Research Article Open Access
Development of Rapid Assay for Ribonucleotide Reduction by Mycobacterium Smegmatis Mc2 155 and their Biochemical Characterisation
Elhachemi Ahmed, Bendaha Mohammed Lamine, Benattouche Zouaoui, Kanoun Khedoudja and Abbouni Bouziane*Laboratoire de synthèse de l’information environnementale, Département de Biologie, Faculté des Sciences, Université Djillali Liabès de Sidi Bel Abbès, Algeria
- Corresponding Author:
- Prof. Dr. Abbouni Bouziane
Laboratoire de synthèse de l’information environnementale
Département de Biologie, Faculté des Sciences
Université Djillali Liabès de Sidi Bel Abbès, Algeria
E-mail: abbounibouziane@yahoo.de
Received date: March 06, 2012; Accepted date: March 24, 2012; Published date: March 26, 2012
Citation: Ahmed E, Lamine BM, Zouaoui B, Khedoudja K, Bouziane A (2012) Development of Rapid Assay for Ribonucleotide Reduction by Mycobacterium Smegmatis Mc2 155 and their Biochemical Characterisation. J Biotechnol Biomaterial 2:132. doi:10.4172/2155-952X.1000132
Copyright: © 2012 Ahmed E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
Mycobacterium smegmatis mc2 155 contains a Ribonucleotide reductases (RNR), which catalyses the irreversible reduction of ribonucleotides to the corresponding 2 �?deoxyribonucleotides required for DNA replication and cell proliferation.
The aim of this work was the development of a rapid assay for ribonucleotide reduction by Mycobacterium smegmatis mc 2 155 and their biochemical characterisation.
For this purpose, the cells of Mycobacterium smegmatis mc 2 155 were permeabilized with two organic solvents toluene and ether for two times (2,5 min) to develop a new assay for ribonucleotide reduction. Due the importance of the growth phase in determining the yield of biomass and ribonucleotide reductase activity of Mycobacterium smegmatis mc 2 155, a correlation between Ribonucleotide reductase activity and growth of Mycobacterium smegmatis mc 2 155 in modified seed medium has been investigated.
For the enrichment of the Ribonucleotide reductase, different purification procedure has been achieved by using fast protein liquid chromatography (FPLC) with superdex G-200 chromatography and Phenyl-Superose HR 5/5 and the enzyme activity was assayed by using (HPLC).
Ribonucleotide reductase activity was detectable in the 40-60% ammonium sulphate fraction. A further purification procedure by gel filtration on the superdex G-200 led to a dissociation of the both subunits. Therefore, a biochemical complementation assay was necessary to identify ribonucleotide reductase activity. The obtained specific activity of the purified protein was 1790 pmol per mg per min with an overall yield of 10%. The purified small subunit of MS2- protein was detected on SDS-PAGE, which was showed a strong band that corresponds to an apparent molecular weight of 38.5 KDa.
The obtained results of ribonuleotide reduction activity with ether permeabilized cells of Mycobacterium smegmatis mc 2 155 presented a comparable enzyme activity for both times, while with toluene permeabilized cells indicated a low enzyme activity. Furthermore, the obtained results of the correlation between ribonucleotide reductase activity and the growth showed that Ribonucleotide redutase is a peak enzyme.
Finally, the permeabilisation of the cells of M. smegmatis mc 2 155 with ether and toluene for short time facilitated us to develop a rapid assay for ribonucleotide reductase activity of others gram positive bacteria.
Keywords
Mycobacterium smegmatis mc2155; Ribonucleotide reductase; Metallo-cofactor; Small subunit MS2; A large subunit MS1
Introduction
Mycobacterium tuberculosis is an aerobic, gram-positive bacterium, non motile containing a high G+C-content genome of approximately 4 x 106 bp with a long generation time 24 hours and is belonging to the family of Actinomycetales [1]. Mycobacterium tuberculosis remains in all of its manifestations important causes of human morbidity and mortality worldwide. The most important mycobacterial disease is leprosy, caused by Mycobacterium leprae; tuberculosis caused by Mycobacterium tuberculosis and déterminated infection with M. avium in individuals with acquired immune deficiency syndrome [2-4]. Mycobacterium tuberculosis can remain in a dormant, non replicating state in the human host for years and can be reactivated to cause disease under certain immunological and/or environmental conditions [5].
