Research Article Open Access
Development of Molecular Identification of Nitrifying Bacteria in Water Bodies of East Kolkata Wetland, West Bengal
| Mousumi Saha1*, Agniswar Sarkar2 and Bidyut Bandhophadhyay1 | |
| 1Department of Biotechnology, Vidyasagar University, West Bengal, India | |
| 2Department of Biotechnology (Recognized by DBT, Govt. of India), The University of Burdwan, Burdwan, West Bengal, India | |
| Corresponding Author : | Mousumi Saha Department of Biotechnology Vidyasagar University West Bengal, India Tel: +91 9830736189 E-mail: mou.cute@gmail.com |
| Received: September 17, 2013; Accepted: October 30, 2013; Published: November 07, 2013 | |
| Citation: Saha M, Sarkar A, Bandhophadhyay B (2013) Development of Molecular Identification of Nitrifying Bacteria in Water Bodies of East Kolkata Wetland, West Bengal. J Bioremed Biodeg 4:211. doi:10.4172/2155-6199.1000211 | |
| Copyright: © 2013 Saha M, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Nitrifying bacteria plays a major role in converting the waste water to valuable renewable resources for the society. Ammonia is the toxic excretory product of most aquatic organisms and nitrite formed by the oxidation of ammonia is also toxic. Ammonia oxidizing bacteria converts ammonia to nitrite and Nitrite oxidizing bacteria convert nitrite to nitrate. Among them, Nitrosomonas sp. and Nitrobacter sp. has been the most widely studied organisms. An advanced molecular technique made it possible to explore the nitrifying bacteria in the environment and to enhance our knowledge of its functioning. In view of this it would be of prime importance for rapid detection of these strains/ isolates using molecular techniques. The present study was undertaken to develop and validate 16S rDNA sequence analysis of Nitrosomonas sp. and Nitrobacter sp. collected from different bheries in East Kolkata Wetland, West Bengal.
| Keywords |
| Ammonia oxidizing bacteria; Nitrite oxidizing bacteria; 16S rDNA; East Kolkata Wetland |
| Introduction |
| East Kolkata Wetland (EKW) is the world’s largest sewage-fed aquaculture and natural water recycling system and contains a series of wetland. It is renowned not simply as the sensitive ecosystem of biodiversity value, but more specifically as an example of best practice in the ‘wise use’ of wetlands (from 1879 onwards) for garbage farming (2500 metric ton of solid waste is dumped every day), sewage-treated fisheries (bheris) and sewage farming [1]. EKW located in the eastern part of the city (22024/- 22036/N to 88023/-88032/E). In addition, organic fertilizer produced by processing the solid waste from this area is found to be commercially important [2-4]. The wetland also supports the livelihoods of around 60,000 residents through the fisheries and other socio-economic activities. |
| It serves as a vast area for research purpose and environment protection through bioremediation or phytoremediation [3]. EKW declared as a Ramsar site on November 2002 as per “Ramsar Guidelines”. Wastewater in EKW flows through various channels into the fish ponds covering about 4000ha, where a wide range of physical, biological and chemical processes take place which helps in improving the quality of the water [4,5]. Consequently this wetland system is popularly known as the “kidney of the city”. |
