FS, Chou SP, Dufour MC, et al. (2004) The 12-month prevalence and trends in DSM-IV alcohol abuse and dependence: United States, 1991-1992 and 2001-2002. Drug Alcohol Depend 74: 223-234.
- Schuckit MA (2009) Alcohol-use disorders. Lancet 373: 492-501.
- Miranda RC, Pietrzykowski AZ, Tang Y, Sathyan P, Mayfield D, et al. (2010) MicroRNAs: master regulators of ethanol abuse and toxicity? Alcohol Clin Exp Res 34: 575-587.
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102-114.
- Breving K, Esquela-Kerscher A (2010) The complexities of microRNA regulation: mirandering around the rules. Int J Biochem Cell Biol 42: 1316-1329.
- Liu J, Githinji J, Mclaughlin B, Wilczek K, Nolta J (2012) Role of miRNAs in neuronal differentiation from human embryonic stem cell-derived neural stem cells. Stem Cell Rev 8: 1129-1137.
- Smalheiser NR, Lugli G (2009) microRNA regulation of synaptic plasticity. Neuromolecular Med 11: 133-140.
- Wibrand K, Pai B, Siripornmongcolchai T, Bittins M, Berentsen B, et al. (2012) MicroRNA regulation of the synaptic plasticity-related gene Arc. PLoS One 7: e41688.
- Abe M, Bonini NM (2013) MicroRNAs and neurodegeneration: role and impact. Trends Cell Biol 23: 30-36.
- Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, et al. (2008) Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron 59: 274-287.
- Tang Y, Banan A, Forsyth CB, Fields JZ, Lau CK, et al. (2008) Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin Exp Res 32: 355-364.
- Sathyan P, Golden HB, Miranda RC (2007) Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J Neurosci 27: 8546-8557.
- Guo Y, Chen Y, Carreon S, Qiang M (2012) Chronic intermittent ethanol exposure and its removal induce a different miRNA expression pattern in primary cortical neuronal cultures. Alcohol Clin Exp Res 36: 1058-1066.
- Laufer BI, Mantha K, Kleiber ML, Diehl EJ, Addison SM, et al. (2013) Long-lasting alterations to DNA methylation and ncRNAs could underlie the effects of fetal alcohol exposure in mice. Dis Model Mech 6: 977-992.
- Lewohl JM, Nunez YO, Dodd PR, Tiwari GR, Harris RA, et al. (2011) Up-regulation of microRNAs in brain of human alcoholics. Alcohol Clin Exp Res 35: 1928-1937.
- American Psychiatric Association (1994) Diagnostic and Statistical Manual of Mental Disorders, (4thedn), Washington, DC.
- Bahi A, Dreyer JL (2013) Striatal modulation of BDNF expression using microRNA124a-expressing lentiviral vectors impairs ethanol-induced conditioned-place preference and voluntary alcohol consumption. Eur J Neurosci.
- Dweep H, Sticht C, Pandey P, Gretz N (2011) miRWalk--database: prediction of possible miRNA binding sites by "walking" the genes of three genomes. J Biomed Inform 44: 839-847.
- Xiao F, Zuo Z, Cai G, Kang S, Gao X, et al. (2009) miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res 37: D105-110.
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102: 15545-15550.
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, et al. (2003) PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267-273.
- Sauguet L, Howard RJ, Malherbe L, Lee US, Corringer PJ, et al. (2013) Structural basis for potentiation by alcohols and anaesthetics in a ligand-gated ion channel. Nat Commun 4: 1697.
- Newton PM, Zeng L, Wang V, Connolly J, Wallace MJ, et al. (2008) A blocker of N- and T-type voltage-gated calcium channels attenuates ethanol-induced intoxication, place preference, self-administration, and reinstatement. J Neurosci 28: 11712-11719.
- Johnson C, Drgon T, Liu QR, Walther D, Edenberg H, et al. (2006) Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. Am J Med Genet B Neuropsychiatr Genet 141B: 844-853.
- Nikpay M, Seda O, Tremblay J, Petrovich M, Gaudet D, et al. (2012) Genetic mapping of habitual substance use, obesity-related traits, responses to mental and physical stress, and heart rate and blood pressure measurements reveals shared genes that are overrepresented in the neural synapse. Hypertens Res 35: 585-591.
- Pedrosa E, Ye K, Nolan KA, Morrell L, Okun JM, et al. (2007) Positive association of schizophrenia to JARID2 gene. Am J Med Genet B Neuropsychiatr Genet 144B: 45-51.
- Liu Y, Chen G, Norton N, Liu W, Zhu H, et al. (2009) Whole genome association study in a homogenous population in Shandong peninsula of China reveals JARID2 as a susceptibility gene for schizophrenia. J Biomed Biotechnol 2009: 536918.
Relevant Topics
- Addiction Recovery
- Alcohol Addiction Treatment
- Alcohol Rehabilitation
- Amphetamine Addiction
- Amphetamine-Related Disorders
- Cocaine Addiction
- Cocaine-Related Disorders
- Computer Addiction Research
- Drug Addiction Treatment
- Drug Rehabilitation
- Facts About Alcoholism
- Food Addiction Research
- Heroin Addiction Treatment
- Holistic Addiction Treatment
- Hospital-Addiction Syndrome
- Morphine Addiction
- Munchausen Syndrome
- Neonatal Abstinence Syndrome
- Nutritional Suitability
- Opioid-Related Disorders
- Relapse prevention
- Substance-Related Disorders
Recommended Journals
Article Tools
Article Usage
- Total views: 14954
- [From(publication date):
September-2013 - Dec 22, 2024] - Breakdown by view type
- HTML page views : 10286
- PDF downloads : 4668