Clearly, new approaches to the development of anti-tuberculosis therapy are necessary. Inactivation of ribonucleotide reductase (RNR); the cell cycle-regulated, allosteric enzyme that catalyses the reduction of nucleoside diphosphates (NDPs) to the corresponding 2´-deoxy nucleoside diphosphates (dNDPs) [6]; may be attractive target for new anti-tuberculosis agents. The enzymatic activity is the first step in DNA synthesis and has therefore been recognized as primary target in the design of cancer chemotherapeutic agents. Furthermore, RNR is gaining wide acceptance as a target for antiviral agents and properly as anti-parasites chemotherapy [7]. There are very important for identification of Mycobacterium tuberculosis RNR as potential drug target: the inhibition of RNR altered the growth patterns of these bacteria. Studies in the 1960s and 1970s showed that Mycobacterium smegmatis cultured in iron-depleted media displayed altered, elongated morphology with decreased DNA synthesis and increased activity of DNA repair enzymes [8]. When grown in the presence of the radical scavenger hudroxyurea Mycobacterium smegmatis contained a decreased DNA/protein ratio, with an increase in DNA polymerase and ATP-dependent DNAase activities, measured in crude extracts [9]. The authors speculated that their results were consistent with the inhibition of mycobacterial activity of RNR. As part of an investigation of the regulation of mycobacterial growth and DNA synthesis by RNR, we report in this work the development of new assay for ribonucleotide reduction, the correlation between ribonucleotide reduction and growth and the purification of this enzyme by chromatography methods.
Materials and Methods
All regents, and reagents and nucleotides were of the high purity. Unlabeled substrate and effectors nucleotides were from Boehringer, Mannhein (Germany), [5-3H] CDP, ammonium salt (0.37-1.1 TBq/ mmol, 10-30 Ci/mmol) was purchased from Amersham-Buchler (Braunschweig, Germany), Dithiothreitol was obtained from Sigma Chemical Co, St,. Louis, MO, USA. Hydroxyurea, 2-mercaptoethanol, streptomycin sulfate and bovine serum albumin were purchased from Serva Feinbiochemica GmbH & Co., KG (Heidelberg, Germany), Alkaline phosphatase was purchased from Boehringer, Mannheim. All others chemicals agents were obtained from Merck, Darmstadt (Germany). 2’, 5’-ADP Sepharose, 4B Superose 12, UNOTM Q12 and Phenylsuperose were obtained from Pharmacia LKB (Freiburg, Germany). ViskingR dialysis tubes were obtained from Serva Feinbiochemica GmbH & Co., KG (Heidelberg, Germany). All others chemicals agents were obtained from Merck, Darmstadt (Germany). Protein content of cell-free extracts was determined according to the procedure of Lowry and co-workers [10] using bovine serum albumin (Sigma, St. Louis) as standard.
Bacterial strains and culture conditions
The strain of Mycobacterium smegmatis mc2155 was supplied by institute of microbiology Hannover-Germany. Mycobacterium smegmatis mc2155 was grown in modified seed media described by Thaler and Diekmann [11], which was containing the following components: glucose 2%, pepton of casein 1%, yeast extract 1%, NaCl 0.25% (pH 7, 2-7, 6) and modified by the adding of 1M MgSO4 2% (v/v) and 0.2 % (v/v) tween 80 to prevent the clumping of the cells of Mycobacterium smegmatis mc2155.
Large scale-incubation of the strains Mycobacterium smegmatis mc2155 was carried out according the method of Griepenburg and coworkers [12], which was grown in modified seed media described above at 37°C in 10 L fermentor with a aeration of 4 L/min and an agitation 250 rpm using fermentor Drive Assembley (New Brunswick Scientific Co. Inc. NJ., USA). Because Ribonucleotide reductase (as cell-cycle controlled enzyme) is present only in proliferating cells and previous investigations had showed highest activity at the late logarithmic phase of the growth curve [13]. Mycobacterium smegmatis mc2 155 was grown for 10 hours and harvested at an optical density at 578 nm 16-20.