| Various bacterial species distributed in different site or bheries in EKW and involved in bioremediation of heavy metals. These diverse microbial resources also produce some commercially important enzymes like protease, oxidase etc. Streptococcus macedonicus produces extracellular protease and food grade bacteriocine [6]. Pseudomonas sp. are involved in the production of extracellular protease [7]. Klebsiella sp. can help to overcome cadmium toxicity [8]. Actinobacteria and Fermicutes participate in the degradation of nitrophenol, nitroaromatic compound, pesticide and herbicide [3]. The metal accumulating or removal property of these microbes can be exploited for treatment of contaminants rich in metals in various sites. Along with the above microbes, proteobacteria like Nitrosomonas sp. present in EKW and enable to oxidize ammonia into nitrite [3]. |
| Ammonia is the principal excretory product of most aquatic organisms, but is toxic, acute and chronic to fish. Nitrite is formed either by the oxidation of ammonia (Nitrification) or the reduction of nitrate. Nitrite is also toxic to fish and another critical water quality factor. Ammonia affects on the central nervous system by causing “acute ammonia intoxication” which includes convulsions and death [9]. Acute concentrations of ammonia may cause loss of equilibrium, hyperexcitability, increased breathing, cardiac output, and oxygen uptake; in extreme cases convulsions, coma, and death. Lower concentrations of ammonia can cause a reduction in hatching success; reduction in growth rate and morphological development; pathologic changes in tissues of gills, livers, and kidneys [10]. Brown Blood Disease occurs in fish when water contains high nitrite concentrations. Nitrite enters the bloodstream through the gills and turns the blood to a chocolate-brown color. Hemoglobin, which transports oxygen in the blood, combines with nitrite to form Methemoglobin, which is incapable of oxygen transport [11]. Warm water fishes like Carps, Catfishes, and Tilapia are fairly sensitive to nitrite, and trout and other cool water fish are sensitive to extremely small amounts of nitrite. Nitrites also react directly with hemoglobin in human blood and other warm-blooded animals to produce methemoglobin. |
| Thus, it is very important to understand the dynamics of nitrification, where may be some microbe like ammonia oxidizing bacteria (AOBNitrosomonas sp.) and nitrite oxiding bacteria (NOB-Nitrobacter sp.) plays the major role in converting the waste water to valuable renewable resources for the society [12-14]. The genera Nitrosomonas and Nitrobacter both are chemolithoautotrophic and members of the Proteobacteria class [15]. Genus Nitrosomonas form a distinct group within the β subdivision where as nitrite-oxidizing bacteria like Nitrobacter occurring in α, δ, and γ subdivisions. Therefore, to maintain the water quality and fish health it’s become very important to keep the ammonia and nitrite within the safe tolerance limit and these can be done some nitrifying bacteria. Various researchers described that only about 1 to 4% of the entire microbial community of EKW has been screened following the traditional and the conventional methods and about 96 to 99% are yet to be isolated and identified [16,17]. EKW, which can prove to be important from point of bioremediation and biotechnology and in this respect, it is necessary to develop the rapid and accurate technology for isolation and identification of these microbes using modern molecular techniques. Therefore, to gather more information about these diverse microbial communities, it is very important to use molecular technologies like 16S rRNA gene sequence, phylogenetic