Preparation of nucleotide of permeable cells
The fresh cells has been harvested in the late-exponential growth phase that displayed higher Mn-RNR activity were washed with 85 mM Potassium phosphate buffer and osmotically adjusted with 0.6 M sorbitol. The pH of this buffer was adjusted 7.2 for the cells grown in seed medium. The cells were collected by low speed centrifugation and adjusted to defined cell number, gently dispersed (30s x 6) in ultrasonic bath (Sonorex RK 102 H, Bandelin). The cells were permeabilized in teflon FEP tubes (Nalge, Rochester, N. Y., USA) by treatment with organic solvent (1% toluene, 10 min, 3 × 109 CFU/ml or 501% (v/v) ether, 1 min, 1 × 109 CFU/ml]. The permeabilized cells were washed twice with the same buffer as above omitting sorbitol in the last washing prior to assay of robonucleotide reductase activity. The suspension of the permeabilized cells were either immediately assayed or divided into small aliquots and quickly frozen liquid nitrogen for storage at temperature (-80°C). Such frozen cells were thawed only once.
Purification of ribonucleotide reductase
All procedures were carried out at temperature of 4°C. 15 g of frozen cells were thawed and suspended in 22,5 ml buffer A (85 mM potassium phosphate buffer, containing 2 mM dithiothreitol, pH, 6.6) and the cells were broken by sonification using a Branson sonifier B-12 at 50 % power for 6 × 10s. The final disrupture was achieved by a threefold passage through prechilled French pressure cell (40 ml, SLM Instruments, Inc., Urbana, USA) at 1500 psi. The homogenate was centrifuged at 19000 rpm for 30 min and the cells debris was discarded. The supernatant was precipitated by addition of 5% streptomycin sulphate in buffer A to the supernatant to the final concentration of 1.5%. The resulting suspension was stirred for an additional 10 min, and the precipitate was removed by centrifugation at 19000 rpm for 30 min. Solid ammonium sulfate was slowly added to the supernatant to 0-40% saturation with stirring. After the addition was completed, the suspension was stirred for further 10 min and the precipitate was collected by centrifugation (19000 rpm for 30 min). Ammonium sulfate was slowly added to the supernatant to obtain 40-60% saturation with stirring. After the addition was completed, the suspension was stirred for 30 min and the precipitate was collected by centrifugation (19000 rpm for 30 min) and resuspended in 10 ml of buffer. The suspension was dialyzed against the same buffer for 2 hours with one buffer change [14].
Assay ribonucleotide reductase activity
The optimum condition of pH, ionic strength, temperature and time utilized for a assaying the RNR activity of Corynebacterium ammoniagenes [15] was used to display the RNR activity of Mycobacterium smegmatis mc2155. Assay mixtures contained, in a total volume of 100 μl 85 mM potassium phosphate buffer, pH 6.6, 0.5 μCi [5-3H] CDP (spec. act 10-20 Ci/ mmol), free from ethanol in vacuum; 10 nmol CDP (50 μM), 10 nmol ATP (50μM) as positive effector; 1.2 μmol dithiothreitol (6 mM); 0.2 μmol MgCl2 (1 mM); and up to 1.5 mg enzyme protein. The mixture was incubated for 20 min at 30°C and the reaction was stopped by boiling for 3 min. Assays with crude protein or with fractioned protein were treated with 50 μg pronase (from Streptomyces griseus, Boehringer) for 90 min at 37°C to destroy heatstable nucleoside N-glycosylase, followed by briefly boiling to destroy the pronase. The nucleotides in the assay solution were converted to the corresponding nucleosides by the addition of 10 units of alkaline phosphatase (from calf intestine, Boehringer) in 10 μl 1 M Tris/HCL buffer, pH 9.9, and incubated at 37°C for 90 min. The sample was boiled for 3 min and centrifuged. The supernatant was decanted and used for automatically nucleoside analysis. The system consists of an automatic sample injector (ASI 120 from Ismatec :sample capacity, 120) which injects 20 μl aliquots onto 25 × 0.4-cm cation-exchange HPLC column (Aminex A-9, Biod Rad) operated in a isocratic liquid chromatography system (model 330 from Beckman). The column is thermostated at 37°C and eluted with 0.1 M borate buffer, pH 8.2. The ribonucleoside, deoxyribonucleoside and free base present in the assay are separated with a flow rate of 0.8 ml/min and are fractioned directly into scintillation vials. One assay can be analysed with this system in 15 min.
Molecular weight determination
The molecular weight of the native ribonucleotide reductase was estimated by gel filtration on SuperoseTM12column (HR 10/30), equilibrated with 85 mM potassium phosphate buffer, containing 2 mM DTT, pH 6.6. For FPLC a flow rate 0.3 ml /min was used and 3 min-fractions were collected. The chromatography, fractions collection and recording of column runs were controlled automatically (gradient programmer GP-250 plus, Pharmacia LKB Freiburg, FRG). The superposeTM 12column was calibrated with reference proteins of known molecular weight; 158 KDa: aldolase, 45.5 KDa : ovalbumin, 35 KDa : ß-lactoglobulin and 12.5 KDa: cytochrome C (Pharmacia). The three fractions containing 100 KDa dimeric CG2 subunit were immediately identified by biochemical complementation with CA1- protein of C. ammoniagenes for recovery of the enzymatic activity.
SDS-PAGE
Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-Page) was performed according the method [16] in a 16% polyacrylamide gel. The gel was stained with Coomassie blue. Apparent relative molecular weights were obtained by comparison to a mixture of reduced reference proteins from molecular weight standard kit (Bio-rad). Relative mobility values (Rf) for ribonucleotide reductase and calibration proteins has been calculated from their position after electrophoresis.
Results
Development of a rapid assay for examination of ribonucleotide reductase from Mycobacterium smegmatismc2155
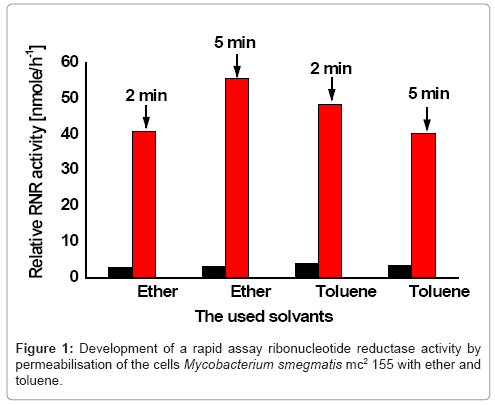
To make the cells of Mycobacterium smegmatis mc2155 permeable to the nucleotide, the cells were permeabilized with toluene and ether two solvents because. For optimization of ribonucleotide reduction assay, the enzyme activity of the toluene and ether permeabilized cells was tested for two times (2, 5 minutes) respectively.
The obtained ribonuleotide reduction with ether permeabilized cells of Mycobacterium smegmatismc2155 presented a comparable enzyme activity for both times (Figure 1). While the ribonucleotide reduction with toluene permeabilized cells indicated a low enzyme activity. This greatly facilitated assay of ribonucleotide reductase in Mycobacterium smegmatis mc2155.
Correlation between Ribonucleotide reductase activity and growth of Mycobacterium smegmatis mc2155 in modified Seed medium
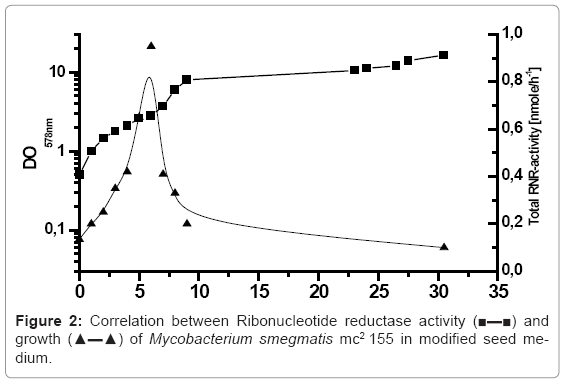
The purification of ribonucleotide reductase of Mycobacterium smegmatis mc2155 required the production of the sufficient biomass with higher ribonucleotide reductase activity. For this purpose, the cells of Mycobacterium smegmatis mc2155 were inoculated in modified seed medium with Tween 80, MgSO4 to prevent the clumping of the cells, incubated in 10 litre bioreactor, at 37°C with aeration 250 rpm. Due the importance of the growth phase in determining the yield of biomass and ribonucleotide reductase activity of Mycobacterium smegmatis mc2155, the cells were harvested in regulatory range time of 1 hour, and the growth was monitored by measuring the optical density at 578 nm. Furthermore, the cells were washed twice or more and permeabilised with ether according to the protocol described above. The correlation between Ribonucleotide reductase activity and growth of Mycobacterium smegmatis mc2155 in modified medium is presented in figure 2. The bioreactor was inoculated with initial optical density of 0.5 of Mycobacterium smegmatis mc2155, the cells concentration has increased during the exponential growth phase OD 578 nm of 8. The cells of Mycobacterium smegmatis mc2155 required 9 hours to reach this growth phase and after that the cells were reached the stationary growth phase. In conclusion, the determined generation rate of incubated Mycobacterium smegmatis mc2155 in modified seed medium was 2.25 hours. The correlated ribonucleotide reductase activity and growth of Mycobacterium smegmatis mc2155 presented in figure 2 showed that the maximum total Ribonucleotide reductase activity of Mycobacterium smegmatis mc2155 (0.95 n mole/h) was obtained after 6 hours incubation by optical density of 2.8. After that, the activity of RNR decreased drastically and reached within half hour value of (0.41 n mole/h). The obtained results indicated that ribonucleotide reductase of Mycobacterium smegmatis mc2155 manifested as a peak enzyme, which the maximum RNR activity reached in the last logarithmic growth phase.