analysis etc [18,19]. Study of AOB and NOB by conventional cultivation techniques is also very difficult because of their long generation times and low growth rates [20]. Therefore, a rapid, culture-independent detection technique for nitrifiers would be useful to provide a record about the natural availability and role of these microbes in bioremediation of various bheries located in the East Kolkata Wetland. In view of this economic importance to aquatic animals, human and animal health hazard problems, it would be of prime importance for rapid detection of these strains/isolates using molecular techniques. With this in view, the present study was undertaken with following objective: To develop and validate different genotyping techniques viz 16S rDNA sequence analysis, Ribotyping and bioinformatics analysis for Nitrosomonas and Nitrobacter sp. collected from different bheries in West Bengal. To develop and standardize PCR technique, for accurate and prompt diagnosis of the species, to create a genomic fingerprint database for surveillance and monitoring genetic variability of isolates and also to establish a biosecurity protocol for the Bheries located in different areas of West Bengal. |
| Materials and Methods |
| Site selection |
| In West Bengal both sewage fed fisheries and floodplain wetlands plays an important fishery resources. The commonly cultivated fish includes Catla (Catla catla), Rohu (Labeo rohita), Mrigal (Cirrhinus mrigala), Silver carp (Hypothalmichthys molitrix), Grass carp (Ctenopharyngodon idella) and Common carp (Cyprinus carpio) are considered to be the best culturable species. |
| Flood plain wetland in West Bengal covers an area of about 42000 ha. Within the West Bengal, water bodies for fish culture are present in North and South 24 Praganas and Nadia Districts. Waste water fed fish ponds in East Kolkata Wetland, West Bengal, India (called Bheris) have a distinctly different architecture resulting in extensive purification of waste along with integrated resource recovery [3]. Bheri, the shallow (50-150 cm), flat bottom, waste water fed fish pond. Kolkata, Nadia, South 24 Parganas and Burdwan were selected for the present study because; they are important districts of West Bengal with respect to both aquaculture and agriculture. East Kolkata Wetland in Kolkata; Kulia, Bhomra in Nadia; Haruabhanga, Lakshmipur in Burdwan and Malancha, Jaynagar in South 24 parganas were identified and surveyed for our study. But for this present study EKW was chosen as it is a waste water fishery. Preliminary physico-chemical and biological survey indicates the presence of nitrifying bacteria in EKW as compared to other sites of West Bengal. Among which research work was initiated under East Kolkata Wetland at Nalban Fisheries (owned by Government of West Bengal and runed by State Fisheries Development Co-operation) and Jhagrasisa bhery (runed by private organization). |
| Collection of samples and process |
| Water samples (25 ml) were collected from Nalban Fisheries and Jhagrasisa bhery. A total 6 fish ponds or tanks were selected for the study and marked as N1, N2, N3 for Nalban Fishery and J1, J2, J3 for Jhagrasisa bhery in East Kolkata Wetland. Samples (two aliquots from each place) were also collected from Topsia (Sewage Pumping Station) and from different channels like Ambedkar Bridge, Bantala, Bamunghata, Ghosher Khal which is connected to the fish pond and Ghusighata (outlet). Samples from each site was collected at around 6.30-7.00 in the morning in sterile plastic bottle (Tarson) from a depth of 1-2.0cm below the surface of the water, kept in ice box (Milton, India) and then taken in laboratory for immediate culture. Water samples were collected in different seasons as winter, summer, post monsoon and monsoon. In each seasons two aliquots were collected from each tanks and a total of 96 aliquots were collected and studied. |