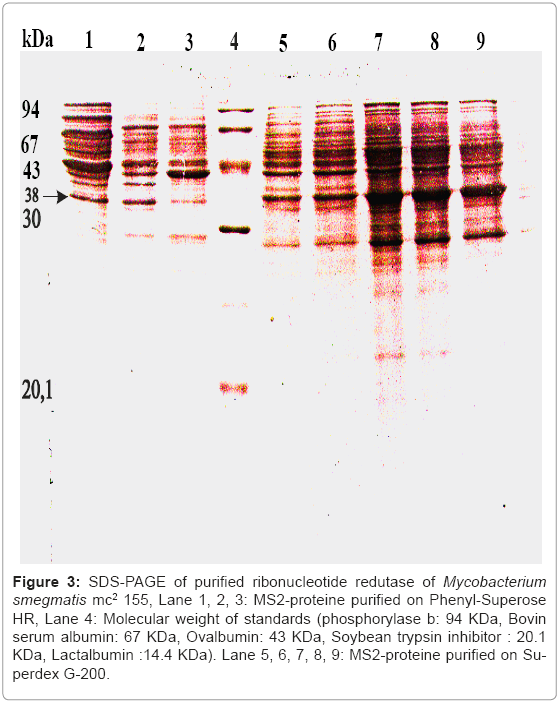
Figure 3: SDS-PAGE of purified ribonucleotide redutase of Mycobacterium smegmatis mc2 155, Lane 1, 2, 3: MS2-proteine purified on Phenyl-Superose HR, Lane 4: Molecular weight of standards (phosphorylase b: 94 KDa, Bovin serum albumin: 67 KDa, Ovalbumin: 43 KDa, Soybean trypsin inhibitor : 20.1 KDa, Lactalbumin :14.4 KDa). Lane 5, 6, 7, 8, 9: MS2-proteine purified on Superdex G-200.
Purification of ribonucleotide reduction of Mycobacterium smegmatis mc2155
A rapid and efficient purification procedure of ribonucleotide reductases from several sources has been described using several methods of chromatography especially by gel filtration on superdex G-200 [17]. Ribonucleotide reductase activity was not detected in the crude extract; however, it was detectable in the 40-60% ammonium sulphate fraction. Furthermore, it was purified by gel filtration on the superdex G-200. The enzyme activity was stable throughout the purification procedure. However, the activity was decreased after 1 month of storage at -70°C if the concentration of protein was lower than 1 mg/ml. The partially purified enzyme was stable throughout 1.5 hour incubation during the activity assay in the presence of substrates and effectors. In order to avoid the loss of the total enzymatic activity a sorbitol was added to purification buffers. Despite the addition of protein stabilizers, total enzyme activity decreased during the purification and this decrease seems to be dependant of protein concentration. The protein solution was applied on Superdex G-200 column and eluted with buffer A. This step has lead to a dissociation of the both subunits of Ribonucleotide reductase of Mycobacterium smegmatis mc2155. Therefore, a biochemical complementation assay was necessary to identify ribonucleotide reductase activity. In the fact, that each used procedure for purification of ribonucleotide redutase of Mycobacterium smegmatis mc2155 affects the structure and the biological activity of this enzyme, this observations leads to confirm that both subunits were not able to form functionally active without biochemical complementation assay.