| Samples were collected and processed on Winogradskyi medium for the isolation of nitrifying bacteria. Nitrosomonas sp. was isolated using Winogradsky phase 1 medium, which contains (NH4)2SO4, 2.0 g; K2HPO4, 1.0 g; MgSO4, 7H2O, O.5 g; NaCl, 2.0 g; FeSO4,7H2O, 0.4 g; CaCO3, 0.01g, and miliQ water upto 1000 ml, pH 7.8). Nitrobacter sp. was isolated using Winogradsky phase 2 medium (KNO2, 0.1 g; Na2CO3, 1.0 g; NaCl, 0.5 g; FeSO4, 7H2O, 0.4 g, and miliQ water upto 1000 ml, pH 7.8) [21-23]. Tubes were incubated at 28°C for 90 hrs aerobically [21,24,25]. Cultures were initially identified by gram bacteria [26]. Suspected cultures were evaluated for presence of ammonia and nitrite and vice versa for production of nitrite and nitrate. These tests were performed and confirmed by a series of reaction. Selected cultures were preserved for further studies at -80°C with 30-40% glycerol stock. |
| Isolation of genomic DNA |
| Bacterial biomass from 10 ml of culture was collected by centrifugation for 5 min at 10000×g and resuspended in a 2 ml polypropylene tube (Eppendrof) containing glass beads with 500 ml buffer containing 20 mM sodium acetate, 1 mM EDTA (pH 5.5), 50 ml 25% SDS and 600 ml phenol: chloroform: isoamyl alcohol (25: 24: 1). Cells were lysed by the addition of 200 μl of 10 mg ml-1 lysozyme and incubation at 37°C for 60 min. 20% Sodium dodecyl sulfate was added to a final concentration of 1% and incubated at 37°C for 60 min. Proteinase K solution was added (10 mg/ml) to a final concentration of 2 mg/ml and incubated at 50°C for 35 min and the mixture was then centrifuged at 10,000 g×10 min at 4°C. The aqueous phase was transferred carefully to a fresh tube, mixed with 600 ml chloroform: isoamyl alcohol (24: 1) and centrifuged 10000 g×10 min. The aqueous phase was transferred to a fresh tube and, after the addition of 0.1 vol. 3 M sodium acetate. DNA was precipitated by incubation with 0.6 volume of isopropanol and 5 ml glycogen (5 mg/ml) for 1 h and subsequently pelleted by centrifugation at 10 000 g×20 min at 4°C. Pellets were washed with 1 ml ice-cold 70% alcohol and dried it by Maxi Vacuum Dryer and resuspended in 500μl TE buffer (pH 8.0). The DNA concentration was estimated by visual comparison with the standard DNA size markers after electrophoresis through 0.8% agarose gel, stained with 0.5 mg ml-1 ethidium bromide (Sigma Chemicals Co.). |
| 16S rDNA-PCR amplification |
| The 16S rRNA genes were amplified by PCR using the universal primers: forward primer 27F 5’-AGA GTT TGA TCC TGG CTC AG- 3’ and reverse primer 1492R 5’- GGT TAC CTT GTT ACG ACT T-3’ (Chromous Biotech Ltd., Bengaluru, India) [27]. PCR reactions has been carried out by following the series of standardizing experimental protocol as annealing temperature, concentration of MgCl2, template DNA, Taq DNA polymerase, dNTP’s and primers. The PCR reaction components consists of 200 mm dNTP, 20 pico moles of primer, 2 units of Taq DNA polymerase enzyme, assay buffer with working concentration of 1.5 mM MgCl2, 20-30 ng template DNA in an assay volume of 25 μl. These concentrations were determined by a series of preliminary standardizing experiments. |
| A cycling program was performed using a Thermal Cycler (Thermo Scientific) with an initial denaturation step at 95°C×3 min, 35 cycles of denaturation at 94°C×1 min, annealing at 55°C×1 min, and extension at 72°C×2 min, and final extension at 72°C×3 min. The presence of the PCR products were examined and visualized by electrophoresis in 1.5% agarose gel in TBE buffer. The gel was stained with EtBr (Sigma) and viewed in Gel Doc System (Biorad Gel Doc. 2000 system). |
| Sequencing of 16S rDNA amplicons |
| The PCR products were purified by the Exosap treatment in the Departmental Molecular Biology Laboratory. For the validation and identification, chosen strains were identified by 16S rDNA PCR pattern analysis on 1.5% agarose gel electrophoresis and those were selected for sequencing. In total, the PCR amplified products were sequenced by Ion torrent Sequencer from the Xcelris Lab Ltd, Ahmedabad, India. |
| Computer assisted pattern analysis |
| Chromatogram of sequence data confirmed the peak, reproducibility, quality and gene sequences of 16S rRNA amplicons. Raw sequence data are arranged as complementary and consensus through online Reverse- Complement tools (www.bioinformatics.org/sms/rev_comp.html) and Genefisher 2, the BiBiServ (Bielefeld BioInformatics Service) is a unit of the Institute for Bioinformatics (bibiserv.techfak.uni-bielefeld.de/ genefisher2/submission.html). Identification of species and similarity calculations were performed, comparing sequences of approximately 1500 bases with sequences available in GenBank using BLAST network services (blast.ncbi.nlm.nih.gov/). Multiple Sequence Alignments (MSA) are carried out by Clustal W following all the sequence pairs were aligned separately for the calculation of distance matrix and constructed a guide tree and sequences are aligned according to the hiearchy. |
| Phylogenetic analysis between Ammonia Oxidizing and Nitrite Oxidizing Bacteria |
| Phylogenetic relationship, similarity index, distance between all isolates and maximum likelihood pattern analysis outlined and phylogenetic tree generated by Mega v5.05. The 16S rDNA sequences in this study were submitted to the GenBank database by using Sequin software (www.ncbi.nlm.nih.gov/projects/Sequin/) under nucleotide accession number. Analysis of the 16S rRNA gene proved as useful taxonomic tools with discriminatory properties for identification up to the species level. |
| Results and Discussion |
| Identification of strains |
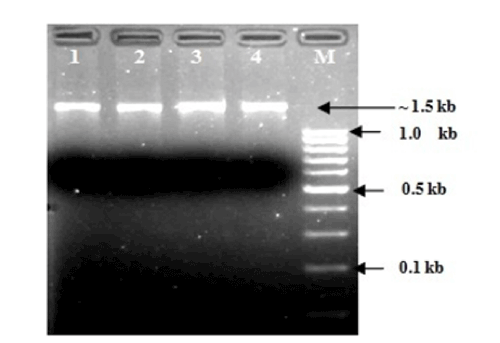
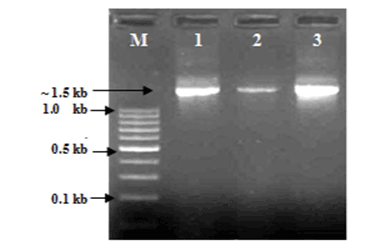
| AOB and NOBs were initially identified by gram staining and produced gram negative characteristics. The nitrite accumulation test was performed to confirm AOBs and the nitrite consumption test was performed to confirm NOBs by following the standard protocol of APHA, (1998) [28]. A comparative 16S rRNA sequence analysis finally confirmed the genus and species (Table 1). Out of total seven strains were sequenced and tested, two were found as Nitrosomonas sp. and another two Nitrosomonas includes eutrtopha and euroraea species; on the other hand, two strains were found as Nitrobacter sp. and another single strain was identified as Nitrobacter vulgaris (Figures 1 and 2). |
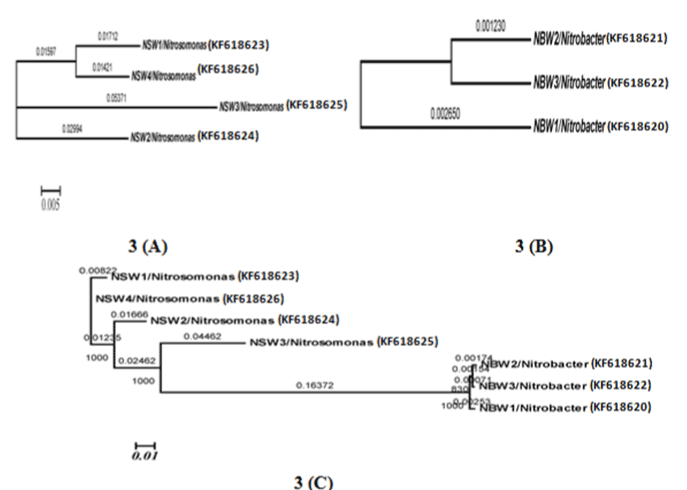
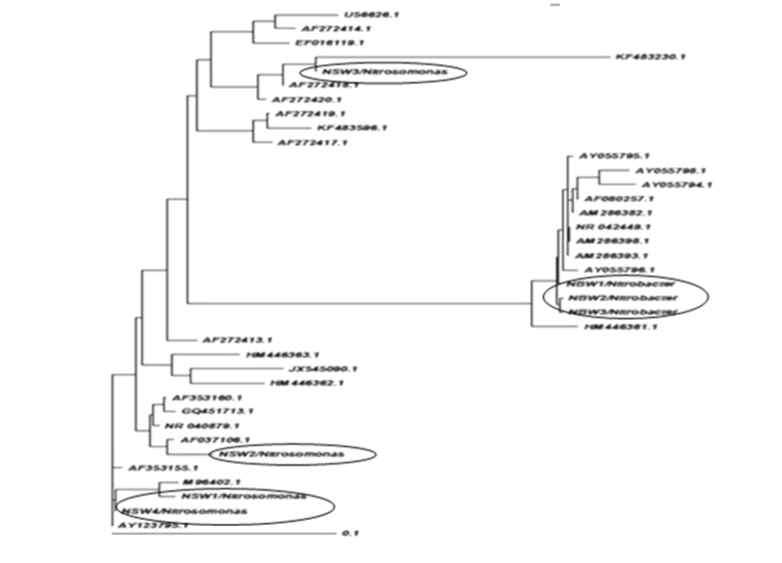
| Phylogenetic analysis |