Functionally, The specific activity of the purified ribonucleotide reductase was 1790 pmol per mg with an overall yield of 10% summarized in table 1 by using the experimental procedure described in Materials and Methods. The purity of enzyme purification was tested by SDS-PAGE (Figure 3). The Purified ribonucleotide reductase showed a strong band with a mobility that corresponds to a apparent molecular weight of 38.5 KDa. As shown in (Figure 3) two apparent peaks corresponding to the MS1-protein and MS2-protein of ribonucleotide redutase of Mycobacterium smegmatis mc2155 activity after the complementation assay of MS2-protein with a large subunit MS1-protein of ribonucleotide reductase, and the MS1-protein with MS2-protein.
| Step | Protein mg/ ml |
Specific activity pmole. mg-1.h-1 |
Total activity pmole. h- |
Yield % | Purification factor |
|---|---|---|---|---|---|
| Sulfate ammonium precipitation (40-60 %) |
64.0 | 540 | 45 | 100 | 1,0 |
| MS2-Protein (Superdex G-200) |
2.1 | 1790 | 4.47 | 10 | 3.31 |
| MS2- Protein(Phenyl- SuperoseHR |
1.06 | 1150 | 2.87 | 6.3 | 2.12 |
Table 1: Purification of ribonucleotide reductase from Mycobacterium smegmatis mc2 155.
The Development of a rapid assay for ribonucleotide reduction by Mycobacterium smegmatis mc2155 was necessary to screen a large series of bacteria. To make the membrane of the cells of Mycobacterium smegmatis mc2155 permeable to the nucleotide, the cells were permeabilized with toluene and ether two solvents because this approach was applicable for both gram-negative bacteria [18]; grampositive bacteria [19-21]. The obtained ribonuleotide reduction activity with ether permeabilized cells of Mycobacterium smegmatis mc2155 presented a comparable enzyme activity for both times (2, 5 minutes), while with toluene permeabilized cells indicated a low enzyme activity.
One crucial point for successful development of an assay of allosterically regulated ribonucleotide reduction in nucleotidepermeable cells of Mycobacterium smegmatis mc2155 was the removal of the endogenous inhibitory or inactivating substances [22,23] by suitable washing procedure. While the cells lost roughly 20% protein through permeabilization, they did not appear to be damaged in the examined with scanning electron microscopy (Data no shown). Furthermore, when the permeabilized cells sample were rapidly frozen in liquid nitrogen and stored at –80°C, the RNR activity retained for several months. The obtained results of the relationship between ribonucleotide reduction and the growth of Mycobacterium smegmatis mc2155 showed a strict correlation and Ribonucleotide redutase is a peak enzyme. The Purification procedure of ribonucleotide reductase of Mycobacterium smegmatis mc2155 led to dissociation of both subunits. The facile dissociation of ribonucleotide reductase of Mycobacterium smegmatis mc2 155 in their subunits during purification by using several methods of chromatography especially by gel filtration on superdex G-200, and the existence of the small subunit and the large subunit of ribonucleotide reductase in inactive form, suggests a biochemical complementation of the both subunits for enzyme assay. The obtained specific activity of the purified protein was 1790 pmol per mg per min with an overall yield of 10%. The purified small subunit of MS2- protein was detected on SDS-PAGE, which was showed a strong band that corresponds to a apparent molecular weight of 38.5 KDa. Finally, the permeabilisation of the cells of M. smegmatis mc2 155 with ether and toluene for short time facilitated us to develop of a rapid assay for ribonucleotide reduction of others gram-positive bacteria.
Acknowledgements
The authors are thankful to Prof. Dr. Auling G. Institute of Microbiology Hannover-Germany to provide us with the strain of Mycobacterium smegmatis mc2155 and the necessary facilities to conduct this research work in their laboratory.
References
- Hiriyanna KT, Ramakrishnan T (1986) Deoxyribonucleic acid replication time in Mycobacterium tuberculosis H37 Rv. Arch Microbiol 144: 105-109.
- Bloom BR, Small PM (1998) The evolving relation between humans and Mycobacterium tuberculosis. N Engl J Med 338: 677-678.