| Grimont and Grimont (1986) described the importance of the 16S rRNA-based probes as taxonomic tools [29]. In the present study, we approached the use of the properties of the 16S rDNA for phylogenetic identification from a different perspective and results apparently covered the majority of NOBs and AOBs can be discriminated by 16s rDNA targeted primers therefore need to take into account the intragenomic diversity of the 16S rRNA gene. Nitrosomonas europaea cell are capable of doing nitrification in wastewater with high efficiency. A recent study was developed by Niranjan Uemoto and Saiki and Kumar Shrestha et al. [12,30], where nitrogen removed using Nitrosomonas europaea and Paracoccus denitrificans from waste water. Nitrobacter population can be detected by using PCR- analysis [13]. |
| As waste water is introduced into the bheries or the fish pond with high load of organics or heavy metals, the bacterial populations neutralizes or break down the organics or heavy metals to achieve maximum benefit and also enhance fish production. Thus, the application of probiotics in sewage fed bheries along with the details molecular characterization of microbes present in bheries can enhance the fish production and can reduce the level of ammonia, H2S stress. Analysis of different samples of Nitrosomonas spp. found in freshwater was done using 16S ribosomal DNA and PCR technique [18]. Classification of any bacterial group, taxonomically as well as molecular characterization is required, thus for studying ammonia- oxidizing bacteria polygenic and molecular analysis was done [19]. Along with this molecular characterization of the bacteria is also important for further investigation and to analyze these bacterial communities. |
| The isolation of samples collected from the distribution network in this study could be attributed to the effective strategy employed in the system, which may increase the survival of the organism as suspended or as part of the biofilm community. The similarity of isolated 16S ribosomal RNA gene, partial sequences were identical and ranging between 94 to 97% (Figures 3 and 4). To draw a proper biosecurity protocol, molecular diagnosis and phylogenetic relationships between the species in EKW, the present technique used in this study can be able to overcome threats and control the disease outbreaks. These data and methodology has been proved in this study as advance molecular tool. Our results emphasize the need to take into account the intragenomic diversity of the 16S rRNA gene. This method has shown discriminatory properties in the identification up to the species level. The study provided a quick identification tool. |
| Conclusion |
| Views were recognition of the heterogeneity detected in the sequences is important to avoid strain misidentification and faithful to the acknowledgement of diversity. Our study aimed towards prompt and accurate identification of members of the genus AOBs and NOBs as indicators in environment and as the most prevalent bacterium in the waterways of EKW in West Bengal, India. The method provided in this work, apart from being a reliable identifier at reasonable cost. |
| Acknowledgements |
| The author acknowledges to Dr. Bidyut Bandhophadhyay for his intellectual input. We also acknowledge Mr. S. D. Ghosh for his kind permission to work at EKW. |
References
|
Tables and Figures at a glance
| Table 1 |
Figures at a glance
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 15731
- [From(publication date):
January-2014 - Dec 02, 2025] - Breakdown by view type
- HTML page views : 10840
- PDF downloads : 4891