- Chaisson RE, Slutkin G (1989) Tuberculosis and human immunodeficiency virus infection. J Infect Dis 159: 96-100.
- Pitchenik AE, Cole C, Russell BW, Fischl MA, Spira TJ, et al. (1984) Tuberculosis, atypical mycobacteriosis, and the acquired immunodeficiency syndrome among Haitain and non-Haitain patients in South Florida. Ann Intern Med 101: 641-645.
- Wayne LG (1994) Dormancy of Mycobacterium tuberculosisand latency of disease. Eur J Clin Microbiol Infect Dis 13: 908?914.
- Reichard P (1993) From RNA to DNA, Why so many ribonucleotide reductases? Science260: 1773-1777.
- Rubin H, Salem JS, Li LS, Yang FD, Mama S, et al. (1993) Cloning, sequence determination, and regulation of the ribonucleotide reductase subunits from Plasmodium falciparum: a target for antimalarial therapy. Proc Natl Acad Sci U S A 90: 9280-9284.
- Winder FG, McNulty MS (1970) Increased DNA polymerase activity accompanying decreased DNA content in iron-deficient Mycobacterium smegmatis. Biochim Biophys Acta 209: 578-580.
- Winder FG, Barber DS (1973) Effects of hudroxyurea, nalidixic acid and zinc limitation on DNA polymerase and ATP-dependant deoxyribonuclease activities of Mycobacterium smegmatis. J Gen Microbiol 76: 189-196.
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193: 265-275.
- Thaler M, Diekmann H (1979) The effect of manganese deficiency on lipid content and composition in Brevibacterium ammoniagenes. Applied Microbiol Biotechnol 6: 379-387.
- Griepenburg U, Lassmann G, Auling G (1996) Detection of a stable free radical in the B2 subunit of the manganese ribonucleotide reductase (Mn-RRase) of Corynebacterium ammoniagenes. Free Radic Res 24: 473-481.
- Luo C, Hansen J, Auling G (1997) Temperature-sensitive mutants of Corynebacterium ammoniagenes ATCC 6872 with a defective large subunit of the manganese-containing ribonucleotide reductase. Arch Microbiol 167: 317-324.
- Griepenburg U, Blasczyk K, Kappl R, H?ttermann J, Auling G (1998) A divalent metal site in the small subunit of the manganese-dependent ribonucleotide reductase of Corynebacterium ammoniagenes. Biochemistry37: 7992-7996.
- Willing A, Follmann H, Auling G (1988) Ribonucleotide reductase of Brevibacterium ammoniangenes is a manganese enzyme. Eur J Biochem 170: 603-611.
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685.
- Abbouni B, Oehlmann W, Stolle P, Pierik AJ, Auling G (2009) Electron paramagnetic resonance (EPR) spectroscopy of the stable-free radical in the native metallo-cofactor of the manganese-ribonucleotide reductase (Mn-RNR) of Corynebacterium glutamicum. Free Radic Res 43: 943-950.
- Moses RE, Richardson CC (1970) Replication and repair of DNA in cells of Escherichia coli treated with toluene. Proc Natl Acad Sci U S A 67: 674-681.
- Matsushita T, White KP, Sueoka N (1971) Chromosom replication in toluenized Bacillus subtilis cells. Nat New Biol 232: 111-114.
- Abbouni B, Elhariry HM, Auling G (2004) Overproduction of NAD+ and 5'-inosine monophosphate in the presence of 10 ?M Mn2+ by a mutant of Corynebacterium ammoniagenes with thermosensitive nucleotide reduction (nrdts) after temperature shift. Arch Microbiol 182: 119-125
- Abbouni B, Elhariry HM, Auling G (2003) Arrest of cell cycle by inhibition of ribonucleotide reductase induces accumulation of NAD+ by Mn+2-supplemented growth of Corynebacterium ammoniagenes. Biotechnol Lett 25: 143-147.
- Auling G, Follmann H (1994) Manganese-dependent ribonucleotide reduction and overproduction of nucleotides in coryneform bacteria. New York, Marcel Dekker Inc.
- Lammers M, Follmann H (1983) The ribonucleotide reductase-A unique group of metalloenzymes essential for cell proliferation. Inorg Elem Biochem 54: 27-91.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 14421
- [From(publication date):
March-2012 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 9869
- PDF downloads : 4552